Open Access Article
Open Access ArticleBisulfite-free, single base-resolution analysis of 5-hydroxymethylcytosine in genomic DNA by chemical-mediated mismatch†
Yafen
Wang‡
a,
Xiong
Zhang‡
a,
Fan
Wu
a,
Zonggui
Chen
b and
Xiang
Zhou
 *a
*a
aCollege of Chemistry and Molecular Sciences, Key Laboratory of Biomedical Polymers of Ministry of Education, The Institute for Advanced Studies, Hubei Province Key Laboratory of Allergy and Immunology, Wuhan University, Wuhan, Hubei 430072, P. R. China. E-mail: xzhou@whu.edu.cn; Fax: +86-27-68756663; Tel: +86-27-68756663
bCollege of Life Science, Wuhan University, Wuhan, Hubei 430072, P. R. China
First published on 11th October 2018
Abstract
5-Hydroxymethylcytosine (5hmC) is known as one of the vital players in nuclear reprogramming and the process of active DNA demethylation. Although the development of whole-genome sequencing methods for modified cytosine bases has burgeoned, the easily operated gene-specific loci detection of 5hmC has rarely been reported. Herein, we present a single-base resolution approach, i.e., chemical-assisted mismatch sequencing (CAM-Seq), which, when combined with traditional oxidation and chemical labeling mediation, can be used for mapping 5hmC at base resolution. We employ chemical oxidation to transform 5hmC to 5-formylcytosine (5fC), followed by chemical labeling to induce C-to-T base changes owing to the fact that the loss of the exocyclic 4-amino group of labeled 5fC leads to C to T conversion and subsequent pairing with adenosine (A) in PCR. The feasibility of CAM-Seq is demonstrated in different synthetic oligonucleotide models as well as in part of the genome of 5hmC-rich mouse embryonic stem cells (mESCs). Moreover, the gene fragment containing 5hmC can be easily biotinylated after oxidation, showing high enrichment efficiency. Our method has the potential capability to map 5hmC in genomic DNA and thus will contribute to promoting the understanding of the epigenetic modification of 5hmC.
Introduction
Chemical modifications of DNA bases can profoundly influence various cellular processes, and several epigenetic modifications have been found in a variety of mammalian tissues and organisms.1,2 5-Hydroxymethylcytosine (5hmC) was discovered as an epigenetic mark, and it is produced via an active DNA demethylation process which involves the iterative oxidation of 5-methylcytosine (5mC) driven by ten-eleven translocation (TET) family proteins.3–7 Of particular interest is the fact that 5hmC has been found in embryonic stem cells6,8–11 and brain tissue,12–15 and it affects gene expression through inhibition binding of 5mC binding proteins or recruiting specific binding proteins of 5hmC.16,17 The relative abundance of 5hmC in different tissues may be due to its cell type specificity18,19 and 5hmC is associated with various diseases such as cancers.20–23 Thus, in addition to being an intermediate of active DNA demethylation, 5hmC is involved in regulatory functions in essential biological processes.24To date, an ample variety of methods have been developed to detect 5hmC. For the whole genome-wide mapping of 5hmC, several strategies have been established, such as hydroxymethylated DNA immunoprecipitation (hMeDIP) assisted by antibodies25 and selective chemical labeling with streptavidin beads after the glucosylation of 5hmC and then biotinylation.26 Although these methods can solve the problem of the presence of 5hmC in short genomic regions, the resolution depends on the fragment size of the genomic DNA, because of which the precise sites cannot be identified. A method for single nucleotide resolution of 5hmC will enable the identification of individual modifications on the genome-wide scale and it will help us to deeply understand the role 5hmC plays in epigenetics. For example, to determine 5hmC distribution with respect to transcription factor binding sites, strategies based on the bisulfite treatment of DNA are developed. 5hmC can be protected by glucosylation, followed by Tet-assisted bisulfite sequencing (TAB-Seq), which can realize the base-resolution analysis of 5hmC in the mammalian genome.27 The previous literature reported that KRuO4 could selectively oxidize 5hmC to 5-formylcytosine (5fC) in high yield, and 5fC is deaminated after bisulfite treatment and therefore sequenced as thymine (T).28 All these excellent studies provide diversified methods to uncover the uncharted territory of 5hmC. It is notable that these methods all rely on harsh bisulfite-mediated conversion, which often causes further DNA degradation.29,30
