Open Access Article
Open Access ArticleProgrammable intracellular DNA biocomputing circuits for reliable cell recognitions†
Xue
Gong
a,
Jie
Wei
a,
Jing
Liu
b,
Ruomeng
Li
a,
Xiaoqing
Liu
 a and
Fuan
Wang
a and
Fuan
Wang
 *a
*a
aKey Laboratory of Analytical Chemistry for Biology and Medicine (Ministry of Education), College of Chemistry and Molecular Sciences, Wuhan University, Wuhan, P. R. China. E-mail: fuanwang@whu.edu.cn
bDepartment of Gastroenterology, Wuhan University Zhongnan Hospital, China
First published on 15th January 2019
Abstract
Dynamic nucleic acid-based biocircuits have spurred substantial research efforts for diagnosis or biomedical applications at the molecular level; nevertheless, it still remains a challenge to design programmable molecular circuit devices for autonomous and accurate diagnosis of low abundance biomolecules in a complex intracellular environment. Herein, a reconfigurable hybridization-based chain reaction is introduced to assemble modular biocomputing circuits that include a general sensing module and a versatile processing module. By modular sensing module design, we realized multiple endogenous miRNA-initiated biocomputing operations, including binary logic gates (OR, AND, INHIBIT and XOR), and more advanced concatenated logic circuits (XOR-AND, XOR-INHIBIT, and XOR-OR) in different living cells. The sensing module transduces the primary miRNA sensing event into an intermediate trigger for activating the processing module that further transduces the specific analyte recognition pattern into an amplified fluorescence readout. Based on an appropriate selection of multiple miRNA analytes, various miRNA expression patterns could be utilized for sensitive and selective cell discriminations. The inherent synergistically accelerated recognition and hybridization features of our biocomputing systems contribute to the amplified detection of multiplex endogenous miRNAs in living cells, thus providing an efficient toolbox for more accurate diagnosis and programmable therapeutics.
Introduction
Conventional silicon-based computers are assembled from elementary logic gates which can perform various Boolean logic operations by recognizing and processing one or more Boolean inputs representing True (1, high voltage) or False (0, low voltage) outputs.1 Thus, they can generate a single binary output according to a certain input–output signal correlation pattern. Since the first proof-of-concept demonstration of DNA biocomputation was utilized for solving intricate Hamiltonian paths,2 a myriad of DNA biocomputing systems were engineered by using topological routes and sequence-design strategies.3–5 These DNA-based bottom-up principles could overcome the bottleneck of miniaturization of conventional silicon-based computation devices.6 Due to its desirable prediction of base-pairing, ease of functionalization and spatial addressability, DNA has been increasingly recognized as a powerful candidate to construct synthetic biocomputing circuit devices.7–9 Despite rapid advances in synthetic biocircuit systems, challenges exist in in vivo living cell biocomputation which requires multiple biomolecular inputs as well as sophisticated information processing constructs.Pioneering efforts have been devoted to the rational design of isothermal nonenzymatic entropy-driven DNA circuits that are based on the toehold-mediated strand-displacement reaction (SDR).10–12 Here, a single-stranded oligonucleotide input is designed to interact with a logic gate device through a predictable branch-migration process. A series of synthetic biocomputing systems have thus been realized by integrating multiple synthetic logic gates into complex biocircuits, thus paving the way to intelligent artificial neural networks.13,14 Despite the extensive exploration of these DNA-based biocomputing circuits, challenges still exist in realizing these synthetic biocomputing platforms for simultaneous and sequential transduction of endogenously generated biomolecular inputs in living biosystems.15–18 Thus, obtaining an in-depth understanding of these genes and neural signalling pathways is more feasible by comprehensively elucidating these multiple cooperatively transcribed biomolecules and intermediates.19–21 Moreover, most of the current DNA biocomputations rely on DNA inputs which are rather limited and difficult to integrate into these more practical biocircuits since most of the intracellular DNA stays in an inclusively double-stranded structure in eukaryotes. It is thus highly desirable to explore other kinds of intracellular biomolecules that can be used as alternative inputs to activate these entropy-driven DNA circuits.
MicroRNA (miRNA) is considered as a promising candidate and is mainly present in the cytoplasm with a single-stranded structure.22 More importantly, miRNA has been demonstrated as a new type of oncogene or tumor suppressors in cancer initiation, progression, and metastasis processes.23,24 The abnormal up- or down-regulated expression of miRNA is associated with various diseases, as compared with their normal counterparts.25–28 These characteristics make miRNA a promising biomarker for early disease diagnosis and therapy, and also an appropriate input for intracellular biocomputing circuits. However, the shortness of miRNA makes accurate miRNA analysis difficult in complex intracellular environments where paragenesis nucleic acids might bring inevitable interference.29 Meanwhile, a moderately high concentration of the input is always needed for efficiently triggering conventional DNA circuits for diagnosis purposes.30–33 Thus, it is highly important to develop more versatile and robust signal amplification strategies to evaluate particular miRNAs in living cells and to facilitate the early diagnosis of miRNA-related diseases. In addition, the dysregulation of one miRNA is demonstrated to be associated with multiple diseases, thus easily leading to false diagnosis.34 Meanwhile, a latent relationship of multiple miRNA indicators is reported to be associated with specific diseases in different states.35 Hence, the implementation of multiple endogenous miRNA-initiated biomolecular circuits is highly desirable in living cells where these biocircuit devices can analyze and modulate key biomolecular information in complex biological environments. More importantly, these miRNA-involved biocircuits can ultimately make valuable contributions to early diagnosis and treatment of key diseases. The hybridization chain reaction (HCR) provides a rapid and facile nonenzymatic amplification strategy that can be performed under simple conditions (constant temperature),36–39 which contributes to its broad application on cell surfaces and even inside living cells.40–43 It provides a general and efficient amplification principle for assembling dsDNA copolymeric nanowires via trigger-initiated successive cross-opening of multiple hairpin substrates. The HCR has been successfully utilized for detecting various nucleic acid analytes44–47 while little attention has been given to HCR-based DNA biocircuits especially in the complex intracellular environment.48–50 The facile design and intrinsic amplification features of the HCR are expected to promote the integration of the HCR into intracellular DNA computing circuits for diagnosis purposes.
