Biochemistry instructors’ use of intentions for student learning to evaluate and select external representations of protein translation
Thomas J.
Bussey
 *a and
MaryKay
Orgill
*a and
MaryKay
Orgill
 b
b
aDepartment of Chemistry and Biochemistry, University of California, San Diego, USA. E-mail: tbussey@ucsd.edu; Tel: +1-858-822-6665
bDepartment of Chemistry and Biochemistry, University of Nevada, Las Vegas, USA
First published on 7th June 2019
Abstract
Instructors draw on their intentions for student learning in the enactment of curriculum, particularly in the selection and presentation of external representation of scientific phenomena. These representations both create opportunities for students to experience non-experiential biochemical phenomena, such as protein translation, and constrain the possibilities for student learning based on the limited number of features depicted and the visual cues used to draw viewers attention to those features. In this study, we explore biochemistry instructors’ intentions for student learning about protein translation and how those intentions influence their selection of external representations for instruction. A series of instructor interviews were used to identify information that students need to know in order to develop a biochemically accurate understanding of protein translation. We refer to this information as the “critical features” of protein translation. Two dominant themes of critical features were identified: (1) components/structures of protein translation and (2) interactions/chemistry of protein translation. Three general components (the ribosome, the mRNA, and the tRNA) and two primary interactions (base pairing and peptide bond formation) were described by all instructors. Instructors tended to favor simpler, stylized representations that closely aligned with their stated critical features of translation for instructional purposes.
Introduction
Students’ beliefs and intuitions about the physical world are often based more on common sense and experience than on scientific reasoning (Halloun and Hestenes, 1985). However, many aspects of science are non-experiential. For example, students (and everyone else, for that matter) cannot directly observe a biochemical reaction on the molecular level as it occurs in the body. As such, scientific knowledge is often expressed as external representations: e.g., graphs, flow charts, models, illustrations, or animations (Trumbo, 1999; Wellington and Osborne, 2001). In fact, scientists often use such representations in their own work to, among other things, visualize a phenomenon (Trumbo, 1999). The importance of representations can be seen in “the exponential growth […] in the number and range of visualization tools now available to the biochemist for teaching, learning, and research” (Schönborn and Anderson, 2006, p. 94). As noted by Schönborn and Anderson (2006, 2010) and reiterated more recently by Arneson and Offerdahl (2018), visual literacy is and continues to be a necessary skill for biochemists and is, therefore, a foundational skill for biochemistry students to develop.As with any instructional strategy, external representations are only as useful as students’ ability to understand them. Biochemistry students must be able to comprehend, evaluate, and construct representations of biochemical phenomena and concepts (Stanley, 1996). Unfortunately, students often find it difficult to work with these representations, which are varied in their formatting, conventions, and styles (Schönborn and Anderson, 2006).
While visual literacy is a necessary skill for science students, it should be noted that students can only learn from representations to which they are exposed. It is, therefore, important to consider how instructors and instructional materials designers constrain the range and types of representations to which students are exposed. Instructional decision making pertaining to the selection and enactment of particular external representation requires just as much scrutiny as the quality and range of information conveyed by that representation.
The use of external representations in science teaching
External representations have gained wide use as pedagogical tools over the past several decades (Lowe, 2003). A 1993 study of science textbooks found that nearly 55% of the printed page was composed of illustrations (Mayer, 1993), thereby making representations a significant instructional medium through which information, in particular science information, is conveyed to students. In 1997, Kozma and Russell described the modern chemistry classroom, noting thatclassroom whiteboards and modern chemistry textbooks are filled with diagrams, charts, graphs, equations, and formulas along with words, photographs, and illustrations. Increasingly, newer educational technologies—video and computers—are also being used in the classroom to represent chemical phenomena in new ways. (p. 950)
Similarly, Lowe (2003) observed that “there has been a growing trend across a range of media to use highly illustrated materials for instruction rather than relying on largely text-based presentations of information” (p. 157).
Two-dimensional representations have long been considered the bedrock of communication both in scientific discourse and education. While technological advances have supported the rapid development and implementation of animated, interactive, and simulated representations, 2D representations remain a popular form of currency in peer-to-peer communication (Goodsell and Jenkinson, 2018, p. 3979).
However, a recent national survey showed that the types of representations used by biochemistry instructors’ varied significantly depending on the type of course being taught or the purpose of the representational use (i.e., during instruction or on an assessment) (Linenberger and Holmes, 2014).
Students’ interpretations of external representations
While all types of external representations create a shared, common experience, students may still develop varied understandings of a shared image based on variations in their level of prior knowledge of the material (e.g., Cook, 2006; Cook et al., 2008) and the specific subset of features to which they attend (e.g., Bussey and Orgill, 2015). As individuals read external representations, they use their prior knowledge in two ways. First, prior knowledge is used to identify relevant information within the external representation (to the neglect of other aspects of the representation). Second, prior knowledge is added to the information gleaned from the external representation to construct a more coherent meaning of the depicted concept. In this way, individuals are able to construct a working mental model of the concept depicted by the external representation by integrating elements of the external representation with their prior knowledge (Braune and Foshay, 1983).Several studies have explored the influence of prior knowledge on students’ development of visual literacy skills (Lowe, 1996; Kozma and Russell, 1997; Lowe, 2003; Schnotz and Lowe, 2003; Lowe, 2004; Cook, 2006; Cook et al., 2008; Harle and Towns, 2012; Sim and Daniel, 2014). The majority of these studies have focused on the differences between the visual literacy skills of novices and those of experts. Novice learners are generally assumed to have less prior knowledge than experts for use in constructing mental representations from instructional materials (Johnson and Lawson, 1998). Additionally, it is thought that what knowledge they do have is not interconnected or hierarchically organized enough to allow for proficient interpretation and integration of new information (Johnson and Lawson, 1998). As a result, novice learners tend to focus on superficial aspects of external representations (Heyworth, 1999; Lowe, 2003, 2004); and the mental models they develop tend to be overly simplistic (Snyder, 2000). For example, Lowe (2004) found that students “tended to use low level strategies that addressed isolated spatial and temporal aspects of the [representation] to the neglect of more inclusive dimensions” (p. 270). That is to say, the students tended to focus on specific features of the external representation rather than the cumulative meaning of or relationships between those features. As a result, the novices in Lowe's study were unable to make meaningful connections based on their superficial and piecemeal approach to understanding the representation.
Experts, on the other hand, have more prior knowledge than novices; but, perhaps more importantly, their knowledge is more highly organized. Their intricate network of prior knowledge and depth of experiences allows expert learners to better relate their mental models to generally agreed upon scientific principles (Geelan, 1997). Because their understanding is not simply superficial and descriptive, experts develop deeper and more meaningful comprehension of material as compared to novices (Snyder, 2000). For example, Snyder (2000) found that the experts categorized the concepts according to their model- and theory-based attributes, thereby describing a coherent view of the concepts (Snyder, 2000). Similarly, Stieff and colleagues (2010) found that students with “more experience in [a] discipline display [problem solving] strategies similar to those seen among experts” (p. 124). This suggests that students with more experience in a discipline are becoming more like experts in their thinking. However, which experiences are essential in developing expertise?
When an instructor selects a representation to depict a particular concept or phenomenon to a student, the instructor has used their more expert-like knowledge to select a representation with a specific intended meaning. However, students do not know what they are looking at or what it is intended to mean. In order for novices to attend to the appropriate information, they must be made aware of it. It is here that external representations present the ability to significantly influence student learning (Lindgren and Schwartz, 2009).
External representations have the potential to cue students to discern particular pieces of information (Jenkinson et al., 2016). Some studies have shown that the shape, color, or orientation of components within the external representation (e.g., Koch and Ullman, 1985; Patrick et al., 2005), the use of arrows (e.g., Hommel et al., 2001), supporting text (e.g., Mayer et al., 1996), the mode of presentation (e.g., Lewalter, 2003), or the amount of variation (e.g., Bussey and Orgill, 2015) can all be used to direct students’ attention to particular aspects of the representation. Patrick and colleagues claim that “[v]isual representations should purposefully use cueing strategies to attract attention and influence what information learners will extract” (2005, p. 364). Thus, if an external representation can be designed in a certain way, students can potentially be cued to attend to information in an expert-like manner. However, if instructors do not select and present these representations to students, then the affordances are moot. Therefore, both the design and selection—and presentation—of external representations can influence what and how students are able to learn. It is especially important to examine these issues in a discipline like biochemistry in which most processes and concepts are not otherwise easily accessible to the senses.