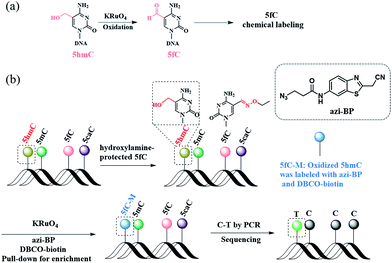
Herein, we describe a bisulfite-free method for the single-base resolution sequencing of 5hmC and demonstrate its feasibility in genomic DNA sequencing (Fig. 1). Malononitrile and an azido derivative of 1,3-indandione were used for labeling 5fC, which realized the identification 5fC without bisulfite treatment at single-base resolution by CLEVER-seq31 and the fC-CET method,32 respectively. We previously reported some chemical molecules for the selective labeling of 5fC,33–35 and some of these chemical molecules can also realize the goal of single-base analysis of 5fC. For instance, our group reported that 5fC can be labeled with azi-BP34 (the structure of azi-BP is shown in Scheme 1), and the azide group of azi-BP-fC can react with DBCO-biotin reagents through a copper-free click reaction26,36 The conceptual basis for the single-base sequencing of 5hmC involves chemical-assisted mismatch sequencing (CAM-Seq). First, 5hmC was selectively oxidized to 5fC by KRuO4. Then 5fC was labeled with the chemical molecule previously reported by our group, namely, azi-BP.34 As mentioned in the reported literature,31,32,34 owing to the original 4′ amino group of 5fC participating in cyclization, which is no longer a competent proton donor for the canonical base pairing with guanine (G), matching with adenine (A) and misreading as thymidine (T) occurs during the PCR process for realizing the single-base resolution analysis of 5hmC. Moreover, the click chemistry reaction to install a biotin tag can selectively enrich the 5hmC-containing fragment. Thus, the design can easily identify 5hmC in a bisulfite-free manner.
Results and discussion
Evaluating the protection of ODN-5fC with hydroxylamine
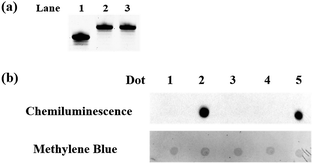
To eliminate the influence of the original 5fC in the tested samples, hydroxylamine was used for protecting 5fC37 to prevent it from reacting with azi-BP. First, we verified that the protected 5fC could not react with azi-BP, and ODN-5fC was chosen for the test. As shown by denaturing polyacrylamide gel electrophoresis (PAGE) analysis (Fig. 1a), the retarded migration of ODN-5fC indicated that it was successfully protected by hydroxylamine. When the protected ODN-5fC was further incubated with azi-BP, no new band appeared in the gel. These results suggested that hydroxylamine could effectively protect 5fC from being labeled with azi-BP.Evaluating the reactivity of azi-BP with oxidized ODN-5hmC
Next, we evaluated the reactivity of azi-BP towards the labeling of oxidized 5hmC. Because the hydroxyl groups of both the 5′ and 3′ ends of DNA can be oxidized by KRuO4,38 a universal next-generation sequencing (NGS) adapter bearing 5′-O-methyl groups and 3′-O-phosphate groups was used to alleviate potential interference from the oxidation of the hydroxyl groups on the ends of the DNA fragment. Double-stranded DNA containing 5hmC (E-DNA-hmC) was ligated with the 5′-O-methyl group and 3′-O-phosphate group modified adapters using the standard protocol, and then the purified adapter-ligated DNA samples were subjected to oxidization by KRuO4. After purification, the mixture was incubated with azi-BP and DBCO-biotin for the dot blot assay. The dot indicated that the oxidized 5hmC can be effectively labeled by azi-BP (Fig. 1b, dot 2), and the 5fC protected by hydroxylamine could not be further labeled with azi-BP (Fig. 1b, dot 4), which suggested that after being protected by hydroxylamine, the original 5fC could not affect the detection of 5hmC.Sanger sequencing for single-base resolution analysis of 5hmC in synthesized DNA
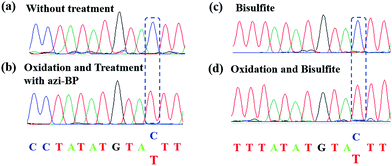
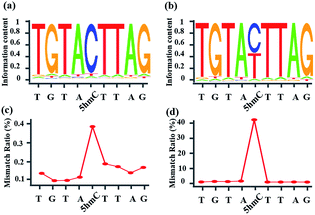