Here, we introduce an isothermal non-enzymatic reconfigurable hybridization chain reaction-based biocomputing system for sensitively and simultaneously analyzing multiple endogenous miRNAs in living cells. The various miRNA expression patterns were then utilized for sensitive and selective cell discriminations. The present DNA computing platform is composed of a lead-in sensing module and a powerful processing module. The sensing module recognizes different patterns of miRNA inputs to produce an intermediate trigger for the next layer of the processing module, which is composed of a cascaded HCR procedure. The processing module amplifies the primary recognition event and facilitates efficient signal transduction, leading to the selectively amplified detection of miRNA through a Förster resonance energy transfer (FRET) readout. Note that the use of the cascaded HCR strategy in living cells enables the formation of high-molecular-weight branched dsDNA copolymers with low cellular diffusibility, accelerating the precise and accurate positioning of targets in situ. By manipulating the library of biocomputing elements, a series of Boolean logic gates and concatenated logic circuits were constructed to promote the reliability of clinical diagnosis in different living cells, where multiple cell-specific miRNAs were chosen as endogenous biocomputing inputs. The cascaded HCR-based biocomputing system enables successively guaranteed recognitions and synergistically accelerated signal amplifications, which can improve the reliability of in situ diagnosis and thus may enable an effective treatment of key miRNA-related diseases.
Results and discussion
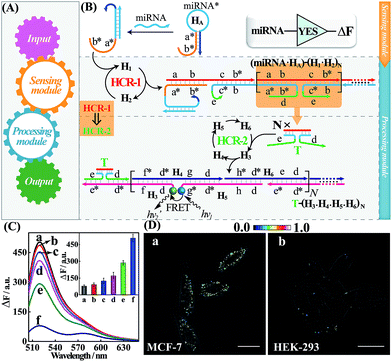
The general architecture of the miRNA-involved biocomputing system is schematically shown in Fig. 1. The biocomputing platform is composed of two DNA computing modules, a sensing module and a processing module (Fig. 1A). The sensing module is comprised of a library of caged initiator subunits that, upon their exposure to external miRNA inputs, self-assemble into an activated initiator via the selection of the corresponding subunits from the sensing module. The generated initiator can then trigger the subsequent processing module that is composed of a library of HCR hairpin reactants. The processing module immediately recognizes the as-formed initiator and amplifies the primary hybridization events of the sensing module through an autonomous and successive hybridization of these hairpin reactants. The simplest YES gate was first constructed as depicted in Fig. 1B. Note that domains X and X* represent complementary base pair regions throughout the paper (for detailed sequences, see Table S1†). In the presence of a given miRNA target (miR-21), sensing hairpin HA recognizes miR-21 and releases an initiator sequence b*-a* in the sensing module, leading to the activation of the subsequent processing module that was composed of a cascaded HCR (HCR-1/HCR-2). The as-generated initiator initiates autonomous and continuous cross-hybridizations between H1 and H2, resulting in the formation of HCR-1 dsDNA nanowires and the concomitant assembly of numerous tandem trigger (T) nanochains. Clearly, the intermediate trigger T can only be assembled through an upstream HCR-1 approach for activating the downstream HCR-2 layer of the processing module. The tandem repeated T can specifically mediate the subsequent alternating hybridizations between H3, H4, H5 and H6, yielding a long HCR-2 dsDNA concatamer. Here, H3 is functionalized at its 3′-end with a fluorescence acceptor (TAMRA) while H5 is modified at its 5′-end with a fluorescence donor (FAM). Only through the downstream HCR-2 approach the two fluorophores (FAM and TAMRA) can be successively brought into close proximity that enables the efficient Förster resonance energy transfer (FRET) process and generates an amplified FRET signal (for details, see Fig. S1A†).To evaluate the sensing performance of our YES gate, the sensitivity was first evaluated by fluorescence experiments. Kinetic traces of the proposed system revealed an increasingly amplified fluorescence response with the elevated concentrations of the miR-21 analyte ranging from 0 to 50 nM (Fig. S1B†), indicating the miR-21-triggered effective generation of FRET readouts using our YES gate device. The most appropriate reaction time-interval was chosen as 3 h for acquiring the respective fluorescence spectra in the subsequent experiments. A good linear relationship is obtained between the fluorescence intensity changes and the concentrations of miR-21 (Fig. S1C†), demonstrating the high performance of our cascaded HCR strategy for amplified miRNA assay. By modular design of the sensing module, a sensitive miR-155-initiated YES gate was similarly designed and constructed for sensing purposes (for details, see Fig. S2A and S2B†), indicating the remarkable amplification and universal applicability features of our designed platform. The present miR-21-sensing platform depicted in Fig. 1B is not only sensitive, but also selective. Fig. 1C shows the fluorescence spectra of the system upon incubating the gate constructs with 25 nM miR-21 and its control counterparts, including let-7a, and one-, two- and three-base miR-21 mutants (for the corresponding kinetics analysis, see Fig. S3†). A clear miRNA discrimination is thus demonstrated in all biological interfering sequences, implying the high selectivity of our sensing platform against other control sequences. The proposed strategy can further be applied for monitoring miRNA targets under more practical and complicated biological conditions, e.g., human serums. Even 10% serum has negligible effect on the performance of the system (Fig. S4†), indicating an acceptable accuracy of our logic gate in complex biological fluids. This highly robust sensing performance makes the utilization of our biocomputing devices feasible in the subsequent intracellular environment. In addition, the morphological features of the HCR reactants and these different HCR-assembled products were also characterized by atomic force microscopy (AFM, Fig. S5†). Many long-branched dsDNA polymeric nanowires were acquired with a height of ∼2 nm for the cascaded HCR product while only linear dsDNA nanowires were observed for the conventional HCR product, validating the successful implementation of our sensing principle.