External representation of protein translation
In biochemistry, educators often rely on external representations in order to convey the structure and function of proteins and their associated biochemical pathways, including protein translation (Linenberger and Holmes, 2014).The complexity of proteins with thousands of atoms presents a challenge for the depiction of their structure. Several different types of representations are used to portray proteins, each with its own strengths and weaknesses. [These types include] space-filling models, ball-and-stick models, backbone models, and ribbon diagrams. (Berg et al., 2007, p. 61)
To address the variety of biochemical structures and processes, graphic artists employ a range of different types of representations. Visual modes of representation “can be static (e.g., illustrations, graphs, charts, photos, or maps) or dynamic (e.g., animation, video, or interactive illustration)” (Mayer and Moreno, 2003, p. 43). “Biochemists [also] make use of a wide range of [external representations…] which can be placed on a continuum from abstract, to more stylized, to more realistic representations of phenomena” (Schönborn and Anderson, 2006, p. 95). More recently, Towns and colleagues (2012) have identified six representational types in their Taxonomy of Biochemistry External Representations (TOBER): (1) particulate, (2) symbolic, (3) microscopic, (4) montage, (5) schematic, and (6) animation. They note that a “student must know what each [external representation] encodes and how that information is presented before they can connect multiple [representations] in the way [instructors] hope to facilitate learning” (Towns et al., 2012, p. 303).
The variety of representations used to depict biochemical phenomena poses both a potential obstacle to and opportunity for learning in biochemistry (Schönborn and Anderson, 2006). For example, Schönborn and colleagues (2002) concluded that students had difficulty decoding the symbols used in a diagram of a protein. Schönborn and Anderson (2006) went on to speculate that biochemistry students’ ability to translate between representations of varying levels of abstraction may not be well developed. Therefore, it is important to continue to explore how the learning of biochemistry is affected by external representations. To do so, it is first important to explore which representations students are exposed to by their instructors and the factors instructors are using in order to select those representations.
Purpose of the current study
Protein translation represents a fundamental, dynamic, stochastic, biomolecular event, an event that is not directly experiential for the learner. The only access students have to this information is through the representations to which they are exposed and the mental models they construct to make sense of and navigate these representations.The study reported here is part of a larger project about what students can learn about protein translation from a variety of common external representations of the process (Bussey, 2013). We have defined common external representations as representations to which instructors would easily and regularly have access, e.g., figures from the best-selling textbooks or animations from published or supplemental sources. It was assumed that biochemistry instructors were not designing their own external representations but were instead selecting instructional materials from the resources around them. In selecting the external representations to use in their course(s), instructors have a significant amount of control over the specific aspects of the content to which students are exposed and the way in which this information is presented. Therefore, it is important to investigate how instructors evaluate and select instructional materials which, in turn, allows students to perceive the appropriate and necessary characteristics of the scientific concepts, i.e., the critical features of the content, while at the same time constraining the range and format of those features. For the purposes of this project, we have defined critical features to be those features necessary to promote a scientifically accurate conception of protein translation.
Instructors’ intentions for student learning; their assumptions about what students can, should, and do know; and their perceptions about how best to represent and convey this information to students inform their selection of instructional content and materials, the adaptation of those materials to align with the critical features of the content, and the enactment of their intentions to create the potential for student learning, i.e., a space of learning. External representations constitute a space of learning as they constrain the potential for student learning about a given topic based on the features of that topic depicted by the representation. What is intended for students to learn and how those intentions are enacted inform the broader conversation around instructional decision making in biochemistry education. Therefore, the focus of the current study is to answer the following research questions:
• What do instructors intend for undergraduate biochemistry students to learn from external representations of protein translation?
a. What are the critical features of protein translation?
b. How do the critical features identified by instructors align with their selection of external representations of protein translation?
Theoretical framework
Instructors’ intentions for student learning directly impact the space of learning and, therefore, the curriculum enacted in the classroom (Bussey et al., 2013). As such, instructors’ articulation of core concepts is vital to the enactment of a coherent curriculum in which content in contextualized and interconnected (Cooper et al., 2017). Variation theory guided the design of this study, which had the goals of (1) capturing the range of instructor perceptions of the core concepts students need to know to understand protein translation (the “critical features” of protein translation), and (2) examining the alignment between the critical features of protein translation identified by the instructors with their selection of external representations for teaching protein translation concepts.As a phenomenographic framework, variation theory allows for the exploration of the variety of experiences of a particular object of learning, i.e., what is to be learned (Marton and Booth, 1997). In this way, variation theory is particularly useful for studies that are intended to directly inform practice. In this project, the object of learning was defined as protein translation (Bussey, 2013). What instructors want students to learn about protein translation, what actually happens in the classroom, and what students actually learn about protein translation are three separate things. Therefore, variation theory also identifies three perspectives from which to examine instructors’ and students’ perceptions of the object of the learning: (1) the intended object of learning (i.e., what should students learn?), (2) the enacted object of learning (i.e., what is possible for students to learn based on the resources they are exposed to in a learning environment?), and (3) the lived object of learning (i.e., what did student actually learn?). It should be noted that research utilizing a variation theory framework must include all three perspectives of the object of learning in order to fully characterize the object of learning (Fig. 1).
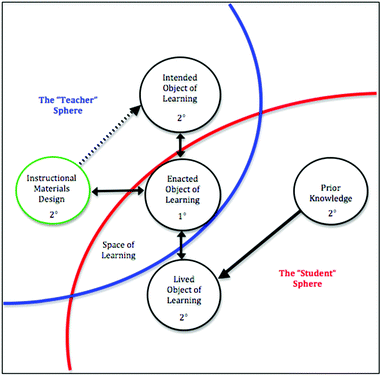 | ||
| Fig. 1 The relationship between Instructional materials design and the objects of learning within variation theory (Bussey et al., 2013). | ||
The work presented here represents the first of these perspectives, i.e., the intended object of learning from a larger study (Bussey, 2013). Instructors have intentions for student learning, but that those intentions are not always enacted (or completely enacted) in the classroom. What happens in the classroom is mediated, in part, by the materials and resources an instructor chooses to present to the students and how they choose to present them. Our interest in the current study is on the external representations instructors choose to use in their classrooms. In a field like biochemistry, where the topics are complex, instructors are more likely to choose from a variety of representations designed by others rather than to construct their own (Fig. 1). The designers of these instructional materials may have different intentions for student learning than the instructors. Therefore, it is important to explore whether and how instructors are choosing representations (from those available) that coordinate with their intentions for student learning. In this study, we examine (1) the critical features of protein translation identified by the instructors and (2) how those critical features align with instructors’ selection of external representations of protein translation.
