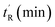Open Access Article
Open Access ArticleStereoisomerism of stapled peptide inhibitors of the p53-Mdm2 interaction: an assessment of synthetic strategies and activity profiles†
Tsz Ying
Yuen
 *a,
Christopher J.
Brown
b,
Yuezhen
Xue
b,
Yaw Sing
Tan
c,
Fernando J.
Ferrer Gago
b,
Xue Er
Lee
b,
Jin Yong
Neo
a,
Dawn
Thean
b,
Hung Yi Kristal
Kaan
d,
Anthony W.
Partridge
*a,
Christopher J.
Brown
b,
Yuezhen
Xue
b,
Yaw Sing
Tan
c,
Fernando J.
Ferrer Gago
b,
Xue Er
Lee
b,
Jin Yong
Neo
a,
Dawn
Thean
b,
Hung Yi Kristal
Kaan
d,
Anthony W.
Partridge
 d,
Chandra S.
Verma
d,
Chandra S.
Verma
 c,
David P.
Lane
c,
David P.
Lane
 b and
Charles W.
Johannes
b and
Charles W.
Johannes
 a
a
aInstitute of Chemical and Engineering Sciences, Agency for Science, Technology and Research, 8 Biomedical Grove, Neuros, #07-01, Singapore 138665. E-mail: yuenty@ices.a-star.edu.sg
bP53 Laboratory, Agency for Science, Technology and Research, 8A Biomedical Grove, #06-06, Immunos, Singapore 138648
cBioinformatics Institute, Agency for Science, Technology and Research, 30 Biopolis Street, #07-01, Matrix, Singapore 138671
dMSD Translational Medicine Research Centre, 8 Biomedical Grove #04-01, Neuros, Singapore 138665
First published on 30th May 2019
Abstract
All-hydrocarbon, i, i+7 stapled peptide inhibitors of the p53-Mdm2 interaction have emerged as promising new leads for cancer therapy. Typical chemical synthesis via olefin metathesis results in the formation of both E- and Z-isomers, an observation that is rarely disclosed but may be of importance in targeting PPI. In this study, we evaluated the effect of staple geometry on the biological activity of five p53-reactivating peptides. We also present strategies for the modulation of the E/Z ratio and attainment of the hydrogenated adduct through repurposing of the metathesis catalyst.
Introduction
Short peptides often lack well-defined secondary structures when excised from the native protein environment. In order to retain the native conformation of the peptide, chemists have rationally developed a myriad of cyclisation strategies. One such method relies on the use of distance-matching tethers that covalently link two amino acid side chains (e.g. Cys,1–9 Lys,10–15 Trp,16–18 and Met19). Alternative stabilisation strategies such as alkene,20–25 oxime,26,27 1,2,3-trizole,28–30 hydrazide,31 alkyne,32 spiropyran,33 as well as mixed cross-linkages have also been published.34,35 The resultant macrocycles often possess enhanced proteolytic resistance, specificity and potency, making them ideal pharmacological agents.36–38 Chief among these targeted therapeutic advances is the ability of cyclic peptides to modulate the previously believed “undruggable” protein–protein interactions (PPI) because of enhanced cellular uptake.36The disruption of the protein–protein interaction between p53 and its negative regulator Mdm2 comprises one of the most studied areas in cancer therapy. The E3 ubiquitin ligase Mdm2 is overexpressed in a number of cancer cell lines, leading to loss of p53 function and uncontrolled cellular proliferation.37–39 Compounds targeting the p53-Mdm2 complex may restore p53 transcriptional activity by blocking the ubiquitination and proteasomal degradation functions of Mdm2.38,40–44 Several classes of molecules have since been developed, mimicking the conserved α-helical region of the Mdm2 binding domain of p53.45–48
We recently published a series of stapled peptides guided by computer simulations that specifically targeted the p53-Mdm2 binding groove.46 These peptides all contained an i, i+7 staple and a conserved Phe-Trp-Leu triad which docked inside the hydrophobic cleft of Mdm2. The peptides were synthesised and tested in a T22 p53 reporter assay as a mixture of E/Z isomers. Helically stabilised Mdm2 binding peptide sMTide-02 in particular had demonstrated promising levels of p53 transcriptional response. We reasoned an isomeric mixture was not a requirement for biological activity and that a targeted synthesis of the more active isomer would serve as a more economical manufacturing route.
To our knowledge, an assessment of optimal E/Z isomer ratio has not been critically examined for bioactive stapled peptides. In fact, the lack of available characterisation data has made cross-comparisons and reproduction of biochemical and biological activities difficult.49,50 Amongst the growing body of research on α-helix stabilisation via i, i+7 hydrocarbon stapling, only a handful of studies have looked at the influence of geometric isomerism on α-helicity and biological activity.49,51,52 The collective published results highlight sequence and target-dependent activity. Wallbrecher et al. reported comparable cellular activity for both E- and Z-ATSP-7041 in SJSA-1 cells.49 However in the case of stapled MEP-N, a 2-fold difference in antimicrobial activity was observed against Staphylococcus aureus and E. coli, with the Z-isomer being more effective against the former but less active against the latter.51 In this paper, we assess the effects of metathesis conditions (catalyst structure, solvent and temperature) on E/Z selectivity of p53-reactivating peptides and provide initial analysis of biophysical and biological properties of the diastereopure, stapled peptide isomers. A synthetic strategy for the attainment of saturated all-hydrocarbon stapled peptides is also presented.
Results and discussion
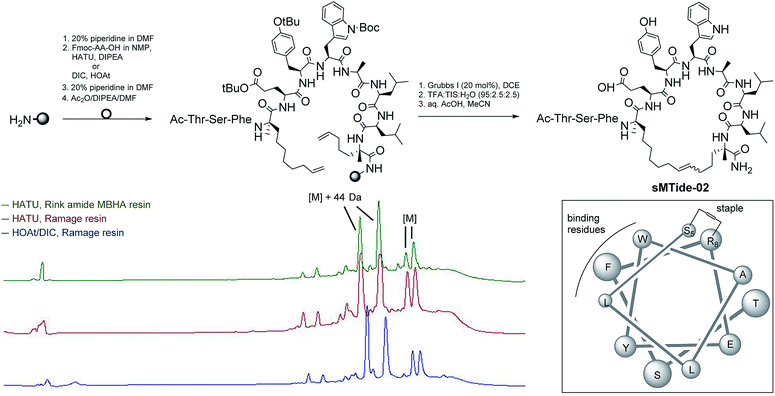
Solid-phase peptide synthesis (SPPS) of sMTide-02
Our first objective was to synthesise the parent peptide sMTide-02 using conventional Fmoc solid phase peptide synthesis.23,53 Three SPPS conditions were trialled (Fig. 1), exploring different combinations of PEG-based resins (H-Rink Amide ChemMatrix® or H-Ramage ChemMatrix®) and activation methods (HATU/DIPEA or HOAt/DIC). Following iterative rounds of amino acid coupling and Fmoc deprotection, the peptide was N-capped and subjected to several metathesis cycles before TFA-mediated cleavage from the resin. Overall, higher yields of crude peptide were obtained with the utilisation of the Ramage resin (0.53 mmol g−1 loading, 50% yield) as opposed to the recommended Rink Amide resin (0.59 mmol g−1 loading, 36% yield).23,53 HOAt was found to be more soluble in NMP relative to HATU, hence use of HOAt in conjunction with DIC has an added advantage of achieving higher concentrations of the reacting solutions. Despite the same peptide active ester being formed under both coupling conditions, we noted a cleaner reaction profile with the use of HOAt/DIC.In all cases, 2 pairs of stapled peptide adducts with approximately 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio could be observed in the HPLC chromatograms. The 2 most intense peaks had identical masses ([M] + 44 Da), which corresponded to the unexpected, incomplete decarboxylation of the tryptophan protecting group. Conversely, the masses of the more hydrophobic peaks corroborated well with the expected m/z of the desired E/Z olefin isomers. Complete decarboxylation was successfully achieved by overnight treatment of the peptide with aqueous acetic acid followed by lyophilisation. Alternatively, use of side-chain unprotected Trp during SPPS also led to the desired product without oxidation or alkylation of the indole group.