Having demonstrated the robust performance of our system, we then explored the YES gate device inside living cells by using an intracellular imaging method. Human breast cancer (MCF-7) cells are known to express relatively high levels of miR-21 while human embryonic kidney (HEK-293) cells are associated with relatively low miR-21 expression profiles.26 Then, MCF-7 and HEK-293 cells were chosen as model systems to investigate the feasibility of our logic gate for monitoring endogenous biomarkers by using confocal laser scanning microscopy (CLSM). To ensure the sufficient biostability of our hairpin probes in a cell culture medium, all DNA probes were synthesized with phosphorothioate bonds for intracellular imaging experiments (for details, see Table S2†). The phosphorothioated DNA probes showed improved tolerance for nuclease-involving degradation (Fig. S6†), thus facilitating their subsequent intracellular applications. The as-designed miR-21-mediated YES logic constructs were transfected into these two different cells using lipofectamine 3000 and incubated with the respective cells at 37 °C for 3 h. As shown in Fig. 1D, an intense FRET imaging signal was obtained in MCF-7 cells but not in HEK-293 cells, indicating a relatively higher miR-21 expression in MCF-7 cells than in HEK-293 cells (for details, see Fig. S7†). Note that the fluorescence emission ratio of FRET to FAM (FFRET/FFAM) was utilized as the readout signal to avoid undesired interference from the surrounding complex intracellular environment and to acquire a more reliable fluorescence readout signal in living cells. The present YES biocomputing device exhibited distinct miRNA expression profiles in different cells, indicating the great potential of the system for accurately detecting cancer-related biomarkers. Meanwhile, a conventional HCR imaging strategy was established for differentiating miRNAs in MCF-7 cells by subtracting H6 from the present cascaded HCR system. No significant FRET signal was observed in the conventional HCR imaging strategy while apparent FRET signals can be seen in the cascaded HCR scheme (Fig. S8†), suggesting the significant signal amplification nature of the cascaded HCR compared to the conventional linear HCR system. Furthermore, the localized endogenous miR-21 profile was demonstrated by a series of z-section analyses of the entire MCF-7 cells (for details, see Fig. S9 and Movie S1†). Thus the programmable HCR-based logic gate can localize and visualize miRNA biomarkers in living cells and may offer new opportunities in intracellular biocomputing and bioanalysis. In short, a reconfigurable HCR was introduced to integrate the enzyme-free modular biocomputing system that is composed of a sensing module and a processing module. Both of these two modules are modular and can be reprogrammed to be encoded with more endogenous miRNA recognitions for the subsequent sophisticated biocomputing processes. By reprograming the sensing module, we can construct a series of Boolean logic devices, such as OR, AND, INHIBIT, and XOR gates, which can then be integrated with the programmed processing module for assembling more advanced and versatile concatenated DNA biocomputing circuits.
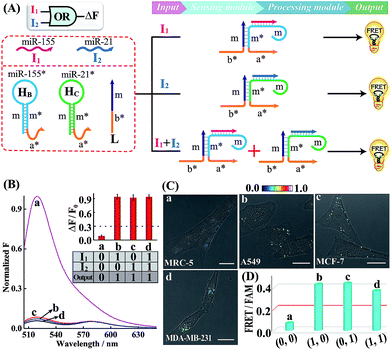
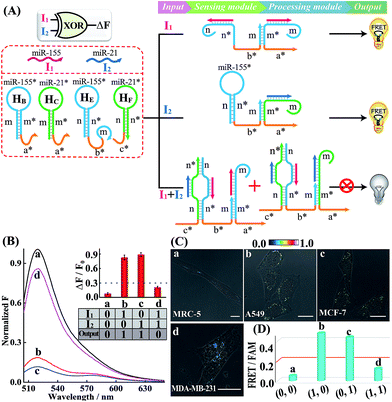
A simple OR Boolean logic gate was initially designed by merely engineering the sensing module that includes two metastable hairpins (HB and HC) and a single-stranded DNA L (Fig. 2A). Each hairpin of the sensing module contains three domains: the loop domain that is complementary to the specific miRNA input (miR-155 or miR-21), stem domain m* that is complementary to region m of L, and the initiator subunit region (orange) that is partially blocked in the stem region. Here, the initiator sequence of the cascaded HCR-comprising processing module is split into two segments a* and b* that are, respectively, conjugated to the 3′-end of two hairpins and 5′-end of L. The limited binding affinity of these split initiator subunits with the corresponding hairpin reactants prohibits the successive cross-opening of H1 and H2 in a HCR-1 fashion, leading to the blockage of the processing module and FRET transduction. However, inputs I1 (miR-155) and I2 (miR-21) could open the respective hairpins HB and HC, exposing the single-stranded domain m* to hybridize with domain m of L. The as-achieved duplex domain m/m* connects these two split initiator subunits to generate the b*-a* initiator for triggering the cascaded HCR amplifier of the processing module. As a result, a significant fluorescence change was observed upon the introduction of either one or both of the inputs I1 (miR-155) and I2 (miR-21) into the system (Fig. 2B), which is in good accord with the characteristic OR logic operation. The successful operation of the OR gate was further confirmed by PAGE experiments where any of these miRNA inputs could activate the assembly of high molecular weight copolymeric dsDNA products (Fig. S10†).