Encouraged by the result of the dot blot assay, we proceeded to perform CAM-Seq to analyze 5hmC at single-base resolution. As reported previously,34 azi-BP labeled 5fC generates a cyclic reaction, which leads to the loss of the 4-amino group of 5fC and thus the failure to pair with G in a canonical base pairing manner. The disabling of the proton donor by the original 4-amino group causes modified 5hmC to pair with A. As a proof-of-concept, an 80 bp-ds DNA containing one site of 5hmC (seq-1 ds-ODN-5hmC, ESI Table S1†) was oxidized and further labeled by azi-BP. After amplification by PCR with MightyAmp DNA polymerase, the products were further applied in single cloning analysis. The Sanger sequencing results showed that modified 5hmC is read as T during sequencing (Fig. 2a and b), indicating that the designed strategy can realize the single-base resolution analysis of 5hmC. To further prove the results of Sanger sequencing, the presence of 5hmC at seq-1 ds-ODN-5hmC was confirmed by conventional bisulfite sequencing (Fig. 2c and d). Whether CAM-Seq can recognize multiple 5hmC sites simultaneously is vital to generalize its application in the genome owing to the fact that 5hmC may also be present continuous. A model DNA strand containing two 5hmC sites (seq-2 ds-ODN-5hmC, ESI Table S1†) was chosen for testing the feasibility. To our delight, the C to T conversions in 5hmC sites were all observed by Sanger sequencing, which showed that the sites of 5hmC can be accurately located by CAM-Seq (Fig. S1†).Sanger sequencing for the single-base resolution analysis of 5hmC in genomic DNA from mESCs
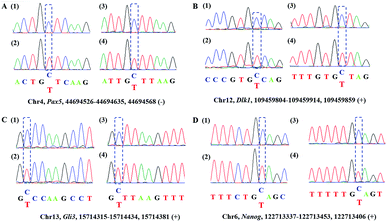
Encouraged by the above results, we then asked whether CAM-Seq could be applied to 5hmC detection in some specific genomic DNA regions. To this end, several specific regions from mESCs39 reported to contain 5hmC were selected for the test. For example, the gene of PAX5 as a B cell transcription factor was chosen for the test. First, we extracted the genomic DNA from mESCs. Then the genomic DNA was incubated with hydroxylamine and subjected to oxidation as described by the protocol reported previously,28,40 followed by labeling with azi-BP and PCR. The PCR products were verified by agarose gel electrophoresis analysis (Fig. S2†). As measured by CAM-Seq, the precise loci of 5hmC in the tested regions can be identified. These results were further confirmed by conventional bisulfite sequencing (Fig. 3). And several other regions were also tested (Fig. S3†). Collectively, this design will be a potential approach for 5hmC detection in the genome.Illumina sequencing of synthesized DNA containing 5hmC
Whether CAM-Seq can be used for high-throughput sequencing is vital for the genome-wide analysis of 5hmC at single-base resolution. A synthetic oligonucleotide with one 5hmC at a defined position (seq-1 ds-ODN-5hmC) was applied in NGS; we also spiked in unmodified DNA (seq-1 ds-ODN-C, seq-1 ds-ODN-5hmC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) seq-1 ds-ODN-C = 1
seq-1 ds-ODN-C = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10) to increase the complexity of the system. The DNA without treatment was used as the control sample. First, the model DNA was ligated to the modified NGS adapter. Then the adapter ligated DNA sample was denatured in NaOH followed by oxidization with KRuO4. After labeling with azi-BP and DBCO-S-S-PEG3-biotin, a pull-down assay was carried out to enrich the labeled 5hmC. The enriched sample was quantified using Qubit and then applied in the amplification with minor modifications. Briefly, the libraries were amplified using the MightyAMP DNA polymerase (Takara). These libraries were sequenced on a HiSeq X Ten platform in 150 bp paired-end mode. As demonstrated by bioinformatics analysis, the NGS data showed significant C-to-T conversion. The ratio of T read is ∼88% (Fig. 4). It is worth mentioning that the C read counts in the treated samples, which are comparable to the T read counts, come from the complementary strand of the 5hmC containing sequence. These inspiring NGS results with single-base resolution encouraged us to implement this CAM-Seq for precise 5hmC mapping in the whole genome in the near future.