1 ratio could be observed in the HPLC chromatograms. The 2 most intense peaks had identical masses ([M] + 44 Da), which corresponded to the unexpected, incomplete decarboxylation of the tryptophan protecting group. Conversely, the masses of the more hydrophobic peaks corroborated well with the expected m/z of the desired E/Z olefin isomers. Complete decarboxylation was successfully achieved by overnight treatment of the peptide with aqueous acetic acid followed by lyophilisation. Alternatively, use of side-chain unprotected Trp during SPPS also led to the desired product without oxidation or alkylation of the indole group.
(E)-sMTide-02 is more active than (Z)-sMtide-02
Ring closing metathesis (RCM) is a reversible, thermodynamic process that constitutes the key step of stapled peptide synthesis. First propounded by Villemin and Tsuji in 1980,54,55 it is now routinely used in macrocycle formations owing to the exceptional functional group compatibility of the olefin metathesis catalysts. Stereoselectivity of the reaction is largely dependent on the degree of conformational strain introduced by the all-hydrocarbon linker. In the case of sMTide-02, stapling led to both E- and Z-isomers in almost equal ratios, which is consistent with a previous report.23 To further explore the role of olefin geometry on the bioactivity of the stapled peptide, separation of the geometric isomers was attempted by conventional reverse-phase HPLC. After multiple rounds of purification, the two closely-eluting isomers were fully separated.The geometric configuration of the 2 isomers was determined by 1H NMR spectroscopy. The earlier-eluting isomer of sMTide-02 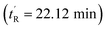 gave an olefinic proton signal at 5.21 ppm with a distinctive, centrosymmetric doublet of triplets splitting pattern [3J(–CH
gave an olefinic proton signal at 5.21 ppm with a distinctive, centrosymmetric doublet of triplets splitting pattern [3J(–CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH–) = 11.0 Hz, Fig. S11, ESI†] and was assigned the Z-configuration. Conversely, the later-eluting sMTide-02 isomer
CH–) = 11.0 Hz, Fig. S11, ESI†] and was assigned the Z-configuration. Conversely, the later-eluting sMTide-02 isomer 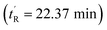 showed olefinic protons at 5.31 ppm with a 3J coupling constant of 15.4 Hz, which corresponded to an E-configuration.
showed olefinic protons at 5.31 ppm with a 3J coupling constant of 15.4 Hz, which corresponded to an E-configuration.
The peptides were assessed for their α-helical content and binding affinity to Mdm2. P53 reporter and LDH release assays were also conducted to measure the ability of the peptides to activate p53 in T22 cells and to disrupt cell membranes. The latter assays were performed in the presence of 2% serum to mimic physiological conditions. As seen from Table 1, cell membranes remained intact after treatment with both geometric forms of sMTide-02 (entries 1–2). In comparison to the Z-isomer, (E)-sMTide-02 showed approximately 2-fold improvement in both binding and p53 activation despite similar levels of α-helicity.
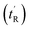 determined using the formula
determined using the formula  helicity determined by circular dichroism spectroscopy; kd determined by competitive fluorescence anisotropy titrations; biological activity determined by T22 p53 reporter assay and cell membrane perturbation effects measured using an LDH assay
helicity determined by circular dichroism spectroscopy; kd determined by competitive fluorescence anisotropy titrations; biological activity determined by T22 p53 reporter assay and cell membrane perturbation effects measured using an LDH assay
| Entry | Peptide | Net charge | α-Helicity | k d (nM) | p53Act (μM) | LDH50 (μM) | |
|---|---|---|---|---|---|---|---|
| 1 | (Z)-sMTide-02 | 22.12 | −1 | 70.8 | 113.1 ± 0.01 | 8.29 ± 0.19 | ND |
| 2 | (E)-sMTide-02 | 22.37 | −1 | 72.7 | 63.19 ± 0.01 | 3.64 ± 0.22 | ND |
| 3 | (Z)-ATSP-7041 | 23.96 | −1 | 77.6 | 32.68 ± 0.01 | 1.80 ± 0.57 | ND |
| 4 | (E)-ATSP-7041 | 24.44 | −1 | 68.7 | 11.03 ± 0.01 | 2.77 ± 0.23 | ND |
| 5 | Early-VIP82 | 18.64 | 0 | 28.6 | 94.45 ± 0.01 | 4.13 ± 0.10 | ND |
| 6 | Late-VIP82 | 18.88 | 0 | 38.5 | 65.47 ± 0.01 | 3.38 ± 0.73 | ND |
| 7 | Early-VIP116 | 19.69 | −1 | 42.1 | 84.44 ± 0.01 | ∼5.63 | ND |
| 8 | Late-VIP116 | 19.96 | −1 | 49.4 | 52.39 ± 0.01 | 3.14 ± 0.38 | ND |
| 9 | Early-VIP115 | 18.09 | +1 | 52.6 | 95.81 ± 0.01 | ∼3.16 | 80.99 ± 1.80 |
| 10 | Late-VIP115 | 18.25 | +1 | 36.7 | 84.48 ± 0.01 | ∼2.24 | 129.8 ± 15.39 |
| 11 | Reduced-sMTide-02 | 23.66 | −1 | 71.2 | 209.5 ± 0.02 | 1.58 ± 0.45 | ND |
| 12 | Reduced-ATSP-7041 | 25.94 | −1 | 69.5 | 112.9 ± 0.01 | 2.77 ± 0.30 | ND |
| 13 | Reduced-VIP82 | 19.68 | 0 | 41.4 | 215.9 ± 0.02 | 3.13 ± 0.73 | ND |
| 14 | Reduced-VIP116 | 20.88 | −1 | 52.8 | 113.7 ± 0.01 | 5.22 ± 0.23 | ND |
| 15 | Reduced-VIP115 | 19.03 | +1 | 45.1 | 227.0 ± 0.03 | ∼3.33 | 82.93 ± 2.17 |
Additional cell permeable, p53-reactivating stapled peptides were synthesised to increase the test set size (Fig. 2). ATSP-7041, a progenitor of the first stapled α-helical peptide entering clinical trials, is a dual inhibitor of MDM2/MDMX developed by Aileron Therapeutics.56 VIP82 was previously identified by our research group as ideal for further development (high p53 induction, negligible LDH release).57 It is modelled after the sMTide-02 template sequence and contains solubilising lysine residues at the N-terminus as well as a C-terminal extension (ENF) for additional binding to a secondary pocket of MDM2.58 Both peptides, together with sMTide-02, were confirmed to be cell permeable and acting through the specific inhibition of Mdm2.57 VIP116 and VIP115, with varying numbers of N-terminal lysine residues, were also synthesised to assess the effects of locally concentrated cationic charges.