Having demonstrated the activation of the OR logic gate by these externally introduced miRNA inputs, the intracellular operation of this gate was investigated by using endogenously generated miRNAs through transfecting the OR gate constructs into different living cells (Fig. 2C). Here, MCF-7, A549 and MDA-MB-231 cells were primarily chosen as model systems to verify the intracellular logic operations of our OR gate. It is reported that A549 displays high expression levels of endogenous miR-155,25 while MDA-MB-231 cells show simultaneously overexpressed miR-21 and miR-155.28 Meanwhile, human normal lung fibroblast (MRC-5) cells was chosen as a noncancerous/negative control for setting an appropriate threshold for Truth and False outputs in living cells for all of the subsequent intracellular imaging experiments.29 The results of the corresponding statistical histogram analysis of the FRET imaging of these different cells are shown in Fig. 2D. A minimal FRET imaging signal was obtained in noncancerous MRC-5 cells (sample a of Fig. 2C and D), demonstrating that both of miR-21 and miR-155 were lowly expressed in MRC-5 cells. However, a significant FRET imaging signal was obtained in A549, MCF-7 and MDA-MB-231 cells (samples b–d of Fig. 2C and D, respectively), validating the successful utilization of our OR logic gate for monitoring multiple endogenous miRNA patterns in living cells (for details, see Fig. S11†). These intracellular imaging results are in good accordance with the conventional qRT-PCR-determined miRNA expressions of these different cells (Fig. S12†), demonstrating the accuracy of the cascaded HCR biocomputing system for quantitative miRNA analysis in living cells.
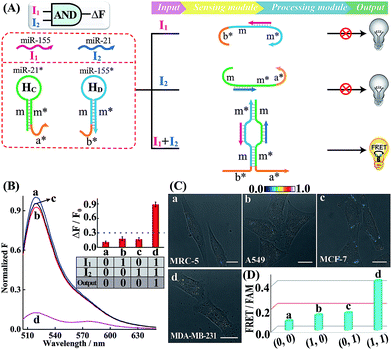
Similarly, further construction of the AND logic gate is demonstrated for enzyme-free amplified analysis of multiple miRNAs (Fig. 3A). Here, the sensing module contains two hairpins HC and HD for recognizing miR-21 (I2) and miR-155 (I1), respectively. The split initiator sequences a* and b* were, respectively, grafted into the 3′-end of HC and 5′-end of HD. Both of these two initiator fragments are partly caged into the stem region of the corresponding hairpins to prevent a possible signal leakage. Even the miR-155 or miR-21 input could hybridize with HD or HC, yet the released single-stranded sequences m and m* are unable to continuously hybridize with HD and HC. These split initiators stay the same with input miR-155 or miR-21, thus prohibiting the processing module and resulting in a False output. In contrast, in the presence of both inputs, I1 and I2, the released single-stranded sequences m and m* allow the synergistically favored inter-hybridization of HC and HD, leading to the efficient connection of these two split initiators to activate the amplified FRET readout (Truth output). Fig. 3B depicts the normalized fluorescence spectra generated by the AND gate system, and Fig. S13† shows the corresponding PAGE characterization, confirming that the logic gate can only deliver a Truth output signal with both I1 and I2 inputs.
Subsequently, the operation of this AND gate was carried out in four different cells (A549, MCF-7, MDA-MB-231 and MRC-5), which display different endogenous expression levels of miR-155 and miR-21. The acquired intracellular FRET imaging of these different cells and the corresponding statistical histogram analysis results are shown in Fig. 3C and D, respectively. Similarly, MRC-5 cells were chosen as the noncancerous/negative control (sample a of Fig. 3C and D). After these cells were transfected with the AND gate components by using lipofectamine 3000, hardly any FRET signal was observed in both A549 and MCF-7 cells that display high expression levels of endogenous miR-155 (I1) and miR-21 (I2), respectively (samples b and c of Fig. 3C and D). This suggests that merely one miRNA input could not activate the present AND gate and thus it delivers a low FRET imaging signal (False output). Next, the miRNA-initiated AND gate was operated in MDA-MB-231 cells that were encoded with high expressions of both miR-155 (I1) and miR-21 (I2). As expected, MDA-MB-231 revealed a clear and significant FRET imaging signal (Truth output, sample d of Fig. 3C and D), indicating an excellent information-processing capability of the rationally designed cascaded HCR platform inside living cells (Fig. S14†). To further validate the endogenous miRNA-activated AND gate, the MDA-MB-231 cells were pretreated with the miR-155 or miR-21 inhibitor, which is fully complementary to the corresponding miRNA input, prior to the AND gate operation. Clearly, a suppressed FRET imaging signal was obtained for these inhibitor-pretreated cells (Fig. S15 and S16†). The reliability of our AND logic gate holds great promise for extensive analysis of multiple miRNA patterns in different cancerous cells.