10) to increase the complexity of the system. The DNA without treatment was used as the control sample. First, the model DNA was ligated to the modified NGS adapter. Then the adapter ligated DNA sample was denatured in NaOH followed by oxidization with KRuO4. After labeling with azi-BP and DBCO-S-S-PEG3-biotin, a pull-down assay was carried out to enrich the labeled 5hmC. The enriched sample was quantified using Qubit and then applied in the amplification with minor modifications. Briefly, the libraries were amplified using the MightyAMP DNA polymerase (Takara). These libraries were sequenced on a HiSeq X Ten platform in 150 bp paired-end mode. As demonstrated by bioinformatics analysis, the NGS data showed significant C-to-T conversion. The ratio of T read is ∼88% (Fig. 4). It is worth mentioning that the C read counts in the treated samples, which are comparable to the T read counts, come from the complementary strand of the 5hmC containing sequence. These inspiring NGS results with single-base resolution encouraged us to implement this CAM-Seq for precise 5hmC mapping in the whole genome in the near future.
Inhibition of DNA polymerase replication by DBCO-PEG4-biotin
After modifications, the large group was added to the labeled 5hmC, which may create a barrier against normal enzymatic reactions such as PCR. 80 bp double-stranded DNA strands containing 5hmC and 5fC (E-DNA-hmC, DNA-fC, ESI Table S1†) were chosen for further study. These DNA strands were incubated with hydroxylamine and then subjected to oxidation. After purification, these samples were labeled with azi-BP and DBCO-S-S-PEG4-biotin. Taq DNA polymerase from a Hieff™ PCR SYBR® Green Master Mix was selected to test the effect of this huge additional group on the replication of labeled 5hmC. Quantitative PCR (qPCR) analysis was used to evaluate the polymerase activity. Ct value represents the number of cycles required to achieve the set fluorescence value. A larger Ct value indicates a less effective DNA template, which means that the replication process was hampered. As expected, the oxidized and chemically labeled 5hmC showed a higher Ct value than the untreated sample (Fig. 5, red line). Moreover, after being protected by hydroxylamine, the 5fC containing DNA could not affect the activity of DNA polymerase. The result is also consistent with the PAGE and dot blot analysis (Fig. 1) in that protected 5fC could not be further labeled with azi-BP. These results demonstrate that our design assisted by PCR amplification can detect a low content of 5hmC in a complex environment, and this encourages us to perform further detailed study.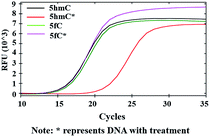 | ||
| Fig. 5 qPCR analysis of 5hmC. Labeled 5hmC inhibits the amplification activity of Taq DNA polymerase. | ||
Enriching 5hmC-containing DNA fragments
In addition to the advantage of 5hmC single-base resolution analysis, the azi-BP can be biotinylated to provide another application for enriching DNA fragments bearing oxidized 5hmC. To evaluate the enrichment efficiency, three 80 bp dsDNA strands (E-DNA-C, E-DNA-hmC, E-DNA-fC, ESI Table S1†) were used as the enrichment models. The process of enrichment efficiency analysis is shown in Fig. 6a. First, these model DNA strands were mixed in equal moles, and incubated with hydroxylamine to protect the formyl group. As mentioned above, the hydroxyl groups of both the 5′ and 3′ ends of DNA can be oxidized by KRuO4 which can induce labeling of non-5hmC containing DNA with azi-BP. Second, the mixture was ligated with a modified adapter and then oxidized by KRuO4. Third, the mixed system was incubated with azi-BP and DBCO-S-S-PEG3-biotin. Finally, the purified ODNs were obtained for affinity enrichment using streptavidin-coated magnetic beads. The enrichment efficiency was calculated using qPCR. 5hmC-DNA was enriched ∼97-fold more than non-modified C-DNA (Fig. 6b and S4†). A similar level of enrichment was observed for 5hmC-DNA compared to 5fC-DNA, indicating that 5fC protected by hydroxylamine could not be captured. The results of enrichment analysis and Illumina sequencing revealed that our strategy can be used as a potential strategy for the single-base resolution analysis of 5hmC in the whole genome without bisulfite treatment.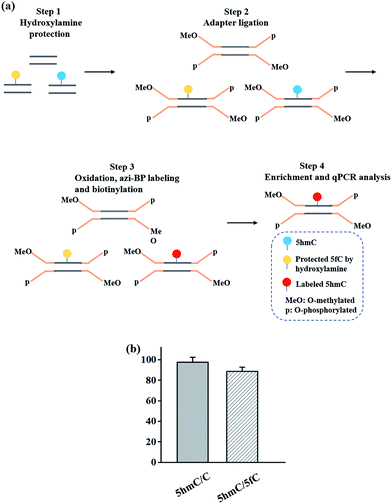 | ||