Methods
Participants
In variation theory, the intended object of learning is defined as what teachers intend their students to learn about a designated object of learning. As such, data on teacher intentions for student learning of protein translation was obtained from interviews with five biochemistry instructors from two universities. All aspects of this project were conducted in accord with policies and practices outlined by our Institutional Review Board. The two universities selected represent both geographical and institutional variability. Two instructors were interviewed from a Department of Biochemistry at large Midwestern university, and three instructors were interviewed from a Department of Chemistry and Biochemistry at large Southwestern university. The instructors had an average of 20.6 years of teaching experience. All instructors interviewed for this project held a PhD in a field related to biochemistry and had taught a college-level biochemistry course that addressed the topic of protein translation. These criteria were used to ensure that the instructor participants had a sufficient level of prior knowledge of the topic and sufficient pedagogical knowledge and experience in order to identify what students should learn about protein translation. All instructors had used external representations to teach biochemical content, and some instructors had previous experience working with designers to create representations for instructional use. A sample size of five instructors is consistent with other studies of teachers’ intentions for student learning utilizing variation theory (e.g., Runesson, 1999; Pang and Marton, 2003). However, additional instructor interviews were considered until it was determined that the sample size allowed for the collected data to reach saturation (Creswell, 2007).External representations
In order to identify the intended object of learning in this study, instructors were interviewed about their understandings of protein translation, their conceptions of what students should learn about protein translation, and their perceptions of specific external representations used to teach protein translation. Due to the variety of representations that have been used to portray the process of protein translation, we set criteria by which to select the external representations used in this study. All representations were selected from common sources (Table 1). Static representations were selected from the top two bestselling biochemistry textbooks. Animations were selected from published or referenced web resources that were freely accessible. All representations were presented in sets of two to provide instructors with a representative range of representations of various types of formats. For the design of this study, it was important that the representations included features that the instructors would potentially identify as being critical for students’ learning of protein translation. As such, all external representations that were selected for use in this project depicted an mRNA strand, a ribosome, and a peptide chain, which Nelson and Cox (2000) identified previously as essential components of translation. Additionally, all representations depicted (at minimum) the elongation stage of translation. Although translation is divided into four stages—(1) activation, (2) initiation, (3) elongation, and (4) termination (Nelson & Cox, 2000)—we decided to focus on representations that primarily depicted aspects of the elongation stage as it is during this stage that the vast majority of the mRNA sequence is translated into the polypeptide sequence by the ribosome (Nelson and Cox, 2000).| Set 1: Textbook diagrams of protein translation | |
|---|---|
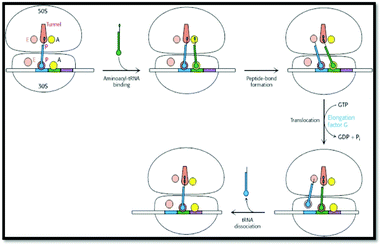
|
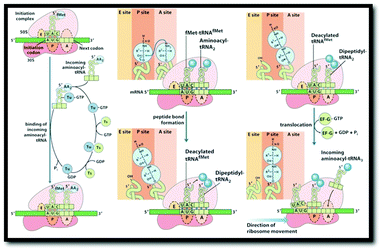
|
| External representation #1: image taken from Berg et al. (2007, p. 872) | External representation #2: image compiled from Nelson and Cox (2008, pp. 1047, 1049) |
Interview protocol
A semi-structured instructor interview protocol (see Appendix A in Bussey and Orgill, 2015) was adapted from Schönborn and Anderson's Three-Phase Single Interview Technique (3P-SIT) (Schönborn et al., 2007; Schönborn and Anderson, 2009). The 3P-SIT structure addresses both participant knowledge of the content (the object of learning) and their understanding external representations depicting the object of learning. The instructor interview protocol used in this study was expanded from the original 3P-SIT model to include initial demographic questions followed by five phases of questions:• Demographics
• Phase 1: The Intended Object of Learning of Translation
• Phases 2–4: Instructor Evaluation/Selection of Common External Representations of Translation
• Phase 5: Instructor Perceptions of External Representations of Translation
General demographic questions, such as “tell me about your current research, or tell me about your teaching experience” allowed the interviewee to share information that was familiar and non-controversial, thereby building rapport so that they could feel more comfortable sharing their thoughts and opinions later on in the interview.
The objective of Phase 1 was to establish what instructors think students should learn about translation, i.e., the intended object of learning. To this end, instructors were asked to explain their understanding of protein translation and how they would or do explain this process to students. Specifically, they were asked to identify the main idea(s) students should come away with following instruction on this topic, i.e., the critical features of the content. Additionally, instructors identified any potential problems students might encounter when trying to understand translation. Note that this line of questioning proceded the instructors’ exposure to the external representations used in this study. This was done to ensure that initial instructor responses were shaped solely by their prior content and pedagogical knowledge of protein translation. Any subsequent deviation in instructor responses following exposure to the external representation could then be attributed to the representations.
In Phases 2 through 4, instructors were asked to evaluate pairs of external representations of protein translation (Table 1). Phase 2 included two textbook diagrams of translation (see Set 1 in Table 1). Instructors described which representation of the pair would best promote student understanding of protein translation. Of the representation they did not choose, instructors described why they did not like it, what could be changed about the representation to make it better for promoting student understanding of protein translation, and what they thought students could learn from and notice about that representation. Similar questions were asked about the representation they did like. Furthermore, instructors were asked to consider what their preferred representation would allow students to learn that would not be allowed by the other representation. Phases 3 and 4 utilized the same procedure as Phase 2 but with Sets 2 and 3, respectively (see Table 1).
Finally in Phase 5, we explored instructors’ perceptions of the various types of representations and how best to depict the process of protein translation to students. Instructors were asked to describe how student learning would be impacted based the format and amount of information presented in the different representations they were shown. The intention of this phase was to determine how instructors went about selecting their ideal representation and how that selection process aligned with what they intended for students to learn.
Data analysis
The instructor interviews were audio-taped and transcribed verbatim. In order to determine the critical features of the object of learning, instructors’ interviews were coded for entities or concepts that were explicitly stated or implied as being important and/or necessary for students to know in order to understand the process of protein translation. For example, Instructor 2 describes the ribosome as both a named feature (“the ribosome”) and an implied feature (“that thing”), both of which were coded as General molecule R, referring to a description or representation of the ribosome as a general entity.Instructor 2: You show the workings of the ribosome […], a mRNA being threaded through that thing […].
In this example, “mRNA” is also identified as a named feature and was coded as General molecule M, referring to a description or representation of the messenger RNA as a general entity. This process was continued for all instructor interviews, and a code list was generated.
This coding scheme was revised using the constant comparative method throughout the coding process (Creswell, 2007). Highly similar codes were collapsed until each defined code described a unique feature of protein translation. The coding scheme was validated using an external coder. The external coder was a postdoctoral researcher with expertise in biochemistry education, experience studying representations, and experience with the use of the 3P-SIT interview strategy. The external coder was independent of the project design, data collection, and initial data analysis. She coded two excerpts from the instructor interviews using the code list. She was asked to apply codes as they were defined on the code list and to identify any areas of confusion or possible new codes. Inconsistent coding and ambiguous code definitions were revised in collaboration with the external coder. A final revised code list containing an operational definition of each code was then created (see Tables 2–4).