As with the case of sMTide-02, all peptides were recovered as E/Z-mixtures and the individual isomers were carefully separated by reverse-phase HPLC. Except for ATSP-7041 (entries 3–4) in which the alkene geometry of the isomers could be assigned (Fig. S12, ESI†), the olefinic protons of remaining peptide pairs (entries 5–10) were not resolved in the 1H NMR spectra. Since we were able to resolve late-sMTide-02 and ATSP-7041 as E-isomers, we believe the late eluting VIP peptides are also E in configuration on the basis that neutralisation of dipole effects would render the E-isomer less polar than the Z-isomer. For the purpose of this paper, the isomers are defined as early- or late-eluting, according to their adjusted retention times 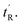
Except for VIP116 where the late isomer was ∼1.8-fold more efficient in p53 reactivation, no significant differences in p53 activation levels or correlation to α-helicity were observed for the remaining peptides pairs. Whilst adjustment of the overall charge from 0 to +2 often leads to enhanced cell permeability of stapled peptides,59 we observed in this study that accumulation of localised positive charges can lead to undesired membrane disruption. VIP115, containing 3 consecutive N-terminal lysine residues and a net charge of +1, gave rise to significant amounts of LDH leakage whereas VIP82 (with 2 N-terminal lysines and a net charge of 0) and other peptides (net charge of −1) activated intracellular p53 with negligible nonspecific cytotoxicity.
Stereoselective synthesis of the late stapled peptide isomer
Few papers in the stapled peptide literature report the E/Z ratio or discuss the implications of isomerism.49,51,52 In our case, and likewise observed by Mangold and Grubbs,52 no apparent relationship between the helicity and staple isomer type could be observed. Although a previous report has demonstrated negligible differences between the binding and cellular activities of E/Z isomers of related MDM2-binding peptides in SJSA-1 cells,49 we observed ∼1.6 to 2.2-fold improvements in binding and p53 activation for the late isomers of VIP116 and sMTide-02. For this reason, we targeted the synthesis of the more hydrophobic, late isomer of these peptides only.A previous report had described the synthesis of stereopure Z-peptide isomers using chelating ruthenium catalysts.60 Stereoselectivity was attributed to steric clash between the catalyst ligands and the approaching olefin. Similarly, by systematically varying different aspects of the existing metathesis protocol, we reasoned that optimum conditions for attaining stapled peptide late isomers could be achieved.
For this study, only ruthenium-based catalysts were considered as molybdenum-based catalysts tend to be air- and moisture-sensitive. Moreover, complete inert conditions are more difficult to achieve in a manual SPPS format.61,62 sMTide-02 was used as the model substrate. As illustrated by Table 2 (entry 1), subjection of the linear, side chain-protected peptide to three rounds of olefin metathesis using Grubbs-I catalyst (1) in DCE resulted in 96% conversion to the desired product with 41% E-selectivity. THF and toluene (entries 2–3) were tolerated by catalyst 1 on resin but resulted in lower conversion and E-selectivity.
| Entry | Peptide | Catalyst | Solvent | Temperature | Conversiona (%) | Selectivityb (early isomer![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) late isomer) late isomer) |
||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 RCMc | 2 RCM | 3 RCM | 1 RCM | 2 RCM | 3 RCM | |||||
| a Percent conversion = product/(product + starting material) as determined by reverse phase HPLC. b Determined by reverse phase HPLC. Early and late isomers of sMTide-02 have Z and E configurations respectively. c Number of RCM cycles. | ||||||||||
| 1 | sMTide-02 | 1 | DCE | rt | 75 | 92 | 96 | 57![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 43 43 |
55![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 45 45 |
59![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 41 41 |
| 2 | sMTide-02 | 1 | THF | rt | 31 | 57 | 72 | 67![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 33 33 |
65![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 35 35 |
66![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 34 34 |
| 3 | sMTide-02 | 1 | PhMe | rt | 45 | 85 | 92 | 67![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 33 33 |
64![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 36 36 |
65![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 35 35 |
| 4 | sMTide-02 | 1 | DCE | 50 °C | 82 | 96 | 97 | 61![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 39 39 |
58![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 42 42 |
55![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 45 45 |
| 5 | sMTide-02 | 2 | DCE | rt | 68 | 94 | 98 | 23![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 77 77 |
23![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 77 77 |
23![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 77 77 |
| 6 | sMTide-02 | 2 | DCE | 50 °C | 98 | >98 | >98 | 24![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 76 76 |
21![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 79 79 |
22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 78 78 |
| 7 | sMTide-02 | 3 | DCE | rt | 20 | 33 | 51 | 61![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 39 39 |
62![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 38 38 |
62![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 38 38 |
| 8 | sMTide-02 | 3 | DCE | 50 °C | 57 | 82 | 91 | 63![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 37 37 |
61![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 39 39 |
59![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 41 41 |
| 9 | sMTide-02 | 4 | DCE | rt | 61 | 77 | 85 | 23![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 77 77 |
24![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 76 76 |
23![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 77 77 |
| 10 | sMTide-02 | 4 | DCE | 50 °C | 87 | >98 | >98 | 27![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 73 73 |
25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 75 75 |
25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 75 75 |
| 11 | ATSP-7041 | 1 | DCE | rt | N/A | N/A | >98 | N/A | N/A | 61![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 39 39 |
| 12 | ATSP-7041 | 2 | DCE | 50 °C | N/A | >98 | N/A | N/A | 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 85 85 |
N/A |
| 13 | VIP116 | 1 | DCE | rt | N/A | N/A | >98 | N/A | N/A | 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50 50 |
| 14 | VIP116 | 2 | DCE | 50 °C | N/A | >98 | N/A | N/A | 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 83 83 |
N/A |
Use of catalyst 2 afforded stapled sMTide-02 in 98% yield after three metathesis cycles, with complete reversal of olefin selectivity (entry 5). In comparison to their Grubbs counterparts, isopropoxy-containing Hoveyda–Grubbs catalysts 3 and 4 had lower reactivity but similar diastereoselectivity (entries 7 and 9). For all catalysts tested, increasing the reaction temperature to 50 °C increased the reaction efficiency but had little impact on stereoselectivity. Notably, the E/Z ratio of the stapled peptide products remained constant from cycle to cycle for all catalyst/solvent/temperature combinations trialled. These results imply that certain steps of the metathesis catalytic cycle had been rendered irreversible, most likely due to ethylene loss from agitation of the resin by gas bubbling.