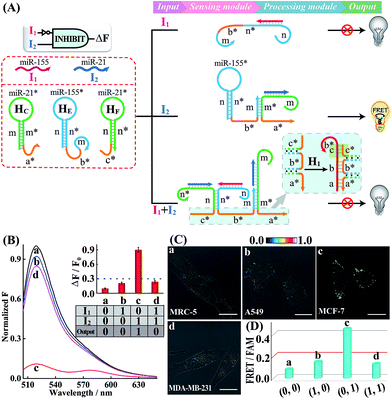
Having demonstrated the successful operation of these simple OR and AND gates, the other two more advanced INHIBIT and XOR gates were then established by using the same miRNA inputs (miR-155 and miR-21). Here, an overlong initiator c*-b*-a* was introduced to block the autonomous HCR-mediated FRET transduction. In fact, the over-hybridization between initiator c*-b*-a* and H1 yields shortened intermediate trigger sequence b* that is too short to hybridize with H2, thus prohibiting the HCR-amplified FRET readout. In the INHIBIT gate, one input carries a NOT gate and can disable the whole system. As shown in Fig. 4A, the sensing module of the INHIBIT gate is composed of three hairpins HC, HE and HF that are, respectively, grafted with initiator fragments a* and b*, and an overhang domain c*. Note that part of sequence c* is also blocked in the stem domain of HF to prevent a possible crosstalk between the sensing module and the processing module. When input I1 (miR-155) was introduced to the system, a False output is obtained, ascribed to the split initiator sequences b* and a* in spite of its hybridization with HE, while input I2 (miR-21) successively hybridizes with HC and HE in the sensing module and colocalizes the two initiator subunits b* and a* to trigger the processing module, resulting in a Truth output. However, in the presence of both inputs, I1 and I2, the hybridization of the inputs to these sensing hairpins (HC, HE and HF) results in the formation of an overlong single-stranded sequence c*-b*-a* to block the processing module (for details, see Fig. S17†). The successful operation of the INHIBIT gate is confirmed by fluorescence measurement (Fig. 4B) and native PAGE characterization (Fig. S18†), which show good agreement with the characteristic operation of the INHIBIT gate. In order to demonstrate the general applicability of our designed logic gate for analyzing trace amounts of miRNAs in cellular environments, we further investigated the present logic system in four different cell lines (Fig. 4C and D; for details, see S19†). As compared with MRC-5 control cells (sample a), the miR-21-rich MCF-7 cells show a highly efficient FRET signal (sample c) while the miR-155-rich A549 cells and the miR-21/miR-155-rich MDA-MB-231 cells show ultralow fluorescence changes (samples b and d, respectively), indicating that the INHIBIT logic gate indeed works well in living cells to monitor the different expression profiles of endogenous miRNAs.
Similarly, an XOR gate that generates the Truth output in the presence of either the I1 or I2 input, but gives the False output with both inputs is developed. The XOR gate includes four hairpins, HB, HC, HE and HF, in the sensing module without changing the processing module as previously described. As depicted in Fig. 5A, either input I1 or I2 can connect the split initiator sequences b* and a* in the sensing module. Then the reconstituted initiator can activate the amplified fluorescence readout (Truth output) in the processing module. However, when both inputs I1 and I2 are present, an over-long initiator sequence c*-b*-a* is assembled to terminate the subsequent HCR transduction as described before. The operation of the XOR gate was identified by fluorescence spectra (Fig. 5B) and native PAGE experiments (Fig. S20†), where the cascaded HCR transduction is triggered only upon introduction of I1 or I2 alone, not in the presence of both I1 and I2. The present XOR gate was further operated in living mammalian cells to trace endogenous miRNA expression patterns (Fig. 5C and D; for details, see S21†). A significantly higher FRET imaging signal was observed in miR-155-rich A549 cells and miR-21-rich MCF-7 cells (samples b and c, respectively) while a relatively weak FRET readout was observed in miR-21/miR-155-rich MDA-MB-231 cells and noncancerous MRC-5 cells (samples a and d, respectively), indicating that these different miRNA-initiated logic gate systems can be employed to discriminate different cell types based on their various intracellular miRNA expression patterns. Furthermore, the intracellular FRET efficiency was evaluated based on a conventional acceptor-photo-bleaching technique. The FRET efficiency was estimated to be 0.54 and 0.57 for A549 and MCF-7 cells, respectively (Fig. S22 and S23†). This demonstrated the programmable and efficient amplification operation of our proposed logic gate, thus offering a versatile and powerful toolbox for tracing miRNAs in living cells.