| Coded feature | Operational definition |
|---|---|
| Aminoacyl (A) site | A description or depiction of the animoacyl (A) site of the ribosome. |
| Anti-codon loop | A description or depiction of the anti-codon loop of a transfer ribonucleic acid. The nucleotide base sequence does not need to be indicated. |
| Codon | A description or depiction of a codon on the messenger ribonucleic acid. The nucleotide base sequence does not need to be indicated. |
| Codon/anticodon base pairing E | A description or depiction of the interaction between start codon of the messenger ribonucleic acid and the complementary anti-codon on the anti-codon loop of the initiator transfer ribonucleic acid. This interaction occurs only during the initiation phase of protein translation. Hydrogen bonding does not need to be indicated. |
| Codon/anticodon base pairing I | A description or depiction of the interaction between a codon of the messenger ribonucleic acid and a complementary anti-codon on the anti-codon loop of a transfer ribonucleic acid. This interaction occurs only during the elongation phase of protein translation. Hydrogen bonding does not need to be indicated. |
| Exit (E) site | A description or depiction of a transfer ribonucleic acid occupying the Exit (E) site of the ribosome. |
| General molecule M | A description or depiction of the messenger ribonucleic acid as a general entity. |
| General molecule P | A description or depiction of the polypeptide chain as a general entity. |
| General molecule R | A description or depiction of the ribosome as a general entity. |
| General molecule(s) AA | A description or depiction of one or several amino acids as a general entity. |
| General molecule(s) T | A description or depiction of one or several transfer ribonucleic acids as a general entity. |
| Large subunit | A description or depiction of the large ribosomal subunit. |
| P-site tRNA | A description or depiction of a transfer ribonucleic acid occupying the Peptidyl (P) site of the ribosome. |
| Peptide bond formation | A description or depiction of the nucleophilic attack by the lone pair electrons on the amine group of the amino acid of the A-site tRNA on the carbonyl carbon of the amino acid of the P-site tRNA resulting in the formation of a peptide bond. |
| Peptidyl (P) site | A description or depiction of the Peptidyl (P) site of the ribosome. |
| Small subunit | A description or depiction of the small ribosomal subunit. |
| Coded feature | Operational definition |
|---|---|
| 3′ end M | A description or depiction of the 3′ hydroxyl end of a messenger ribonucleic acid. |
| 3′ end T | A description or depiction of the 3′ hydroxyl end of a deacylated transfer ribonucleic acid. |
| 3′ poly A tail | A description or depiction of the polyadenylated 3′ tail of a mature eukaryotic messenger ribonucleic acid |
| 5′ end M | A description or depiction of the 5′ phosphate end of a messenger ribonucleic acid. |
| A-site tRNA | A description or depiction of a transfer ribonucleic acid occupying the Aminoacyl (A) site of the ribosome. |
| Evolution | A description or depiction of the impact of the process of protein translation on evolution. |
| Exiting tRNA/ribosome/mRNA | A description or depiction of the relationship between the messenger ribonucleic acid, the ribosome, and the transfer ribonucleic acid that has exited the ribosome. |
| General molecule(s) EF | A description or depiction of one or several elongation factors as a general entity. |
| General molecule(s) RF | A description or depiction of one or several release factors as a general entity. The term termination factor(s) will be included in this code. |
| General process A | A description or depiction of the process of activation of transfer ribonucleic acids as a general entity. This code also includes charging of transfer ribonucleic acids. |
| General process E | A description or depiction of the process of elongation as a general entity. This code includes indications of polypeptide chain growth |
| General process I | A description or depiction of the process of initiation as a general entity. |
| Incoming tRNA/ribosome/mRNA | A description or depiction of the relationship between the messenger ribonucleic acid, the ribosome, and the transfer ribonucleic acid that is about to enter the Aminoacyl (A) site the ribosome. |
| Methionine | A description or depiction of the amino acid, methionine, of the initiator transfer ribonucleic acid in eukaryotes. This code also includes formylmethionine on the initiator transfer ribonucleic acid in prokaryotes. |
| Nucleotide sequence (multiple codons) | A description or depiction of the nucleotide sequence of multiple codons of the messenger ribonucleic acid. This code is inclusive of the start and stop codons and refers to a series of codons on the messenger ribonucleic acid. |
| Primary structure | A description or depiction of the primary sequence of amino acids of the polypeptide chain. The identity of the amino acids does not need to be indicated. |
| Random motion of cellular components | A description or depiction of the random motion of cellular components in the cytoplasm. Indications of how those random motions can lead to component collisions and subsequent chemical reaction may also be included. |
| Reaction kinetics | A description or depiction of the rate or speed of the process of protein translation. |
| Regulation | A description or depiction of various aspects of cellular regulation on the process of protein translation. |
| Ribosomal translocation | A description or depiction of the movement of the ribosome from one codon to the next from the start codon to the stop codon of the messenger ribonucleic acid. |
| Shine–Dalgarno sequence | A description or depiction of the Shine–Dalgarno region of the prokaryotic messenger ribonucleic acid located upstream of the start codon. |
| Tunnel | A description or depiction of the channel through the large ribosomal subunit from the P-site to the cytoplasm though which the growing polypeptide chain exits the ribosome. |
| Coded feature | Operational definition |
|---|---|
| 16S rRNA | A description or depiction of the ribosomal ribonucleic acid component of the small ribosomal subunit of prokaryotes. |
| 2D shape | A description or depiction of the two dimensional cloverleaf shape of a transfer ribonucleic acid. |
| 3D shape | A description or depiction of the three dimensional L-shape of a transfer ribonucleic acid. |
| 5′ end T | A description or depiction of the 5′ phosphate end of a transfer ribonucleic acid. |
| 5′ methylated cap | A description or depiction of the 7-methylguanosine cap at the 5′ end of a mature eukaryotic messenger ribonucleic acid. |
| E-site tRNA | A description or depiction of a transfer ribonucleic acid occupying the Exit (E) site of the ribosome. |
| EF-G | A description or depiction of the prokaryotic elongation factor G. |
| EF-Ts | A description or depiction of the prokaryotic elongation factor Ts. |
| EF-Tu | A description or depiction of the prokaryotic elongation factor Tu. |
| Energetics | A description or depiction of thermodynamic considerations of the process of protein translation. |
| General molecule(s) IF | A description or depiction of one or several initiation factors as a general entity. |
| GTPase activity of EFs | A description or depiction of the process of hydrolysis of guanosine triphosphate to guanosine diphosphate and phosphate as carried out by an elongation factor as a general entity. |
| Hydrogen bonding E | A description or depiction of the hydrogen bonds formed between the codon of a messenger ribonucleic acid and the anti-codon of a complementary transfer ribonucleic acid during elongation. |
| Hydrogen bonding I | A description or depiction of the hydrogen bonds formed between the start codon of the messenger ribonucleic acid and the anti-codon of the initiator transfer ribonucleic acid during initiation. |
| Nucleotide sequence (anti-codon loop) | A description or depiction of the nucleotide sequence of the anti-codon loop of a transfer ribonucleic acid. |
| Nucleotide sequence (start codon) | A description or depiction of the nucleotide sequence of the start codon of the messenger ribonucleic acid (AUG). |
| Regeneration of activated tRNAs | A description or depiction of the process of regenerating transfer ribonucleic acids with an appropriate amino acid that corresponds to the anti-codon sequence. |
| Secondary structure | A description or depiction of the folding of the polypeptide chain as it interacts with the cytoplasm. |
| Sequential AA | A description or depiction of the amino acid attached to the incoming transfer ribonucleic acid. |
| Start codon | A description or depiction of the start codon of the messenger ribonucleic acid. The nucleotide sequence does not need to be indicated. |
| Stop codon | A description or depiction of the stop codon of the messenger ribonucleic acid. The nucleotide sequence does not need to be indicated. |
Results and discussion
In this section, we characterize instructors’ intentions for student learning about protein translation. First, we describe instructors’ perceptions of the educational value and disciplinary perspective of protein translation in the undergraduate biochemistry curriculum. Second, we discuss the range of critical features of translation identified by our participants and the emergent categorization and thematic analysis of these features. Last, we examine instructors’ decision-making processes as they use their intentions for student learning to evaluate and select external representations of translation.A biochemical perspective on protein translation
One of the underlying assumptions of this project is that protein translation is an important object of learning within the undergraduate-level biochemistry education curriculum; hence, biochemistry students should be learning particular critical features of this object of learning in order to develop correct understandings of it. Although limited prior literature has documented the importance of this topic, it was necessary to establish whether or not the instructor participants in this project held a similar view, as their responses to the instructor interview are based on their perceptions of the value of the topic of protein translation and their subsequent intentions for student learning about this object of learning.During the interviews, all instructors described protein translation as an important biochemical topic.
Instructor 3: [Protein translation] is obviously one of the most important concepts […] in biochemistry. It's part of the dogma…
They were also quick to point out that the treatment of this information from a biochemical perspective was different from that of a biological perspective.
Instructor 3: The biochemistry approach is to look at things at more at the molecular level, so we are more interested in looking at […] how the actual mechanisms of this reactions work. […] From a molecular biology point of view, you would be looking more at eh, eh, the interactions between the different […] players, the proteins, the mRNA, but more of those interactions than the actual forces that are involved in protein translation.
Thus, instructors said that biochemistry is focused on the chemical or mechanistic approach to describing the protein translation process rather than a more general overview of the interactions of components of the process as described in molecular biology.
Instructor 4: One of the things that I look for to define my course is, um, non-redundancy with their biology classes. So almost all of my students are also taking molecular biology, immunology, virology, microbiology, genetics […] whereas, this is a chemistry course and so for me, I definitely want to focus on, on chemical concepts, chemical principles, and, and all the things I think they’re not going to get elsewhere.
Although biochemistry was described by the instructors as being conceptually focused on the chemical concepts and principles of the process, this may be due in part to the fact that instructors perceive this as a deficiency in the treatment of the process from a biological perspective. Thus, some of the instructors perceived that a biochemical presentation of the protein translation process complements and bolsters more biological presentations of protein translation to which the students have been exposed.
Researcher: You had mentioned earlier that, ah, you know, you kind of see some of your role as filling in the gaps […]. What are things you feel like you’re filling in that they’re not getting in [a more biology focused] context?
Instructor 5: Just greater structure. Just greater specificity of structure. For instance, um, this is definitely true for high school, but over there in biology, if they’re teaching molecular biology or genetics, they’re, they’re more interested in information flow throughout the cell and diseases and, and how things can be disrupted; whereas, and I always tell them this, what's the prefix for this course, it's CHEM. It's chemistry. […The content is] overlapping for sure, but it's a different treatment of the area completely, and, and it should be. It's, it's, it's chemistry, so it's more of the actual specific structures and chemical reactions.