Interestingly, Mangold and Grubbs also observed some of the highest levels of E-selectivity when bis-mesityl N-heterocyclic carbine-containing ruthenium catalysts 2 and 4 were used for i, i+7 stapling.52 The peptides studied were distinctively different: one was venom-derived and two were based on the helical binding domain of the NOTCH transcription factor complex. Macrocycle ring size and inherent activity of the catalyst appear to be the dominant drivers of stereoselectivity, but we cannot completely rule out sequence dependency since side chain interactions will also influence conformational biasing in the transition state.
Given that second-generation catalyst 2 was highly active towards E-selective metathesis of sMTide-02, additional experiments were performed on related p53-reactivating stapled peptides. As a direct comparison, ATSP-7041 and VIP116 (representative lysine-containing peptide with a −1 net charge) were treated with both Grubbs first-generation catalyst (1) and the optimised Grubbs second-generation catalyst (2) conditions at 50 °C. Use of catalyst 1 led to >98% conversion to stapled ATSP-7041 and VIP116 after 3 metathesis cycles (entries 11 and 13, Table 2), with respective late-isomer selectivity of 39 and 50%. As expected, metathesis with catalyst 2 provided the stapled peptides in >98% yield after 2 metathesis cycles, and with an improved ≥83% selectivity for the late isomer (entries 12 and 14, Table 2).
Hydrogenated sMTide-02 is more active than olefinic sMTide-02
Thus far, we have attempted to address the inherent isomerism of i, i+7 stapled peptides by modulating the product ratio through catalyst control. Perhaps a more straightforward approach would be to remove the isomers altogether. This would also obviate the need for olefin geometry assignment which is often not trivial.Despite demonstrated successes of sequential macrocyclisation and hydrogenation, from the seminal work of Blackwell and Grubbs to on-resin strategies proposed by Schafmeister and Whelan,20,21,63 a structure search of the literature revealed a single incident of an i, i+7 stapled peptide bearing a saturated hydrocarbon bridge.64 Moreover, the staple was introduced into the peptide sequence as a pre-assembled diaminodiacid building block and not formed by intramolecular cyclisation of 2 alkenyl groups.
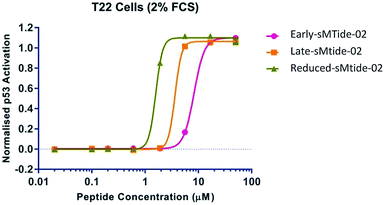
Intrigued by an apparent underutilisation of the two-step metathesis/reduction protocol, we subjected resin-bound (E)-sMTide-02 to a transfer hydrogenation and probed the robustness of the literature conditions. We favoured the method outlined by Schafmeister et al.21 as the experiment could be conducted on-resin using standard laboratory techniques, without the requirement for sealed tubes or pressure equipment. Accordingly, resin-bound, side-chain protected stapled sMTide-02 (synthesised using the optimum conditions from the previous catalyst study) was pre-swelled in NMP then treated with 2,4,6-triisopropylbenzenesulfonyl hydrazide (TPSH) and piperidine at 60 °C for 2 hours, resulting in the formation of a more hydrophobic species. Despite iterative treatments with fresh reagents, the reaction only proceeded to 39% conversion (entry 1, Table 3). Nevertheless, the reaction mixture was subjected to reverse-phase purification and sufficient material was attained for bioactivity validation. To our surprise, further improvement in cellular potency was achieved with the saturated form of sMTide-02 (Fig. 3), prompting us to explore whether the hydrogenation effect is applicable to other peptides.
| Entry | Peptide | Catalyst | Macrocyclisation conditions | Transfer hydrogenation conditions | Conversiona (%) | ||
|---|---|---|---|---|---|---|---|
| T = 2 h | T = 4 h | T = o/n | |||||
| a Percent conversion = product/(product + starting material) as determined by reverse phase HPLC. b Solvent exchange required. c After each HPLC analysis, an additional aliquot of reagents was added to the reaction mixture. | |||||||
| 1 | sMTide-02 | 2 | 2 RCM, DCE, 50 °Cb | TPSH,c pip., NMP, 60 °C | 17 | 29 | 39 |
| 2 | sMTide-02 | 2 | 2 RCM, DCE, 50 °C | NaBH4, MeOH, rt | 0 | 0 | 6 |
| 3 | sMTide-02 | 2 | 2 RCM, DCE, 50 °C | NaBH4,c MeOH, rt | 0 | 0 | 11 |
| 4 | sMTide-02 | 2 | 2 RCM, DCE, 50 °C | Bu4NBH4,c MeOH, rt | 1 | 12 | 42 |
| 5 | sMTide-02 | 2 | 2 RCM, DCE, 50 °C | Bu4NBH4,c MeOH, 50 °C | 16 | 40 | 45 |
| 6 | sMTide-02 | 2 | 2 RCM, DCE, 50 °C | Et3SiH,c rt | 0 | 0 | 23 |
| 7 | sMTide-02 | 2 | 2 RCM, DCE, 50 °C | PhSiH3,c rt | 1 | 1 | 1 |
| 8 | sMTide-02 | 2 | 2 RCM, DCE, 50 °C | Ph2SiH2,c rt | 0 | 0 | 0 |
| 9 | sMTide-02 | 2 | 2 RCM, DCE, 50 °C | PMHS,c rt | 3 | 6 | 11 |
| 10 | sMTide-02 | 2 | 2 RCM, DCE, 50 °C | Et3SiH,c 50 °C | 13 | 45 | 82 |
| 11 | sMTide-02 | 1 | 3 RCM, DCE, 50 °C | Et3SiH,c 50 °C | 44 | 59 | 84 |
| 12 | sMTide-02 | 3 | 3 RCM, DCE, 50 °C | Et3SiH,c 50 °C | 16 | 31 | 75 |
| 13 | sMTide-02 | 4 | 2 RCM, DCE, 50 °C | Et3SiH,c 50 °C | 19 | 57 | 78 |
| 14 | VIP116 | 2 | 2 RCM, DCE, 50 °C | TPSH,c pip., NMP, 60 °C | N/A | N/A | 45 |
| 15 | VIP116 | 2 | 2 RCM, DCE, 50 °C | Et3SiH,c 50 °C | N/A | N/A | 76 |
| 16 | ATSP-7041 | 2 | 2 RCM, DCE, 50 °C | TPSH,c pip., NMP, 60 °C | N/A | N/A | 39 |
| 17 | ATSP-7041 | 2 | 2 RCM, DCE, 50 °C | Et3SiH,c 50 °C | N/A | N/A | 90 |
Synthesis of saturated stapled peptides via a one-pot metathesis/transfer hydrogenation
In light of the experimental findings that saturated stapled peptides may have advantages over the corresponding unsaturated forms, there appears to be an unmet need for an efficient method for their preparation. NHC-containing ruthenium complexes are known to be viable pre-catalysts for the direct and transfer hydrogenation of alkenes, however these systems are generally limited to polarised C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C bonds.65 More recently, a tandem metathesis/hydrogenation reaction was published by Connolly et al. in which the use of ruthenium metathesis catalysts in conjunction with sodium borohydride led to the hydrogenation of unactivated alkenes.66 Besides the broad reactant scope, the reaction could be carried out in a single pot at ambient temperature. In an attempt to expand the utility of the method, the described solution-phase conditions were carried out on resin.