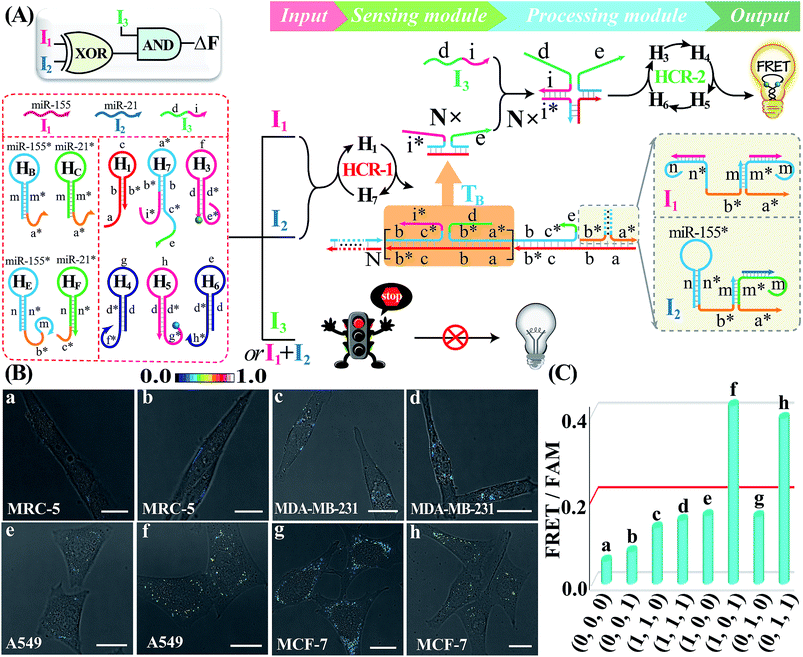
Besides these single-layered logic gate devices obtained through engineering the sensing module, more complicated concatenated logic gate biocircuits could be similarly established by further reprograming the processing module. In fact, the operation of the cascaded HCR-amplified processing module corresponds to a characteristic YES gate operation of all the previous single-layered logic gates. Thus, the processing module could also be similarly reconfigured as the sensing module to assemble more advanced biocomputing circuits. Here, the programmed processing module is mainly based on redesigning one hairpin (H2) of the HCR reactants. An integrated XOR-AND circuit was first established through a further modulation of the information communication between upstream HCR-1 and downstream HCR-2 from the previous XOR logic gate. As depicted in Fig. 6A, the original segment d of H2, a key transducing fragment of the processing module, was replaced with sequence i* to achieve H7. The input I1- or I2-assembled initiator b*-a* activated the upstream HCR-1 to form a linear copolymeric dsDNA product, consisting of tandem adjacent domain i*-e, that acted as transmission input TB for the downstream HCR-2. The other newly introduced input I3 was composed of domains d and i. Obviously, the input I3 or TB alone cannot activate the second layer of the AND gate for limited accessible stages with HCR-2 reactants (False output). However, I3 can hybridize with TB through the i*/I duplex to assemble a colocalized domain d-e, which could activate the downstream HCR-2 to generate an amplified FRET signal (Truth output). The feasibility of our characteristic XOR-AND biocircuit was revealed by the fluorescence spectra (Fig. S24A†) and native gel electrophoresis measurements (Fig. S24B†). This demonstrated that the reconfigurable HCR system was indeed able to construct more complicated DNA logic circuits as dictated by endogenous miRNAs and exogenous interfering DNA. The present XOR-AND circuit was then introduced to simultaneously monitor these multiple endogenous miRNAs in the intracellular environment by choosing these aforementioned four different cells with various miRNA expression profiles. In addition, some of these cells were pretreated with input I3 in order to demonstrate the correct logic operation of the XOR-AND circuit (Fig. 6B and C; for details, see S25†). As expected, all of these four intact cells (samples a, c, e and g), the I3-treated MDA-MB-231 cells (sample h) and the I3-treated noncancerous MRC-5 cells (sample b) show a low fluorescence imaging readout while the I3-treated A549 cells and MCF-7 cells show an obvious FRET imaging signal (samples f and h, respectively). Clearly, the intracellular operation of the present biocomputing system follows the expected truth table of the XOR-AND circuit. Taken together, this miRNA-initiated intelligent HCR system is capable of autonomously and programmably evaluating these relatively overexpressed and down-regulated endogenous miRNAs.
Similarly, an integrated XOR-INHIBIT biocircuit was assembled to demonstrate the versatility of the present biocomputing circuit (Fig. S26†). The present XOR-INHIBIT biocircuit was also operated in four different living cells (Fig. S27 and S28†). A significantly clear FRET imaging signal was only observed in miR-155 (I1)-rich A549 cells and miR-21 (I2)-rich MCF-7 cells (samples e and g, respectively), which is consistent with the function of the XOR-INHIBIT circuit. An XOR-OR biocircuit was also assembled and proved by fluorescence and gel electrophoresis characterization (for details, see Fig. S29†), and was then demonstrated by intracellular imaging experiments (for details, see Fig. S30 and S31†). These results herein significantly demonstrate the versatile and robust features of our reconfigurable HCR system for monitoring the various expressions of multiple endogenous miRNAs in living cells. The programmable HCR logic circuits can simultaneously process an enormous amount of metabolic information in a single living cell, further validating the feasibility of our biocomputing system for detecting multiple biomarkers in intracellular environments.