These quotations suggest that biochemistry instructors have a unique intended object of learning as compared to instructors from other disciplines who may teach about protein translation. Therefore, the results and implications discussed here should be considered in light of the discipline in addition to the object of learning.
Critical features of protein translation
Unique individual or disciplinary emphases may make some features of protein translation more or less important to a particular instructor in promoting a specific understanding of that object of learning. Although all of the instructors who participated in this study had taught undergraduate introductory biochemistry courses, they did not come from the exact same disciplinary training and did not conduct research in the same areas. As such, each instructor described a unique set of critical features. However, we identified 16 common features of protein translation described by all five instructors as being critical for students’ developing a correct understanding of protein translation. An additional 43 features of protein translation were described by at least one of the instructors. All of these features describe a variety of scientifically accurate components and interactions of protein translation. As we have defined critical features to be those features necessary to promote a scientifically accurate conception of protein translation, all 59 features identified from this analysis of the instructor interviews are considered critical features. However, based on further analysis of the instructor interviews, some critical features were deemed to be more important than others based on the number of instructors that described the same features. If all of the instructors discussed the same feature, that feature was determined to be more important than a feature only discussed by a single instructor. Therefore, we have categorized the critical features as primary, secondary, or tertiary critical features indicating whether all (N = 5), a majority (N = 3+), or a minority (N < 3) of instructors identified a given feature (Tables 2–4, respectively). All critical features in Tables 2–4 are listed alphabetically. Thus, the order of listing is not intended to indicate a hierarchy of features outside of their designation of primary, secondary, or tertiary.The primary critical features were organized into two general themes of features: (1) components/structures of protein translation and (2) interactions/chemistry of protein translation (shown in Table 6). Components/Structures were identified as features used to describe or depict individual constituents or a portion of a constituent perceived as being involved in the process of protein translation. Interactions/Chemistry were identified as features used to describe or depict relationships between two or more components of process. Some of these relationships were described or depicted as specific chemical forces or reactions (i.e., peptide bond formation) while others were described as more general component interactions (i.e., codon/anticodon base pairing).
The majority of primary critical features described by instructors were components or structures of protein translation. Thirteen of the 16 primary critical features identified the basic molecular machinery (i.e., the ribosome, mRNA, and tRNA) and significant structural features of those components (i.e., the large and small ribosomal subunits, the A, P, and E sites of the ribosome, the anticodon loop of the tRNA, and the codon structure of the mRNA). In the sections that follow, we will briefly describe what it is that instructors thought students should understand about these general categories of primary critical features.
The ribosome. Instructors identified students’ understanding of the ribosome as essential to their understanding of protein translation. For example, Instructor 2 described the importance of student understanding of the ribosomal structure.
Instructor 2: You’d certainly have to lay out the structure of the ribosome […]
The ribosome, as both a structural component of the translational machinery and as the catalytic component of the peptide bond formation reaction, was highly referenced by all of the instructors. For example, Instructor 3 described the catalytic nature of the ribosome.
Instructor 3: The students need to understand […] the role that, that ribosomes play in, ah, in the whole process especially the ribosomal RNA, how it's actually part of the catalytic machinery […]
As indicated in the description of the ribosome by Instructor 3, many of the features identified by the instructors were described based on their “role” in the process. Therefore, the instructors’ common reference to the ribosome indicates that it plays a central role in the process of protein translation. Overall, the instructors identified knowledge of the ribosome as a general entity, knowledge of the ribosome's structure, and knowledge of the ribosome's catalytic properties to be important to students’ developing a correct understanding of protein translation.
The mRNA. The messenger RNA was also identified by all of the instructors as a key feature of protein translation. Instructor 3 described the role of mRNA as an important intermediary in the information-processing pathway of the central dogma.
Instructor 3: The students need to understand the concept of moving from DNA to mRNA to protein, and understand how the, how the cell uses different types of RNAs to, eh, eh, eh to translate the […] sleeping code of DNA into the functional code that is protein, and that includes the mRNA.
The instructors highlighted the role of mRNA in the production of “functional” proteins. Instructor 2 also identified the mRNA as a necessary component of translation and discussed the importance of student understanding of processing of mRNA prior to translation, i.e., providing a big picture view of translation as a part of a bigger process rather than as a discrete reaction.
Instructor 2: I think, also it would be useful to show how a lot of messenger RNAs have all kinds of regulatory sequences on them. […] There are many proteins that bind to those and those affect translation […] and so, I think it would be useful to let students know that, um, um messenger RNAs aren’t just synthesized and, and um immediately get translated. A lot of them go through a much more complicated process before they get translated.
Instructor 2 elaborated on the information-processing context of this process, noting that as the molecular information progresses from DNA to mRNA to protein, there are regulatory processes involved as well. The messenger RNA does not enter translation solely as the product of transcription but also the product of regulatory elements that can influence and alter the protein outcome of translation.
The tRNA. The big picture application of the context and implications for protein translation is coupled with a micro-scale emphasis on the chemistry of structure as seen in instructors’ descriptions of the transfer RNA. For example, Instructor 5 emphasized the disciplinary differences in the presentation of tRNA structure to students.
Instructor 5: It's chemistry, so it's more of the actual specific structures […] and a good example is probably the transfer RNA. The classic cloverleaf structure, [a biology class] could probably show that. I show the folded structure, the three-dimensional one and try to, at times, indicate the very interesting, um, non-standard Watson–Crick base pairing.
Similar to Instructor 5, Instructor 4 noted the intricacies of transfer RNA structure when describing what students should know about protein translation.
Instructor 4: You know, the nature of the chemical linkage that connects a tRNA molecule to its cognate amino acid, it's an ester linkage. […] My guess is that's maybe something that's rather glossed over more in a, in a biology class.
Interaction features. In addition to individually referencing, in whole or in part, the ribosome, mRNA, and tRNA, instructors also referred to the interactions between these components. For example, Instructor 4 described the mRNA/tRNA interaction during codon/anticodon base pairing.
Instructor 4: I’m emphasizing, you know, chemical issues, a lot like, um, for example, […] the tRNA anti-codon and the messenger RNA codon as being complementary, that's reverse complement. Thinking about strand polarity very carefully.
Instructors also spent considerable time describing the peptide bond formation between adjacent amino acids.
Instructor 2: I think it's necessary to get some chemistry in there just so that students have a feeling for why the growing peptide ends up on the tRNA it ends up on. Um, if you don’t understand who's attacking who, and what the reaction looks like, then it's hard to follow all that.
It is interesting to note that the majority of primary critical features identified by instructors were specific components of the process rather than chemical properties or interactions. When describing the difference between a biochemical and biological treatment of protein translation, the instructors had seemed to relegate the component parts of the process to a biological treatment of this object of learning. This may indicate that the instructors see the ribosome, mRNA, and tRNA as necessary biological prior knowledge that provides a foundation for a more chemically focused discussion of the chemical properties, forces, or reactions involved in protein translation.
Many of these secondary features reinforced or further clarified the primary critical features. In particular, the primary component features related to the mRNA, the ribosome, the tRNA, the amino acids, and the polypeptide chain were expanded and/or clarified by many of these secondary features. For example, the nucleotide sequence of the mRNA (Nucleotide sequence (multiple codons)) was described by four of the five instructors. Each of these descriptions of the nucleotide sequence of the mRNA further expanded on the mRNA component feature. For example, Instructor 1 used the term “sequence” to refer to the mRNA nucleotide sequence, discussing its relation to several other primary critical features, including the ribosome.
Instructor 1: [In response to a representation of protein translation] The individual players are identified with labels that, in the very early part of it where you blew up and showed the sequence and binding of the sequence to the ribosome, that was very nice.
In this quotation, he used the term sequence to refer both to the mRNA as a general feature of the process and the mRNA's relationship with another primary feature, the ribosome. His comments clarified the mRNA component feature in that mRNA is not only a general component of the protein translation process but also a component that interacts with the ribosome during protein translation.