C bonds.65 More recently, a tandem metathesis/hydrogenation reaction was published by Connolly et al. in which the use of ruthenium metathesis catalysts in conjunction with sodium borohydride led to the hydrogenation of unactivated alkenes.66 Besides the broad reactant scope, the reaction could be carried out in a single pot at ambient temperature. In an attempt to expand the utility of the method, the described solution-phase conditions were carried out on resin.
Following 2 metathesis cycles using catalyst 2 at 50 °C, sodium borohydride (2 equivalents) and MeOH (1/20 volume) were added to the same reaction pot (entry 2, Table 3). The reduction of stapled sMTide-02 was monitored by periodic HPLC/MS analysis. After overnight reaction, only trace amounts of the desired product could be detected. Addition of excess reagents (entry 3) had no effect on product conversion whereas use of the more soluble tetrabutylammonium borohydride (entry 4) afforded the alkane product in an improved 42% yield. Increasing the reaction temperature to 50 °C resulted in faster conversion rates but the reaction had plateaued at ∼40–45% conversion (entry 5).
The mechanism of the transfer hydrogenation likely proceeds via a metal–dihydride species, with sodium borohydride acting as the hydride donor.66 Considering that organosilanes are widely employed in SPPS as the reductant in palladium-mediated deallylations, we decided to substitute silanes for tetrabutylammonium borohydride. Our search of the literature revealed precedence for the use of triethylsilane in solid-supported, sequential metathesis/hydrogenation.67,68 The microwave irradiation conditions described by Mata and co-workers were optimised for α,β-unsaturated ester substrates whereas Jida et al. focused on peptides with allylic ethers. To the best of our knowledge, concomitant metathesis/hydrogenation of internal, unactivated alkenes has not been attempted.
Unlike the borohydride conditions where methanol was added to solubilise the reducing agent, organosilanes can be introduced neat. Evaluation of various silanes revealed triethylsilane to be the optimum reductant (entry 6, Table 3). Use of phenylsilane, diphenylsilane or polymeric PMHS in conjunction with catalyst 2 provided low yielding products (entries 7–9). With all else being equal, increasing the reaction temperature of triethylsilane reduction to 50 °C drastically increased product conversion from 23 to 82% (entry 10). To our delight, other common ruthenium metathesis catalysts also demonstrated compatibility with the one-pot metathesis/hydrogenation set up (entries 11–13).
Having identified suitable conditions for the on-resin hydrogenation of sMTide-02, we also examined the utility of the one-pot protocol using stapled VIP116 and ATSP-7041. Both peptides were first subjected to conventional TPSH conditions (entries 14 and 16, Table 3). Again, the reaction had plateaued at ∼40%. Under our optimised conditions, hydrogenated VIP116 and ATSP-7041 were obtained in 76 and 90% yield, respectively (entries 15 and 17).
The peptides, along with reduced VIP82 and VIP115, were subjected to the same biological assays as their unsaturated counterparts (Table 1, entries 12–15). Higher kd values were observed, but their diminished binding to Mdm2 did not lead to an attenuation in their ability to reactivate p53. In all cases, the reduced analogues exhibited similar % helicity, LDH50 values and were no less active than the corresponding alkenyl derivatives.
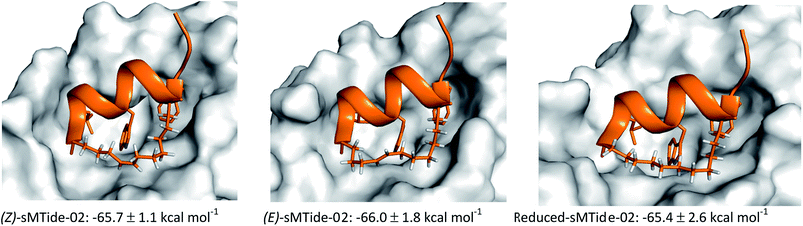
For each peptide trio tested, the reduced form consistently displayed the highest kd value whereas the late-isomer the lowest. In an attempt to understand the structural basis for these binding results, molecular dynamics (MD) simulations of sMTide-02-Mdm2 complexes were performed. The shortest peptide sMTide-02 was chosen to minimise variations in energies due to transient interactions of the flexible peptide regions with Mdm2. The calculated average binding free energies of the isomers were in qualitative agreement with the experimental binding affinity trend (Fig. 4), however the differences were not significant enough for any conclusive structure–activity relationship to be drawn.
Staple effects are more prominent in animal models
Thus far, we have relied on cellular assays to evaluate the p53 activation efficiency of stapled peptide analogues. By transitioning to animal models, we wished to further validate these findings. Animal studies were conducted using (E)-ATSP-7041 (progenitor of the first stapled peptide in clinical trials, the structure of which was determined using the published X-ray data69) as the positive control. The solubility of the representative stapled peptides (sMTide-02, VIP116 and ATSP-7041) was assessed by the addition of Hanks' balanced salt solution (HBSS) to peptide solutions in DMSO. All isomers of ATSP-7041 could be dissolved easily. The other stapled peptides immediately precipitated out from solution but upon standing at room temperature with occasional trituration, VIP116 became solubilised in HBSS buffer after 2–3 hours. Hence VIP116 analogues were assessed in the pharmacodynamics study, with 2% DMSO in HBSS buffer as the vehicle control.C57BL/6 mice bearing allograft tumour (B16F10 melanoma cells) were administered with the controls or VIP116 peptides (40 mg kg−1) via intraperitoneal injection. 6 hours post injection, spleen and tumour tissues were harvested and p53 stabilisation was detected by immunohistochemistry (IHC) staining (Fig. 5). In the absence of peptide, negligible amount of brown staining was detected in the spleen and a small percentage of positive cell population was observed in tumour tissues. Compared to the vehicle control, mice treated with (E)-ATSP-7041 and early-VIP116 showed increased p53 expression levels and p53 positive cell population in both spleen and tumour tissues. Late-VIP116 and reduced-VIP116 led to even stronger p53 immunopositive staining. Overall, the allografts results were in good agreement with the spleen samples.