| Fig. 6 Enrichment of 5hmC. (a) Schematic of the enrichment efficiency analysis. (b) Extent of enrichment of 5hmC in double-stranded DNA towards C and 5fC ODN sequences. | ||
Conclusions
In summary, we demonstrated a new method to detect and sequence 5hmC at single-base resolution by chemical-mediated mismatching. The Sanger sequencing and Illumina sequencing analyses clearly identified 5hmC through C-to-T conversion. We were able to detect the 5hmC signal from mESC genomic DNA. The reactivity inhibition of DNA polymerase caused by the labeled 5hmC can enable quick screening of existing 5hmC. A pull-down analysis indicated that our strategy can selectively enrich fragments containing 5hmC in DNA. During the revision of this study, a bisulfite-free method that detects 5hmC in hESCs and cfDNA has been reported.41 The main advantage of this method is that it is bisulfite-free, yet has a single-base resolution nature. However, the condition of oxidation that we use is harsh. Further efforts should be made to improve this strategy. In general, our findings can provide an alternative for the whole-genome mapping of epigenetic modifications.Methods
Synthesis of 80 bp ODN-hmC and ODN-fC by polymerase chain reaction
To synthesize 5hmC/5fC containing oligodeoxynucleotides, modified nucleotides were incorporated into the PCR. 80 bp ds ODN-hmC (E-DNA-hmC) (see Table S1†) was synthesized using template 2, forward primer 2 and reverse primer 2 in the presence of dATP, dTTP, dGTP and dhmCTP. 80 bp ds ODN-fC (E-DNA-hmC) (see Table S1†) was synthesized using template 3, forward primer 3 and reverse primer 3 in the presence of dATP, dTTP, dGTP and dfCTP. PCR reactions were prepared with 10× DreamTaq buffer (5 μL), 2 μL forward primer (10 μM), 2 μL reverse primer (10 μM), 1 μL template (100 nM), 1 μL dATP (10 mM), 1 μL dTTP (10 mM), 1 μL dGTP (10 mM), 1 μL dXTP (X = hmC or fC) (10 mM), 5 U DreamTaq polymerase and ddH2O to give a final volume of 50 μL. The mixture was then subjected to the following thermal cycle: 95 °C for 3 min, 35 cycles of 95 °C for 10 s, 60 °C for 30 s, and 72 °C for 90 s, and 72 °C for 3 min using a T100™ Thermal Cycler (BioRad). The PCR product was purified using a DNA Clean & Concentrator™-5 kit according to the manufacturer's instructions. The PCR product was confirmed by 4% agarose gel electrophoresis (AGE) analysis (Fig. S3†). The DNA concentration was quantified using a NanoDrop 2000c (Thermo Scientific, USA).Hydroxylamine protection of 5fC
To prevent 5fC from reacting with azi-BP, we use hydroxylamine to protect 5fC. The hydroxylamine protection of 5fC was performed in 100 mM MES buffer (pH 5.0), 10 mM O-ethylhydroxylamine (in ddH2O, Aldrich, 274992), and 2 μL (100 μM) ODN-5fC or 200 ng 80 bp ds ODN-5fC for 2 h at 37 °C. The mixture was purified using Micro Bio-Spin™ P-6 Gel Columns in SSC buffer after being pre-washed with ddH2O (3 × 500 μL) for removing O-ethylhydroxylamine and MES.Dot-blot assay
For the dot-blot assay, different DNA strands were dotted on an Amersham Hybond-N+ membrane (GE Healthcare). After drying, the dots were crosslinked with the membrane using UV light (254 nm) at RT for 5 min twice and washed with 1× TBST twice. Then the membrane was blocked with 5% BSA at 37 °C for 1 h and washed with 1× TBST five times. After labeling with streptavidin–HRP (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1500) (Thermo Scientific) at 37 °C for 1 h and washing with 1× TBST four times, the results were visualized by enhanced chemiluminescence (SuperSignal™ West Pico Chemiluminescent Substrate, Cat: 34077, Thermo Scientific) using a Molecular Imager® ChemiDocTM XRS+ Imaging System (Bio-Rad). Finally, the membrane was steeped in methylene blue (in NaOAc buffer) to verify the existence of DNA in every dot.