Instructor 1 goes on to note that “[this representation] had a very nice, clear representation of how the sequence of the anticodon and the codon matches up.” Here he uses the description of the nucleotide sequence to refer not to the mRNA in general, but to the codon as a functional unit of the mRNA and to the codon/anti-codon interaction as a prominent chemical interaction of protein translation. This example underscores the emergence of the two main themes that were previously identified in the primary critical features: (1) components/structures of protein translation and (2) interactions/chemistry of protein translation; however, among the secondary critical features, the structural features generally described substructures of previously identified components and the interactions generally focused on the sequencing of biochemical events. Moreover, the relationship between these two themes typifies the general nature of the biochemical perspective, i.e., a biochemical understanding of protein translation is not just about knowing about the parts of protein translation, but specifically how those parts interact with each other. This goes beyond a simple retelling of the sequence of events, but focuses on the underlying chemical principles and mechanisms to provide the biochemical rationale for the specific interactions, progression, and outcomes of this process. The additional secondary critical features pertaining to the mRNA, ribosome, tRNA, amino acids, and polypeptide chain serve to further detail or clarify the more general corresponding component feature.
Two other component features were identified as secondary critical features: elongation factors and release factors. Both of these features are notable as they are unique, new structures that had not been previously identified as primary critical features. The majority of instructors described these two factors as general entities rather than specific factors, i.e., instructors named these factors without describing their structure or function in detail. This indicated that although many instructors felt the need to include a discussion of these factors in the description of the process, the lack of common discussion warrants their classification as secondary features. Also, both features were described in terms of their role in facilitating the general process of protein translation. Rather than being a central component or interaction, these features tended to be seen as features helping to support the overall process. For example, Instructor 5 commented on the depiction of the release factor as a good addition to the depiction of the termination of the process.
Instructor 5: [In response to a representation of protein translation] We’re at the end of it now. Now they’re just showing the complete chain. Oh nice, now we’ve got the release factor coming in.
The secondary critical features also included several interaction features. Unlike the secondary component features, these interaction features generally provided new information regarding the interactions occurring at various points in time during the protein translation process, specifically during activation, initiation, and elongation. The majority of references to these general processes indicated the addition of sequence ordering information to the intentions for student learning. For example, Instructor 3 uses the phrase “the actual process” to describe the elongation stage of protein synthesis depicted in a certain portion of External Representation #6. This seems to indicate that he is valuing certain stages of the process more than other, or perhaps he is valuing the chemistry that is occurring during those stages.
Instructor 3: [In response to a representation of protein translation] They isolated that part, and I think that's appropriate because then I can explain that and say ok when, now you have the mRNA, now you have the ribosome, you have all this tRNA floating around, and then basically what they did is basically zoomed in, into the, the actual process, the mechanistic process.
Initiation, although not referred to by name in the quotation above, is identified in the sequential discussion of component interactions, “now you have the mRNA, now you have the ribosome.” This stage ordering of the process is used to categorize interactions based on time, i.e., this happens first, then this, then that. Although several instructors discuss similar stage descriptions of interaction features, other instructors specifically discounted the importance of this information for a biochemical presentation of the topic.
Instructor 1: You know, the trivial stuff is the sequence in which [protein translation] happens, the messenger binds, eh, those things are, are hardly more than rote memorization,
Researcher: MmHmm
Instructor 1: and any kid who's been through freshman biology ought to have a pretty reasonable notion about what happens first and so on […]
This does not imply that students should not know the sequence of events. Instead, it implies that students should already know the sequence of events by the time they are presented with protein translation in a biochemistry course. Thus, the categorization of sequence information and the corresponding interaction features as secondary features is consistent with instructor perceptions.
Also among the secondary critical features are three features not associated with a specific biomolecular structure or local interaction: reaction kinetics, evolution, and regulation. These features were used by instructors to address broad applications of students’ knowledge of protein translation rather than their specific knowledge of the process of translation. For example, Instructor 5 integrates both regulation and evolutionary concepts in the teaching of translation.
Instructor 5: The fun part of teaching [protein translation] is I like regulation. I love talking about how things are regulated and when you’re talking about biochemistry, how things are regulated like pathways of feedback and so forth and pathways that are positive loops and all that. I always tell [the students], look at the big picture. Does this make sense? And it always makes beautiful sense after several billion years of evolution. […] They get kind of an eye opening, oh yeah that just, that just, of course, that's the way it should be, it just makes sense, and when biochemistry, when it all starts to tie in like that back together, and, eh, I kind of understand the big picture, I think then we’ve accomplished something.
These broad applications seemed to reinforce individual instructor preferences within biochemistry and are not exclusive to the process of protein translation yet situate it within the “big picture.” As such, these three secondary critical features were thematically organized as “General Considerations” to acknowledge instructors’ use of these features to invoke a broader contextualization of protein translation.
As with the secondary features, the tertiary components features were generally more detailed descriptions of the previously identified critical features. For example, Instructors 1 and 3 both described the start and stop codons, further clarifying unique substructures of the mRNA (a primary critical feature) and their specific roles. In addition to these types of substructures, the tertiary critical features also included one new component feature: initiation factors. As with the secondary critical features, the majority (16 out of 21) of the tertiary critical features identified components or structures of translation. Interestingly, hydrogen bonding is included among the tertiary interaction features. This feature is notable as it is highly related to the primary feature of codon/anti-codon base pairing; however, its tertiary status indicates that most instructors did not describe the chemical nature of the base pairing interaction. Instead, the chemical focus of protein translation seems to be placed on peptide bond formation over other chemical interactions and properties.
| Critical feature | Primary foci | Example |
|---|---|---|
| Primary (1°) | Major biomolecules/structures | mRNA |
| General component interactions | Base pairing | |
| Secondary (2°) | Substructures | Nucleotide sequences |
| Sequencing of events | Elongation | |
| General conditions | Reaction kinetics | |
| Tertiary (3°) | Unique/named substructures | AUG (start codon sequence) |
| Specific chemical interactions | Hydrogen bond formation |
The primary critical features of protein translation can be categorized into two major groups: (1) the major biomolecules involved in the process and (2) the general interactions between those structures. Secondary critical features can be organized around three primary foci: descriptions of (1) specific structural aspects of the major biomolecules, (2) the timing or sequencing of specific component interactions, and (3) the general conditions one would need to consider when describing the process of translation. Finally, tertiary critical features can be organized around two primary foci: (1) unique substructures and (2) specific chemical activity and interactions of component structures.
Thematic analysis of critical features
In addition to their primary, secondary, or tertiary status, the critical features were also found to be highly interrelated. For example, the large and small ribosomal subunits, the A, P, and E-sites, the tunnel, and the 16S rRNA all referred to components of the ribosome As noted previously, critical features generally fell into two main themes: (1) components/structures of protein translation and (2) interactions/chemistry of protein translation. Within each theme, primary critical features generally described the major biomolecules or interactions, while secondary and tertiary critical features provided more detailed substructures or the sequencing of interactions. As the goal of this thematic analysis is to describe the instructors’ intentions for student learning about protein translation, each theme was sub-categorized into the general components or interactions of protein translation in order to describe the general features instructors intended for student to learn (Table 6).| Theme | General feature | Coded feature | Type of feature |
|---|---|---|---|
| Component/structure | mRNA | General molecule M | 1° |
| codon | 1° | ||
| Nucleotide sequence (multiple codons) | 2° | ||
| Shine–Dalgarno sequence | 2° | ||
| 5′ end M | 2° | ||
| 3′ end M | 2° | ||
| 3′ poly A tail | 2° | ||
| 5′ methylated cap | 3° | ||
| Start codon | 3° | ||
| Stop codon | 3° | ||
| Nucleotide sequence (start codon) | 3° | ||
| Ribosome | General molecule R | 1° | |
| Large subunit | 1° | ||
| Small subunit | 1° | ||
| Aminoacyl (A) site | 1° | ||
| Peptidyl (P) site | 1° | ||
| Exit (E) site | 1° | ||
| Tunnel | 2° | ||
| 16S rRNA | 3° | ||
| tRNA | General molecule(s) T | 1° | |
| Anti-codon loop | 1° | ||
| P-site tRNA | 1° | ||
| A-site tRNA | 2° | ||
| 3′ end T | 2° | ||
| E-site tRNA | 3° | ||
| 3D shape | 3° | ||
| 2D shape | 3° | ||
| 5′ end T | 3° | ||
| Nucleotide sequence (anti-codon loop) | 3° | ||
| Amino acids | General molecule AA | 1° | |
| Methionine | 2° | ||
| Sequential AA | 3° | ||
| Polypeptide chain | General molecule P | 1° | |
| Primary structure | 2° | ||
| Secondary structure | 3° | ||
| Initiation factors | General molecule(s) IF | 3° | |
| Elongation factors | General molecule(s) EF | 2° | |
| EF-Tu | 3° | ||
| EF-Ts | 3° | ||
| EF-G | 3° | ||
| Release factors | General molecule(s) RF | 2° | |
| Interactions/chemistry | Activation | General process A | 2° |
| Regeneration of activated tRNAs | 3° | ||
| Initiation | codon/anti-codon base pairing I | 1° | |
| General process I | 2° | ||
| Hydrogen bonding (codon/anti-codon) I | 3° | ||
| Elongation | Peptide bond formation | 1° | |
| codon/anti-codon base pairing E | 1° | ||
| General process E | 2° | ||
| Incoming tRNA/ribosome/mRNA | 2° | ||
| Exiting tRNA/ribosome/mRNA | 2° | ||
| Ribosomal translocation | 2° | ||
| GTPase activity of EFs | 3° | ||
| Hydrogen bonding (codon/anti-codon) E | 3° | ||
| General considerations | Reaction kinetics | 2° | |
| Evolution | 2° | ||
| Regulation | 2° | ||
| Random motion of components | 2° | ||
| Energetics | 3° | ||
The thematic categorization of the critical features resulted in the identification of eight general component parts of protein translation: the mRNA, the ribosome, the tRNAs, the amino acids, the polypeptide chain, the initiation factors, the elongation factors, and the release factors. Each general component is differentiated by its unique structure and role in protein translation.