Conclusions
To summarize, we successfully established an endogenous miRNA-initiated biocomputing system that is composed of a versatile sensing module and an amplification processing module, allowing the information transduction in a more crowded intercellular environment. Based on a reconfigurable hybridization chain reaction, the sensing module can specifically recognize and transduce primary multiplex endogenous miRNA inputs into an intermediate trigger for activating the amplified processing module. Different cells were encoded with various miRNA expression profiles; thus these DNA computing logic gates and circuits were realized for discriminating different living cells with a remarkably distinct FRET readout. The present biocomputing system allows autonomous information processing and amplification without the involvement of any enzymes or complicated nano-labels for overcoming limitations in engineering biomolecular networks. The isothermal enzyme-free DNA computing circuits enabled successive multiple guaranteed recognitions and synergistically accelerated signal amplifications in living cells, which may greatly improve the reliability of in situ diagnosis and thus enable an effective disease treatment.Experimental
Chemicals and materials
HPLC-purified synthetic miRNAs and DNAs were obtained from Sangon Biotech (Shanghai, China), and the sequences are listed in Tables S1 and S2.† Lipofectamine 3000 and DNase I were purchased from Thermo Fisher Scientific. All other reagents were of analytical grade and used as received, and solutions were prepared with ultrapure water from a Milli-Q purification system.Operation of the HCR-based biocomputing circuits
All of the hairpin DNAs (4 μM) were separately annealed at 95 °C for 5 min, followed by cooling down to 25 °C in HEPES buffer (10 mM HEPES, 1 M NaCl, 50 mM MgCl2, pH 7.2). Then, the inputs and the corresponding logic gates were added to the processing module containing H1, H2, H3, H4, H5 and H6 (100 nM each) to initiate the HCR process. All fluorescence measurements were performed on a Cary Eclipse fluorescence spectrophotometer (Varian Inc.) at room temperature. The mixture was excited at 495 nm with a recording emission range from 505 nm to 650 nm.Cell culture and transfection of the designed biocomputing circuits
MCF-7, MRC-5, HEK-293, A549 and MDA-MB-231 cells were routinely cultured in DMEM (MEM for MRC-5) supplemented with 10% FBS and 1% penicillin/streptomycin at 37 °C in a humidified 5% CO2 atmosphere. The cells were plated in 30 mm glass-bottomed culture dishes and grown overnight to reach ∼80% confluent. Transfection assays were then performed by using the lipofectamine 3000 transfection reagent according to the manufacturer's protocol with minor modification.50 In brief, 200 μL of Opti-MEM supplemented with 5 μL lipofectamine 3000 were mixed with 200 μL Opti-MEM containing 0.2 nmol biocomputing components for 5 min. The cells were then incubated with the 400 μL mixture supplied with 50 μL fetal bovine serum at 37 °C for 3 h. The cells were then taken out and washed three times with cold PBS to remove the excess DNA probe and immersed in 1 mL PBS before imaging. For inhibitor experiments, MDA-MB-231 cells were pre-treated with an inhibitor oligonucleotide (final concentration, 100 nM) for 1 h, followed by transfection and incubation with the AND gate as described above for 3 h. For biocircuit experiments, some of the cells were pre-treated with their respective inputs, respectively, for 1 h and then were transfected with these circuit components as described above for 3 h.Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
This work is supported by the National Basic Research Program of China (973 Program, 2015CB932601), National Natural Science Foundation of China (21874103, 81602610, 81472735 and 21503151), and Jiangsu Provincial Natural Science Foundation of China (BK20161248 and BK20160381), Fundamental Research Funds for the Central Universities (No. 2042018kf0210), and Open Fund of Key Laboratory of Analytical Chemistry for Biology and Medicine (Wuhan University), Ministry of Education (ACBM2017005).Notes and references
- A. P. de Silva and S. Uchiyama, Nat. Nanotechnol., 2007, 2, 399–410 CrossRef CAS PubMed.
- L. M. Adleman, Science, 1994, 266, 1021–1024 CrossRef CAS PubMed.
- F. Wang, C. Lu and I. Willner, Chem. Rev., 2014, 114, 2881–2941 CrossRef CAS PubMed.
- Y. Benenson, B. Gil, U. Ben-Dor, R. Adar and E. Shapiro, Nature, 2004, 429, 423–429 CrossRef CAS PubMed.
- S. M. Douglas, I. Bachelet and G. M. Church, Science, 2012, 335, 831–834 CrossRef CAS PubMed.
- P. Ball, Nature, 2000, 406, 118–120 CrossRef CAS PubMed.
- D. Y. Zhang and G. Seelig, Nat. Chem., 2011, 3, 103–113 CrossRef CAS PubMed.
- W. Engelen, L. H. H. Meijer, B. Somers, T. F. A. De Greef and M. Merkx, Nat. Commun., 2017, 8, 14473–14481 CrossRef CAS PubMed.
- A. A. Green, J. Kim, D. Ma, P. A. Silver, J. J. Collins and P. Yin, Nature, 2017, 548, 117–121 CrossRef CAS PubMed.
- B. Yurke, A. J. Turberfield, A. P. Mills, F. C. Simmel and J. L. Neumann, Nature, 2000, 406, 605–608 CrossRef CAS PubMed.
- K. He, Y. Li, B. Xiang, P. Zhao, Y. Hu, Y. Huang, W. Li, Z. Nie and S. Yao, Chem. Sci., 2015, 6, 3556–3564 RSC.
- P. Yin, H. M. T. Choi, C. R. Calvert and N. A. Pierce, Nature, 2008, 451, 318–322 CrossRef CAS PubMed.
- L. Qian, E. Winfree and J. Bruck, Nature, 2011, 475, 368–372 CrossRef CAS PubMed.
- L. Qian and E. Winfree, Science, 2011, 332, 1196–1201 CrossRef CAS PubMed.
- W. Zhou, R. Saran and J. Liu, Chem. Rev., 2017, 117, 8272–8325 CrossRef CAS PubMed.
- B. Li, Y. Jiang, X. Chen and A. Ellington, J. Am. Chem. Soc., 2012, 134, 13918–13921 CrossRef CAS PubMed.
- H. Wang, C. Li, X. Liu, X. Zhou and F. Wang, Chem. Sci., 2018, 9, 5842–5849 RSC.
- J. Elbaz, F. Wang, R. D. Levine, F. Remacle and I. Willner, Nano Lett., 2012, 12, 6049–6054 CrossRef CAS PubMed.
- J. J. Li, A. A. Green, H. Yan and C. Fan, Nat. Chem., 2017, 9, 1056–1067 CrossRef CAS PubMed.
- B. Groves, Y. J. Chen, C. Zurla, S. Pochekailov, J. L. Kirschman, P. J. Santangelo and G. Seelig, Nat. Nanotechnol., 2016, 11, 287–294 CrossRef CAS PubMed.