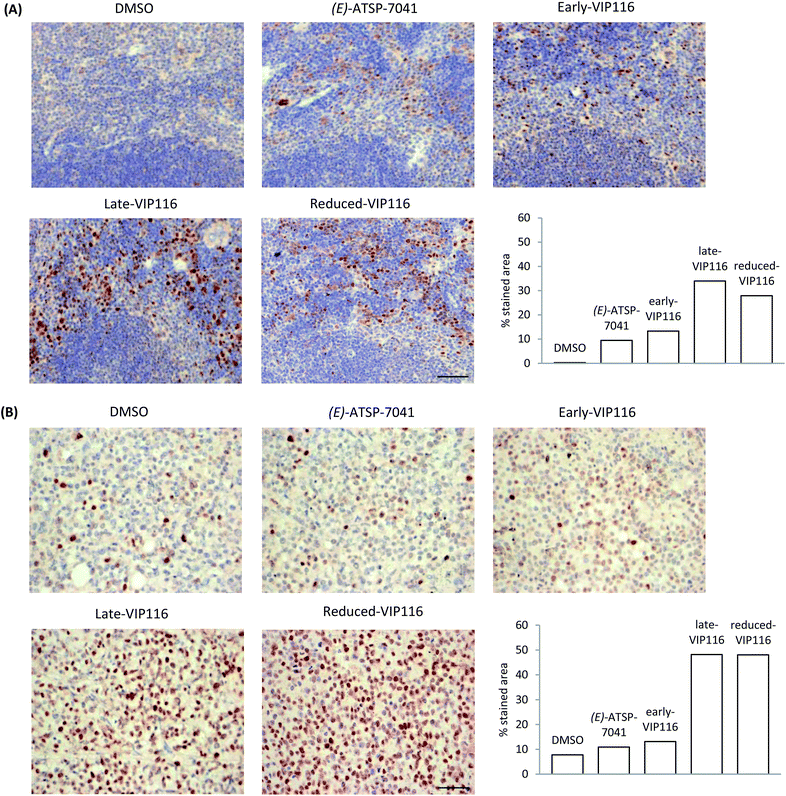 | ||
| Fig. 5 (A) p53 accumulation in mouse spleen tissues after treatment with VIP116 analogues or controls. (B) p53 accumulation in mouse B16-F10 allograft tumours after treatment with VIP116 analogues or controls. Each image is representative of 5 different images taken of each sample (n = 2–4). Scale bars represent 50 μm. Immunostaining quantification was performed using inverted images in ImageJ.70 | ||
Differences between the in vitro and in vivo data could be observed however. For instance, the p53Act values of (E)-ATSP-7041 and late-VIP116 were roughly the same and about 1.8-fold higher than that of early- and reduced-VIP116, but in the allograft tumour model, reduced-VIP116 was found to be as active as late-VIP116 and 4-fold more efficient in reactivating p53 than (E)-ATSP-7041 and early-VIP116. These results cannot be easily explained without further investigation, but are likely due to pharmacokinetics and bioavailability reasons. This also highlights the danger of eliminating compounds based solely on in vitro binding data and the need to develop more predictive tools for cellular uptake and stability for peptide macrocycles.
Conclusions
We hypothesised that changes in the atomic arrangement of the cross-link of p53-reactivating stapled peptides would have an impact on binding since the hot spot residues resided on one helical face and the chemical brace was oriented towards the protein. Differences in biophysical and biological properties were especially apparent between the E/Z-isomers of sMTide-02 and VIP116. We reasoned the late isomer, with improved cellular activity, would be a better candidate to take forward for development. Thus we systematically screened metathesis conditions and found that the use of NHC-containing ruthenium catalysts favoured the formation of the desired, more hydrophobic i, i+7 stapled peptide isomer. Notably, isomer differences in VIP116 had a marked effect in animal experiments.We further established a one-pot metathesis/reduction protocol to generate the corresponding hydrogenated peptides. Introduction of triethylsilane to the metathesis mixture effectively redirected the active ruthenium species to catalyse the hydrogenation of internal, unactivated olefins. Such chemical modifications obviate the often non-trivial tasks of stapled peptide isolation in a fixed ratio or isomer characterisations and might offer a more economical manufacturing route. Despite being the weakest binders, the reduced peptides induced comparable p53 activity to their olefin counterparts in the cellular assays. Again, enhancement in p53 activity was observed in mice for the VIP116 analogue. These observations could not be explained by differences in α-helical content but may be a result of favourable membrane translocation due to increased hydrophobicity,71 enhanced endosomal escape and/or p53 activation through binding to Mdmx.
Preliminary results provide sufficient evidence to warrant further investigation of the therapeutic use of sMTide-02 and VIP116 templates which are actively being explored in our laboratory. Our findings also highlight the importance of identifying the impact of E/Z isomer and saturated analogues of all-hydrocarbon stapled peptides. Although this work focused solely on i, i+7 Mdm2-binding stapled peptides, the chemical strategies presented herein for the modulation of E/Z-ratio and one-pot metathesis/reduction are expected to be amenable to other stapled peptides and should be considered for other pathways modulated by α-helical stapled peptides.
Author contributions
T. Y. Y., F. J. F. G. and C. W. J. designed the chemistry experiments. C. J. B., Y. X. and D. P. L. designed the biology experiments. Y. S. T. and C. S. V. designed the computational modelling experiments. F. J. F. G., X. E. L. and N. J. Y. synthesised the peptides. T. Y. Y. and N. J. Y. performed the metathesis and hydrogenation experiments. D. T. carried out T22 p53 reporter, fluorescence anisotropy competition and LDH release assays. Y. X. performed animal studies. H. Y. K. K. and A. W. P. collected the CD spectra. T. Y. Y., C. J. B., Y. X., Y. S. T. analysed the data. T. Y. Y. wrote the paper and all authors contributed to revising the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This research was funded by the A*STAR JCO Visiting Investigatorship Programme (JCO 1235d00048) and the Industry Alignment Fund (Pre-positioning, HBMS domain H17/01/a0/010).Notes and references
- D. Y. Jackson, D. S. King, J. Chmielewski, S. Singh and P. G. Schultz, J. Am. Chem. Soc., 1991, 113, 9391 CrossRef CAS.
- F. M. Brunel and P. E. Dawson, Chem. Commun., 2005, 20, 2552 RSC.
- A. A. Aimetti, R. K. Shoemaker, C.-C. Lin and K. S. Anseth, Chem. Commun., 2010, 46(23), 4061 RSC.
- A. Muppidi, Z. Wang, X. Li, J. Chen and Q. Lin, Chem. Commun., 2011, 47(33), 9396 RSC.
- C. M. Grison, G. M. Burslem, J. A. Miles, L. K. A. Pilsl, D. J. Yeo, Z. Imani, S. L. Warriner, M. E. Webb and A. J. Wilson, Chem. Sci., 2017, 8(7), 5166 RSC.
- N. Assem, D. J. Ferreira, D. W. Wolan and P. E. Dawson, Angew. Chem. Int. Ed., 2015, 54(30), 8665 CrossRef CAS PubMed.
- Y. Wang and D. H. C. Chou, Angew. Chem., Int. Ed., 2015, 54(37), 10931 CrossRef CAS PubMed.