1500) (Thermo Scientific) at 37 °C for 1 h and washing with 1× TBST four times, the results were visualized by enhanced chemiluminescence (SuperSignal™ West Pico Chemiluminescent Substrate, Cat: 34077, Thermo Scientific) using a Molecular Imager® ChemiDocTM XRS+ Imaging System (Bio-Rad). Finally, the membrane was steeped in methylene blue (in NaOAc buffer) to verify the existence of DNA in every dot.
The modified adapter solution for protecting hydroxyl groups at both the 5′ and 3′ ends of DNA
The modified universal adapter is a 5′-O-methylated ODN with a sequence of 5′-MeO-AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCT-3′ (Takara Biotechnology, China) and a 3′-O-phosphorylated ODN with a sequence of 5′-GATCGGAAGAGCACACGTCTGAACTCCAGTCACNNNNNNATCTCGTATGCCGTCTTCTGCTTG-O-phosphate-3′ (Sangon Biotech, China Aldrich), where NNNNNN corresponds to TruSeq (Illumina) barcoding, made up to a solution (25 μM each) in 50 mM NaCl and 20 mM Tris–HCl (pH 7) and annealed prior to use.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We thank the National Natural Science Foundation of China (21432008, 91753201 and 21721005 to X. Z.) and the China Postdoctoral Innovative Talent Support Program of China (No. BX20180228 to Y. W.). The numerical calculations in this paper have been done on the supercomputing system in the Supercomputing Center of Wuhan University.Notes and references
- D. Han, X. Lu, A. H. Shih, J. Nie, Q. You, M. M. Xu, A. M. Melnick, R. L. Levine and C. He, Mol. Cell, 2016, 63, 711–719 CrossRef CAS PubMed.
- Y. Fu and C. He, Curr. Opin. Chem. Biol., 2012, 16, 516–524 CrossRef CAS PubMed.
- L. Shen and Y. Zhang, Curr. Opin. Cell Biol., 2013, 25, 289–296 CrossRef CAS PubMed.
- Y. Chen, T. Hong, S. Wang, J. Mo, T. Tian and X. Zhou, Chem. Soc. Rev., 2017, 46, 2844–2872 RSC.
- S. Ito, L. Shen, Q. Dai, S. C. Wu, L. B. Collins, J. A. Swenberg, C. He and Y. Zhang, Science, 2011, 333, 1300 CrossRef CAS PubMed.
- M. Tahiliani, K. P. Koh, Y. Shen, W. A. Pastor, H. Bandukwala, Y. Brudno, S. Agarwal, L. M. Iyer, D. R. Liu, L. Aravind and A. Rao, Science, 2009, 324, 930 CrossRef CAS PubMed.
- Y. Wang, C. Liu, T. Wang, T. Hong, H. Su, S. Yu, H. Song, S. Liu, X. Zhou, W. Mao and X. Zhou, Anal. Chem., 2016, 88, 3348–3353 CrossRef CAS PubMed.
- G. Ficz, M. R. Branco, S. Seisenberger, F. Santos, F. Krueger, T. A. Hore, C. J. Marques, S. Andrews and W. Reik, Nature, 2011, 473, 398 CrossRef CAS PubMed.