- J. Hemphill and A. Deiters, J. Am. Chem. Soc., 2013, 135, 10512–10518 CrossRef CAS PubMed.
- S. Tyagi, Nat. Methods, 2009, 6, 331–338 CrossRef CAS PubMed.
- E. Berezikov, V. Guryev, J. van de Belt, E. Wienholds, R. H. A. Plasterk and E. Cuppen, Cell, 2005, 120, 21–24 CrossRef CAS PubMed.
- D. P. Bartel, Cell, 2004, 116, 281–297 CrossRef CAS PubMed.
- C. J. Cheng, R. Bahal, I. A. Babar, Z. Pincus, F. Barrera, C. Liu, A. Svoronos, D. T. Braddock, P. M. Glazer, D. M. Engelman, W. M. Saltzman and F. J. Slack, Nature, 2015, 518, 107–110 CrossRef CAS PubMed.
- C. Liang, P. Ma, H. Liu, X. Guo, B. Yin and B. Ye, Angew. Chem., Int. Ed., 2017, 56, 9077–9081 CrossRef CAS PubMed.
- Z. Cheglakov, T. M. Cronin, C. He and Y. Weizmann, J. Am. Chem. Soc., 2015, 137, 6116–6119 CrossRef CAS PubMed.
- W. Zhou, D. Li, C. Xiong, R. Yuan and Y. Xiang, ACS Appl. Mater. Interfaces, 2016, 8, 13303–13308 CrossRef CAS PubMed.
- S. M. Hammond, Nat. Methods, 2006, 3, 12–13 CrossRef CAS PubMed.
- Y. S. Jiang, S. Bhadra, B. Li and A. D. Ellington, Angew. Chem., Int. Ed., 2014, 53, 1845–1848 CrossRef CAS PubMed.
- F. Wang, J. Elbaz, C. Teller and I. Willner, Angew. Chem., Int. Ed., 2011, 50, 295–299 CrossRef CAS PubMed.
- D. Fan, K. Wang, J. Zhu, Y. Xia, Y. Han, Y. Liu and E. Wang, Chem. Sci., 2015, 6, 1973–1978 RSC.
- J. Feng, Z. Xu, F. Liu, Y. Zhao, W. Yu, M. Pan, F. Wang and X. Liu, ACS Nano, 2018, 12, 12888–12901 CrossRef CAS PubMed.
- S. R. Ryoo, J. Lee, J. Yeo, H. K. Na, Y. K. Kim, H. Jang, J. H. Lee, S. W. Han, Y. Lee, V. N. Kim and D. H. Min, ACS Nano, 2013, 7, 5882–5891 CrossRef CAS PubMed.
- J. Shen, S. A. Stass and F. Jiang, Cancer Lett., 2013, 329, 125–136 CrossRef CAS PubMed.
- R. M. Dirks and N. A. Pierce, Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 15275–15278 CrossRef CAS PubMed.
- F. Wang, J. Elbaz, R. Orbach, N. Magen and I. Willner, J. Am. Chem. Soc., 2011, 133, 17149–17151 CrossRef CAS PubMed.
- M. Liang, M. Pan, J. Hu, F. Wang and X. Liu, ChemElectroChem, 2018, 5, 1380–1386 CrossRef CAS.
- H. M. Choi, V. A. Beck and N. A. Pierce, ACS Nano, 2014, 8, 4284–4294 CrossRef CAS PubMed.
- Z. Wu, G. Liu, X. Yang and J. Jiang, J. Am. Chem. Soc., 2015, 137, 6829–6836 CrossRef CAS PubMed.
- H. M. Choi, J. Y. Chang, A. T. Le, J. E. Padilla, S. E. Fraser and N. A. Pierce, Nat. Biotechnol., 2010, 28, 1208–1212 CrossRef CAS PubMed.
- J. Wang, J. Chao, H. Liu, S. Su, L. Wang, W. Huang, I. Willner and C. Fan, Angew. Chem., Int. Ed., 2017, 56, 2171–2175 CrossRef CAS PubMed.
- R. Lin, Q. Feng, P. Li, P. Zhou, R. Wang, Z. Liu, Z. Wang, X. Qi, N. Tang, F. Shao and M. Luo, Nat. Methods, 2018, 15, 275–278 CrossRef CAS PubMed.
- J. Wang, M. Pan, J. Wei, X. Liu and F. Wang, Chem. Commun., 2017, 53, 12878–12881 RSC.
- R. Deng, K. Zhang and J. Li, Acc. Chem. Res., 2017, 50, 1059–1068 CrossRef CAS PubMed.
- F. Xuan and I. M. Hsing, J. Am. Chem. Soc., 2014, 136, 9810–9813 CrossRef CAS PubMed.
- H. Zhang, F. Li, B. Dever, X.-F. Li and X. C. Le, Chem. Rev., 2012, 113, 2812–2841 CrossRef PubMed.
- S. Bi, S. Yue and S. Zhang, Chem. Soc. Rev., 2017, 46, 4281–4298 RSC.
- E. E. Augspurger, M. Rana and M. V. Yigit, ACS Sens., 2018, 5, 878–902 CrossRef PubMed.
- J. Wei, X. Gong, Q. Wang, M. Pan, X. Liu, J. Liu, F. Xia and F. Wang, Chem. Sci., 2018, 9, 52–61 RSC.
Footnote |
| † Electronic supplementary information (ESI) available: DNA sequences, more characterization, miRNA-initiated imaging systems and FRET efficiencies. See DOI: 10.1039/c8sc05217d |
| This journal is © The Royal Society of Chemistry 2019 |