- Y. Wang, B. J. Bruno, S. Cornillie, J. M. Nogieira, D. Chen, T. E. Cheatham, C. S. Lim and D. H. C. Chou, Chem. - Eur. J., 2017, 23(29), 7087 CrossRef CAS PubMed.
- A. J. Rojas, C. Zhang, E. V. Vinogradova, N. H. Buchward, J. Reilly, B. L. Pentelute and S. L. Buchwald, Chem. Sci., 2017, 8(6), 4257 RSC.
- J. W. Taylor, Biopolymers, 2002, 66, 49 CrossRef CAS PubMed.
- K. Fujimoto, N. Oimoto, K. Katsuno and M. Inouye, Chem. Commun., 2004, 11, 1280 RSC.
- K. Fujimoto, K. M. Kajino and M. Inouye, Chem. - Eur. J., 2008, 14(3), 857 CrossRef CAS PubMed.
- M. Kajino, K. Fujimoto and M. Inouye, J. Am. Chem. Soc., 2011, 133(4), 656 CrossRef CAS PubMed.
- G. Lautrette, F. Touti, H. G. Lee, P. Dai and B. L. Pentelute, J. Am. Chem. Soc., 2016, 138(27), 8340 CrossRef CAS PubMed.
- H. G. Lee, G. Lautrette, B. L. Pentelute and S. L. Buchwald, Angew. Chem., Int. Ed., 2017, 56(12), 3177 CrossRef CAS PubMed.
- K. M. Makwana and R. Mahalakshmi, Org. Lett., 2015, 17(10), 2498 CrossRef CAS PubMed.
- L. Mendive-Tapia, S. Preciado, J. García, R. Ramón, N. Kielland, F. Albericio and R. Lavilla, Nat. Commun., 2015, 6, 7160 CrossRef PubMed.
- E. Y.-L. Hui, B. Rout, Y. S. Tan, C. S. Verma, K.-P. Chan and C. W. Johannes, Org. Biomol. Chem., 2018, 16, 389 RSC.
- X. Shi, R. Zhao, Y. Jiang, H. Zhao, Y. Tian, Y. Jiang, J. Li, W. Qin, F. Yin and Z. Li, Chem. Sci., 2018, 9, 3227 RSC.
- H. E. Blackwell and R. H. Grubbs, Angew. Chem., Int. Ed., 1998, 37(23), 3281 CrossRef CAS PubMed.
- C. E. Schafmeister, J. Po and G. L. Verdine, J. Am. Chem. Soc., 2000, 122(24), 5891 CrossRef CAS.
- Y.-W. Kim, P. S. Kutchukian and G. L. Verdine, Org. Lett., 2010, 12(13), 3046 CrossRef CAS PubMed.
- Y.-W. Kim, T. N. Grossmann and G. L. Verdine, Nat. Protoc., 2011, 6, 761 CrossRef CAS PubMed.
- J. F. Reichwein, B. Wels, J. A. W. Kruijtzer, C. Versluis and R. M. J. Liskamp, Angew. Chem., Int. Ed., 1999, 38(24), 3684 CrossRef CAS PubMed.
- C. M. Haney, M. T. Loch and W. S. Horne, Chem. Commun., 2011, 47(39), 10915 RSC.
- S. Cantel, A. L. C. Isaad, M. Scrima, J. J. Levy, R. D. DiMarchi, P. Rovero, J. A. Halperin, A. M. D'Ursi, A. M. Papini and M. Chorev, J. Org. Chem., 2008, 73, 5663 CrossRef CAS PubMed.
- M. Empting, O. Avrutina, R. Meusinger, S. Fabritz, M. Reinwarth, M. Biesalski, S. Voigt, G. Buntkowsky and H. Kolmar, Angew. Chem., Int. Ed., 2011, 50(22), 5207 CrossRef CAS PubMed.
- Y. H. Lau, Y. Wu, P. de Andrade, W. R. J. D. Galloway and D. R. Spring, Nat. Protoc., 2015, 10, 585 CrossRef CAS PubMed.
- Y. H. Lau, P. de Andrade, N. Skold, G. J. McKenzie, A. R. Venkitaraman, C. Verma, D. P. Lane and D. R. Spring, Org. Biomol. Chem., 2014, 12(24), 4074 RSC.
- Y. H. Lau, P. de Andrade, S.-T. Quah, M. Rossmann, L. Laraia, N. Skold, T. J. Sum, P. J. E. Rowling, T. L. Joseph, C. Verma, M. Hyvonen, M. L. S. Itzhaki, A. R. Venkitaraman, C. J. Brown, D. P. Lane and D. R. Spring, Chem. Sci., 2014, 5(5), 1804 RSC.
- A. A. Vinogradov, Z.-N. Choo, K. A. Totaro and B. L. Pentelute, Org. Lett., 2016, 18(6), 1226 CrossRef CAS PubMed.
- P. A. Cistrone, A. P. Silvestri, J. C. J. Hintzen and P. E. Dawson, ChemBioChem, 2018, 18(10), 1031 CrossRef PubMed.
- K. Fujimoto, M. Amano, Y. Horibe and M. Inouye, Org. Lett., 2006, 8(2), 285 CrossRef CAS PubMed.
- J. C. Serrano, J. Sipthorp, W. Xu, L. S. Itzhaki and S. V. Ley, ChemBioChem, 2017, 18(12), 1066 CrossRef CAS PubMed.
- P. M. Cromm, S. Schaubach, J. Spiegel, A. Fürstner, T. N. Grossmann and H. Waldmann, Nat. Commun., 2016, 7, 11300 CrossRef CAS PubMed.
- E. M. Driggers, S. P. Hale, J. Lee and N. K. Terrett, Nat. Rev. Drug Discovery, 2008, 7, 608 CrossRef CAS PubMed.
- E. Marsault and M. L. Peterson, J. Med. Chem., 2011, 54, 1961 CrossRef CAS PubMed.
- J. Mallinson and I. Collins, Future Med. Chem., 2012, 4, 1409 CrossRef CAS PubMed.
- C. J. Brown, C. F. Cheok, C. S. Verma and D. P. Lane, Trends Pharmacol. Sci., 2011, 32, 53 CrossRef CAS PubMed.
- C. J. Brown, S. Lain, C. S. Verma, A. R. Fersht and D. P. Lane, Nat. Rev. Cancer, 2009, 9, 862 CrossRef CAS PubMed.
- J. K. Murray and S. H. Gellman, Biopolymers, 2007, 88, 657 CrossRef CAS PubMed.
- M. Wallace, E. Worrall, S. Pettersson, T. R. Hupp and K. L. Ball, Mol. Cell, 2006, 23, 251 CrossRef CAS PubMed.
- W. Xue, L. Zender, C. Miething, R. A. Dickins, E. Hernando, V. Krizhanovsky, C. Cordon-Cardo and S. W. Lowe, Nature, 2007, 445, 656 CrossRef CAS PubMed.