- S. Ito, A. C. D'Alessio, O. V. Taranova, K. Hong, L. C. Sowers and Y. Zhang, Nature, 2010, 466, 1129 CrossRef CAS PubMed.
- H. Stroud, S. Feng, S. Morey Kinney, S. Pradhan and S. E. Jacobsen, Genome Biol., 2011, 12, R54 CrossRef CAS PubMed.
- K. E. Szulwach, X. Li, Y. Li, C.-X. Song, J. W. Han, S. Kim, S. Namburi, K. Hermetz, J. J. Kim, M. K. Rudd, Y.-S. Yoon, B. Ren, C. He and P. Jin, PLoS Genet., 2011, 7, e1002154 CrossRef CAS PubMed.
- S.-G. Jin, X. Wu, A. X. Li and G. P. Pfeifer, Nucleic Acids Res., 2011, 39, 5015–5024 CrossRef CAS PubMed.
- S. Kriaucionis and N. Heintz, Science, 2009, 324, 929 CrossRef CAS PubMed.
- K. E. Szulwach, X. Li, Y. Li, C.-X. Song, H. Wu, Q. Dai, H. Irier, A. K. Upadhyay, M. Gearing, A. I. Levey, A. Vasanthakumar, L. A. Godley, Q. Chang, X. Cheng, C. He and P. Jin, Nat. Neurosci., 2011, 14, 1607 CrossRef CAS PubMed.
- T. Wang, Q. Pan, L. Lin, K. E. Szulwach, C.-X. Song, C. He, H. Wu, S. T. Warren, P. Jin, R. Duan and X. Li, Hum. Mol. Genet., 2012, 21, 5500–5510 CrossRef CAS PubMed.
- V. Valinluck, H.-H. Tsai, D. K. Rogstad, A. Burdzy, A. Bird and L. C. Sowers, Nucleic Acids Res., 2004, 32, 4100–4108 CrossRef CAS PubMed.
- O. Yildirim, R. Li, J.-H. Hung, P. B. Chen, X. Dong, L.-S. Ee, Z. Weng, O. J. Rando and T. G. Fazzio, Cell, 2011, 147, 1498–1510 CrossRef CAS PubMed.
- J. A. Hackett, J. J. Zylicz and M. A. Surani, Trends Genet., 2012, 28, 164–174 CrossRef CAS PubMed.
- C. Quivoron, L. Couronné, V. Della Valle, C. K. Lopez, I. Plo, O. Wagner-Ballon, M. Do Cruzeiro, F. Delhommeau, B. Arnulf, M.-H. Stern, L. Godley, P. Opolon, H. Tilly, E. Solary, Y. Duffourd, P. Dessen, H. Merle-Beral, F. Nguyen-Khac, M. Fontenay, W. Vainchenker, C. Bastard, T. Mercher and O. A. Bernard, Cancer Cell, 2011, 20, 25–38 CrossRef CAS PubMed.
- K. Chen, J. Zhang, Z. Guo, Q. Ma, Z. Xu, Y. Zhou, Z. Xu, Z. Li, Y. Liu, X. Ye, X. Li, B. Yuan, Y. Ke, C. He, L. Zhou, J. Liu and W. Ci, Cell Res., 2015, 26, 103 CrossRef PubMed.
- C. J. Mariani, J. Madzo, E. L. Moen, A. Yesilkanal and L. A. Godley, Cancers, 2013, 5, 786–814 CrossRef CAS PubMed.
- E. Kriukiene, Z. Liutkeviciute and S. Klimasauskas, Chem. Soc. Rev., 2012, 41, 6916–6930 RSC.
- T. F. Kraus, D. Globisch, M. Wagner, S. Eigenbrod, D. Widmann, M. Munzel, M. Muller, T. Pfaffeneder, B. Hackner, W. Feiden, U. Schuller, T. Carell and H. A. Kretzschmar, Int. J. Cancer, 2012, 131, 1577–1590 CrossRef CAS PubMed.
- H. Wu and Y. Zhang, Cell, 2014, 156, 45–68 CrossRef CAS PubMed.
- H. Wu, A. C. D'Alessio, S. Ito, Z. Wang, K. Cui, K. Zhao, Y. E. Sun and Y. Zhang, Genes Dev., 2011, 25, 679–684 CrossRef CAS PubMed.