The critical features related to interactions of components were then categorized based on the relative timing of the interaction. This resulted in the identification of three general stages of protein translation: activation, initiation, and elongation. (Although termination is generally identified as the fourth stage of protein translation (Nelson and Cox, 2000, 2008; Berg et al., 2007), it was not among the features identified by the instructors.)
Finally, a category was created for miscellaneous features: general considerations. These miscellaneous features tended to refer to interactions or concepts that were significantly broader than the topic of protein translation such as reaction kinetics. However, the instructors discussed most of these general consideration features as they described how to place translation in a broader context. Moreover, these general considerations were secondary or tertiary critical features, indicating that they described particular perspectives for situating protein translation that were instructor-dependent.
The intended object of learning
Using the primary, secondary, and tertiary categories as well as the general features and themes, we determined that the general intended object of learning was for students to develop a functional conception of the components and sequence of events of protein translation in order to understand the underlying chemical principles and interactions of the biochemical process. It was deemed critical that students should learn the how the component parts of the mRNA, ribosome, and tRNAs assemble to create the translational machinery. Specifically, the codon/anti-codon base pairing interaction was emphasized as an important feature in understanding how the genetic message was transferred from the mRNA into the polypeptide chain. Additionally, it was identified that students should learn how the amino-acylated tRNAs interact in the various sites of the ribosome during peptide bond formation. Variation in instructors’ intentions for student learning was seen outside of these general intentions and centered on a particular area of instructor interest or on the application of protein translation to a broader biochemical context.Selection of external representations
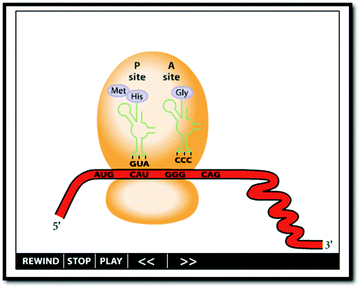
Once we identified the intended object of learning, we then wanted to examine how those intentions influenced the selection and evaluation of external representations of protein translation. As previously mentioned, biochemistry instructors tend to select from existing representations for classroom use rather than creating their own. The designers of these external representations may highlight or depict critical features in a certain manner that either reinforces or challenges instructors’ intentions for students learning. The criteria instructors use to make selections of these instructional materials can then be used to more fully characterize their perceptions of what and how students should be learning about protein translation.Instructor 2: I mean one of things that I think biochemists really like is structures. You know, you’ve got all of these protein structures and they’re beautiful […] and biochemists love them and the temptation is to use bazillions of them in [a] book. Um, the reality is students don’t care about those so much. They want to know what's going on, and they want to know what information do I need to know to get through the next exam. […] If you put an elaborate structure there and you say this is the phosphorylated form [of a protein] and they can’t see the phosphates, that's not a good thing. […] Now if you make a, a simpler form that sort of suggests the shape [of the protein] but allows you to highlight the phosphorylations and, or other modifications or the substrate coming in or whatever it is that you want to show, I think you’re better off.
He goes on to say:
Instructor 2: I think the key thing is trying to design [a representation] so students can get the information that they’re supposed to get out of a figure.
Several of the instructors shared the views of Instructor 2. Some representations were identified as being better than others because they contained many or most of the identified primary critical features and were thought to cue students to notice those critical features. Other representations were viewed less favorably because they either lacked depictions of critical features, inaccurately displayed certain critical features, or were not presented in a manner that would draw students’ attention to the critical features. As such, a key consideration for instructors when selecting an appropriate representation seems to be (1) the alignment of the features presented in the representation with instructors' identified critical features and (2) the accuracy and importance of those features as perceived by the instructor.
The majority of instructors identified External Representation #2 (see Table 1), as the “best” representation of protein translation. This is notable as this representation contains the most depicted critical features, including all 16 of the primary critical features. It should be noted that instructors did not exclusively use of primary critical features when selecting a representation. A representation may have included all or most of the primary critical features but lacked some specific secondary or tertiary feature that the instructor identified as necessary which led them to reject that particular representation. It is also interesting to note that External Representation #2 is a static diagram found in a textbook. Schönborn and Anderson (2006) note that there “is an automatic pedagogical superiority that has been bestowed upon animated [external representations…] as compared with static [external representations]” (p. 97). A recent meta-analysis also concluded that animations generally improved student learning over static representations (Berney and Bétrancourt, 2016). Moreover, protein translation is a dynamic process. As such, one may assume that instructors might favor an animation to best depict a dynamic biochemical processes like translation. However, this was not observed among the majority of instructors that participated in this study. Instructors frequently referred to a lack of information present in the animations as compared to the textbook diagrams.
Beyond the amount of content present, instructors were also concerned about the level of abstraction presented in some of the dynamic external representations.
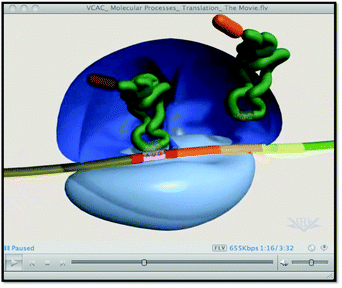
Instructor 3: I think [the 3D appearance] makes it distracting […] Now you’re looking at how pretty, it looks too pretty. […] Students are going to be looking at the shapes and the other shapes coming in […] and out. […] That makes it too distracting.
Interestingly, Instructor 5 noted that animations that depicted more three-dimensional structures were appealing but not from an educational standpoint.
Instructor 5: […] if I’m going to do animation, that's what I want. To me, it's not going to be a […] a learning opportunity so much as sort of a big picture environment where I can say ‘this is what we think in terms of a contour relief in three dimensional structure, we kind of think this is the relationship of the players and we’re trying to get things to scale and so forth.
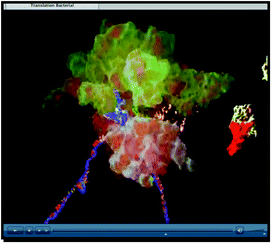
For educational purposes, instructors tended to prefer the more stylized representations over the more realistic representations. They felt that the more realistic, dynamic representations were often more complicated because of the additional structural, spatial, or temporal information presented and the lack of visual cues such as distinctions in color or the naming of a particular structure or interaction.
Instructor 1: [In response to a realistic animation] You know if you asked me what I took away from that, um, I got the idea that its very complicated process involving a number of players, some of which were briefly identified, but none of them stuck. I don’t think I could go back and name you very many of the players. I had, even though the G proteins were shown here, I didn’t get any sense that this was a process in which energy was invested. I didn’t get any connection between the sequence of the codons and the sequence of the amino acids. They, they, they represented the forming polypeptide chain gorgeous, but that's just distracting. A simple schematic showing one to one color matches is much more effective.