- A. Ventura, D. G. Kirsch, M. E. McLaughlin, D. A. Tuveson, J. Grimm, L. Lintault, J. Newman, E. E. Reczek, R. Weissleder and T. Jacks, Nature, 2007, 445, 661 CrossRef CAS PubMed.
- L. T. Vassilev, B. T. Vu, B. Graves, D. Carvajal, F. Podlaski, Z. Filipovic, N. Kong, U. Kammlott, C. Lukacs, C. Klein, N. Fotouhi and E. A. Liu, Science, 2004, 303, 844 CrossRef CAS PubMed.
- C. J. Brown, S. T. Quah, J. Jong, A. M. Goh, P. C. Chiam, K. H. Khoo, M. L. Choong, M. A. Lee, L. Yurlova, K. Zolghadr, T. L. Joseph, C. S. Verma and D. P. Lane, ACS Chem. Biol., 2013, 8, 506 CrossRef CAS PubMed.
- S. Shangary, D. Qin, D. McEachern, M. Liu, R. S. Miller, S. Qiu, Z. Nikolovska-Coleska, K. Ding, G. Want, J. Chen, D. Bernard, J. Zhang, Y. Lu, Q. Gu, R. B. Shah, K. J. Pienta, X. Ling, S. Kang, M. Guo, Y. Sun, D. Yang and S. Wang, Proc. Natl. Acad. Sci. U.S.A., 2008, 105, 3933 CrossRef CAS PubMed.
- Y. S. Chang, B. Graves, V. Guerlavais, C. Tovar, K. Packman, K.-H. To, K. A. Olson, K. Kesavan, P. Gangurde, A. Mukherjee, T. Baker, K. Darlak, C. Elkin, Z. Filipovic, F. Z. Qureshi, H. Cai, P. Berry, E. Feyfant, X. E. Shi, J. Horstick, D. A. Annis, A. M. Manning, N. Fotouhi, H. Nash, L. T. Wassilev and T. K. Sayer, Proc. Natl. Acad. Sci. U.S.A., 2013, 110, E3445 CrossRef CAS PubMed.
- R. Wallbrecher, P. Chène, S. Ruetz, T. Stachyra, T. Vorherr and R. Brock, Br. J. Pharmacol., 2017, 174, 2613 CrossRef CAS PubMed.
- F. Wachter, A. M. Morgan, M. Godes, R. Mourtada, G. H. Bird and L. D. Walensky, Oncogene, 2017, 36, 2184 CrossRef CAS PubMed.
- H. Chapuis, J. Slaninová, L. Bednárová, L. Monincová, M. Buděšínský and V. Čeřovský, Amino Acids, 2012, 2047 CrossRef CAS PubMed.
- S. L. Mangold and R. H. Grubbs, Chem. Sci., 2015, 6, 4561 RSC.
- G. H. Bird, W. C. Crannell and L. D. Walensky, Curr. Protoc. Chem. Biol., 2011, 3, 99 Search PubMed.
- D. Villemin, Tetrahedron Lett., 1980, 21, 1715 CrossRef CAS.
- J. Tsuji and S. Hashiguchi, Tetrahedron Lett., 1980, 21, 2955 CrossRef CAS.
- F. Meric-Bernstam, M. N. Saleh, J. R. Infante, S. Goel, G. S. Falchook, G. Shapiro, K. Y. chung, R. M. Conry, D. S. Hong, J. S.-Z. Wang, U. Steidl, L. D. Walensky, V. Guerlavais, M. Payton, D. A. Annis, M. Aivado and M. R. Patel, J. Clin. Oncol., 2017, 2505 Search PubMed.
- D. Thean, J. S. Ebo, T. Luxton, X. E. C. Lee, T. Y. Yuen, F. J. Ferrer, C. W. Johannes, D. P. Lane and C. J. Brown, Nat. Sci. Rep., 2017, 7, 1763 CrossRef CAS PubMed.
- Y. S. Tan, J. Reeks, C. J. Brown, D. Thean, F. J. Ferrer Gago, T. Y. Yuen, E. T. L. Goh, X. E. C. Lee, C. E. Jennings, T. L. Joseph, R. Lakshminarayanan, D. P. Lane, M. E. M. Noble and C. S. Verma, J. Phys. Chem. Lett., 2016, 7, 3452 CrossRef CAS PubMed.
- L. D. Walensky and G. H. Bird, J. Med. Chem., 2014, 57, 6275 CrossRef CAS PubMed.
- S. L. Mangold, D. J. O'Leary and R. H. Grubbs, J. Am. Chem. Soc., 2014, 136, 12469 CrossRef CAS PubMed.
- R. H. Grubbs, Angew. Chem., Int. Ed., 2006, 45, 3760 CrossRef CAS PubMed.
- G. C. Vougioukalakis and R. H. Grubbs, Chem. Rev., 2010, 110, 1746 CrossRef CAS PubMed.
- A. Whelan, J. Elaridi, M. Harte, S. Smith, W. R. Jackson and A. J. Robinson, Tetrahedron Lett., 2004, 45, 9545 CrossRef CAS.
- F.-L. Wang, Y. Guo, S.-J. Li, Q.-X. Guo, J. Shi and Y.-M. Li, Org. Biomol. Chem., 2015, 13, 6286 RSC.
- S. Horn and M. Albrecht, Chem. Commun., 2011, 47, 8802 RSC.
- T. Connolly, Z. Wang, M. A. Walker, I. M. McDonald and K. M. Peese, Org. Lett., 2014, 16, 4444 CrossRef CAS PubMed.
- A. A. Poeylaut-Palena, S. A. Testero and E. G. Mata, Chem. Commun., 2011, 47, 1565 RSC.
- M. Jida, C. Betti, P. W. Schiller, D. Tourwé and S. Ballet, ACS Comb. Sci., 2014, 16, 342 CrossRef CAS PubMed.
- Y. S. Chang, B. Graves, V. Guerlavais, C. Tovar, K. Packman, K.-H. To, K. A. Olson, K. Kesavan, P. Gangurde, A. Mukherjee, T. Baker, K. Darlak, c. Elkin, Z. Filipovic, F. Z. Qureshi, H. Cai, P. Berry, E. Feyfant, X. E. Shi, J. Horstick, D. A. Annis, A. M. Manning, N. Fohouhi, H. Nash, L. T. Vassilev and T. K. Sawyer, Proc. Natl. Acad. Sci. U. S. A., 2013, 110(36), E3445 CrossRef CAS PubMed.
- J. Schindelin, L. Arganda-Carreras, E. Frise, V. Kaynig, M. Longair, T. Pietzsch, S. Preibisch, C. Rueden, S. Saalfeld, B. Schmid, J.-Y. Tinevez, D. J. White, V. Hartenstein, K. Eliceiri, P. Tomancak and A. Cardona, Nat. Methods, 2012, 9(7), 676 CrossRef CAS PubMed.
- K. Sakagami, T. Masuda, K. Kawano and S. Futaki, Mol. Pharm., 2018, 15(3), 1332 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9sc01456j |
| This journal is © The Royal Society of Chemistry 2019 |