- C.-X. Song, K. E. Szulwach, Y. Fu, Q. Dai, C. Yi, X. Li, Y. Li, C.-H. Chen, W. Zhang, X. Jian, J. Wang, L. Zhang, T. J. Looney, B. Zhang, L. A. Godley, L. M. Hicks, B. T. Lahn, P. Jin and C. He, Nat. Biotechnol., 2010, 29, 68 CrossRef PubMed.
- M. Yu, G. C. Hon, K. E. Szulwach, C.-X. Song, L. Zhang, A. Kim, X. Li, Q. Dai, Y. Shen, B. Park, J.-H. Min, P. Jin, B. Ren and C. He, Cell, 2012, 149, 1368–1380 CrossRef CAS PubMed.
- M. J. Booth, M. R. Branco, G. Ficz, D. Oxley, F. Krueger, W. Reik and S. Balasubramanian, Science, 2012, 336, 934 CrossRef CAS PubMed.
- G. Hayashi, K. Koyama, H. Shiota, A. Kamio, T. Umeda, G. Nagae, H. Aburatani and A. Okamoto, J. Am. Chem. Soc., 2016, 138, 14178–14181 CrossRef CAS PubMed.
- M. Münzel, L. Lercher, M. Müller and T. Carell, Nucleic Acids Res., 2010, 38, e192 CrossRef PubMed.
- C. Zhu, Y. Gao, H. Guo, B. Xia, J. Song, X. Wu, H. Zeng, K. Kee, F. Tang and C. Yi, Cell Stem Cell, 2017, 20, 720–731 CrossRef CAS PubMed.
- B. Xia, D. Han, X. Lu, Z. Sun, A. Zhou, Q. Yin, H. Zeng, M. Liu, X. Jiang, W. Xie, C. He and C. Yi, Nat. Methods, 2015, 12, 1047 CrossRef CAS PubMed.
- C. Liu, Y. Wang, W. Yang, F. Wu, W. Zeng, Z. Chen, J. Huang, G. Zou, X. Zhang, S. Wang, X. Weng, Z. Wu, Y. Zhou and X. Zhou, Chem. Sci., 2017, 8, 7443–7447 RSC.
- Y. Wang, C. Liu, X. Zhang, W. Yang, F. Wu, G. Zou, X. Weng and X. Zhou, Chem. Sci., 2018, 9, 3723–3728 RSC.
- Y. Wang, C. Liu, W. Yang, G. Zou, X. Zhang, F. Wu, S. Yu, X. Luo and X. Zhou, Chem. Commun., 2018, 54, 1497–1500 RSC.
- C.-X. Song, T. A. Clark, X.-Y. Lu, A. Kislyuk, Q. Dai, S. W. Turner, C. He and J. Korlach, Nat. Methods, 2011, 9, 75 CrossRef PubMed.
- C.-X. Song, K. E. Szulwach, Q. Dai, Y. Fu, S.-Q. Mao, L. Lin, C. Street, Y. Li, M. Poidevin, H. Wu, J. Gao, P. Liu, L. Li, G.-L. Xu, P. Jin and C. He, Cell, 2013, 153, 678–691 CrossRef CAS PubMed.
- F. Kawasaki, D. Beraldi, R. E. Hardisty, G. R. McInroy, P. van Delft and S. Balasubramanian, Genome Biol., 2017, 18, 23 CrossRef PubMed.
- C. Zhao, H. Wang, B. Zhao, C. Li, R. Yin, M. Song, B. Liu, Z. Liu and G. Jiang, Nucleic Acids Res., 2014, 42, e81 CrossRef CAS PubMed.
- F. Kawasaki, S. Martínez Cuesta, D. Beraldi, A. Mahtey, R. E. Hardisty, M. Carrington and S. Balasubramanian, Angew. Chem., 2018, 130, 9842–9844 CrossRef.
- H. Zeng, B. He, B. Xia, B. A. I. Dongsheng, X. Lu, J. Cai, L. Chen, A. Zhou, C. Zhu, H. Meng, Y. Gao, H. Guo, C. He, Q. Dai and C. Yi, J. Am. Chem. Soc., 2018 DOI:10.1021/jacs.8b08297.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8sc04272a |
| ‡ These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2019 |