Similarly, Instructor 5 described this type of realistic, dynamic representation as visually distracting.
Instructor 5: It's too much in terms of, um, it would be fine for maybe a research article […] that might work, but it, it seems to be very busy at the very beginning just setting it up, and you would have a very hard time following that. I mean I did actually, to tell you the truth.
He went on to say:
Instructor 5: I found myself just wanting to look at the contours, it was very distracting, actually. […] I couldn’t follow, and I teach it [laughs].
Instructors frequently expressed concerns or frustration that the available representations were never quite fully aligned with their stated intentions for student learning. This seems to suggest a mismatch between the depicted features of available representations and the critical features of instructors’ intended object of learning. In general, the instructors tended to favor simple, stylized representations that most closely aligned with the critical features they identified as being most important (frequently the primary and secondary critical features).
Implications for teaching
Biochemistry is complex. Similarly, external representations of biochemical phenomena are complex. It is unlikely that instructors have the time or technical expertise to design these types of representations for use in their classrooms. Additionally, as an educational enterprise, it seems inefficient for each instructor to create their own instructional materials if the goals for student learning are broadly shared amongst instructors. Thus, it is much more likely that instructors will choose from a range of options (some of which are designed by instructional materials designers without chemical or biochemical content knowledge, but all of whom have their own intentions for student learning).Representational objectives set out by textbook book publishers, graphic artists, and/or instructional materials designers may be aligned or misaligned with instructors’ intentions for student learning. While this misalignment may be unavoidable, there are several steps that the designers of external representations could take so that instructors can make informed choices about their selection of external representations. For example, external representations of scientific phenomena, in particular those representations found in commercially available educational materials, could contain a representational objective statement (i.e., “This representation is designed to highlight the relationship between…”) to help instructors make informed choices about alignment between their intentions for student learning and the intentions for student learning that guided the creation of the available representations. A representation objective statement may also be valuable to the students to further cue them to seek out the primary critical features depicted by an external representation.
Instructors should make an informed choice about what resources best allow students to notice and experience the critical features necessary to promote a particular perception of an object of learning. It is also worth noting that in the process of evaluating a variety of external representations, instructors may be making explicit (to themselves and others) their intentions for student learning. It may be that the critique of available representations is actually helping instructors to think about what the critical features of the object of learning are.
In selecting an appropriate representation for promoting student learning about protein translation, instructors should first consider the alignment between their intentions for student learning and the possibilities for learning created by the external representation. While students can learn from a variety of representations, not all representations depict the same features. Moreover, not all features are depicted in the same manner. Additionally, some representations may present additional, non-critical features that may distract students from noticing and learning about the critical features. Therefore, instructors should carefully consider which features are critical for students to learn and then select a representation or representations that cue students to notice those features. The current study suggests that biochemistry instructors have a unique intended object of learning as compared to instructors from other disciplines who may teach about protein translation. Therefore, a clear disciplinary perspective may be necessary when identifying the specific critical features for an object of learning.
Limitations
In accordance with variation theory, we have assumed that instructors’ verbal descriptions represent their salient knowledge of the content. In doing so, it is possible that we have not been able to fully capture instructors’ intended object of learning. However, we have been able to capture the critical features of protein translation as expressed by the participating instructors. While potentially incomplete, the expressed critical features are those that can be communicated to students; and, therefore, we have assumed that they are the most important critical features of the intended object of learning.It should also be reiterated that the instructors interviewed in this study represent two different institutional structures (a Department of Biochemistry and a Department of Chemistry and Biochemistry) and pursue several different research interests. While this phenomenographic study was designed intentionally to draw upon the range of instructor perceptions, a broader survey of instructors, particularly those from a molecular biology context, may help to further characterize and clarify the critical features of protein translation.
With regard to the representations used in this project, we have also identified potential limitations. Although the instructors were able to evaluate the external representations used in this study, those representations constitute a small, pre-selected pool of possible representations of translation. It is quite possible that, left to their own devices, biochemistry instructors would choose different representations based on the familiarity and accessibility of available representations and the alignment of those representations with their intentions for student learning. Therefore, instructors’ reactions to the external representations of translation described above should be contextualized to the specific representations used in this study.
Comparing representations is often a comparison of apples and oranges. No two representations used in this study depicted the same set of features. Therefore, it is not surprising that instructors favored some representations over others. Is the difference in the amount of depicted features, the manner in which those features are being presented, or the visual strategies that are cueing viewers to notice those features actually causing the perception that some external representations better than others? Any future studies should attempt to identify or create representations that contain features that might be more aligned in order to better assess the specific impacts of a given representation on student learning.
Conflicts of interest
There are no conflicts to declare.References
- Arneson J. B. and Offerdahl, E. G., (2018), Visual literacy in bloom: using bloom's taxonomy to support visual learning skills, CBE—Life Sci. Educ., 17(1), ar7.
- Ayres P. and van Gog T., (2009), State of the art research into Cognitive Load Theory, Comput. Human Behav., 25, 253–257.
- Braune R. F. and Foshay W. R., (1983), Towards a practical model of cognitive information processing: task analysis and schema acquisition for complex problem-solving situations, Instr. Sci., 12, 121–145.
- Berg J. M., Tymoczko J. L. and Stryer L., (2007), Biochemistry, 6th edn, New York, NY: W.H. Freeman.
- Berney S. and Bétrancourt M., (2016), Does animation enhance learning? A meta-analysis, Comput. Educ., 101, 150–167.
- Bussey T. J., (2013), What can biochemistry students learn about protein translation? Using variation theory to explore the space of learning created by some common external representations, Doctoral Dissertation, University of Nevada, Las Vegas, Las Vegas, NV.
- Bussey T. J. and Orgill M., (2015), What do biochemistry students pay attention to in external representations of protein translation? The case of the Shine–Dalgarno sequence, Chem. Educ. Res. Pract., 16(4), 714–730.
- Bussey T. J., Orgill M. and Crippen K. J., (2013), Variation theory: a theory of learning and a useful theoretical framework for chemical education research, Chem. Educ. Res. Pract., 14(1), 9–22.
- Cook M. P., (2006), Visual representations in science education: the influence of prior knowledge and cognitive load theory on instructional design principles, Sci. Educ., 90, 1073 – 1091.
- Cook M. P., Wiebe E. N. and Carter G., (2008), The influence of prior knowledge on viewing and interpreting graphics with macroscopic and molecular representations, Sci. Educ., 92, 848–867.
- Cooper M. M., Posey L. A. and Underwood S. M., (2017), Core Ideas and Topics: Building Up or Drilling Down? J. Chem. Educ., 94(5), 541–548.
- Creswell J. W., (2007), Qualitative inquiry and research design: Choosing among five approaches, 2nd edn, Thousand Oaks, CA: SAGE Publications.
- Geelan D. R., (1997), Prior knowledge, prior conceptions, prior constructs: what do constructivists really mean, and are they practicing what they preach? Aust. Sci. Teach. J., 43(3), 26–29.
- Goodsell D. S. and Jenkinson J., (2018), Molecular Illustration in Research and Education: Past, Present, and Future, J. Mol. Biol., 430(21), 3969–3981.
- Halloun I. A. and Hestenes D., (1985), The initial knowledge state of college physics students, Am. J. Phys., 53(11), 1043–1055.
- Harle, M. and Towns, M. H., (2012), Students' understanding of external representations of the potassium ion channel protein, part I: affordances and limitations of ribbon diagrams, vines, and hydrophobic/polar representations, Biochem. Mol. Biol. Educ., 40, 349–356.
- Heyworth R. M., (1999), Procedural and conceptual knowledge of experts and novice students for the solving of a basic problem in chemistry, Int. J. Sci. Educ., 21(2), 195–211.
- Hommel B., Pratt J., Colzato L. and Godijn R., (2001), Symbolic control of visual attention, Psychol. Sci., 12(5), 360–365.
- Jenkinson J., Jantzen S., Gauthier A. and McGill G., (2016), The effect of attention cueing in molecular animation to communicate random motion, Learning from Text and Graphs in a World Diversity, EARLISIG, Geneva, pp. 96–98.
- Johnson M. A. and Lawson A. E., (1998), What are the relative effects of reasoning ability and prior knowledge on biology achievement in expository and inquiry classes? J. Res. Sci. Teach., 35(1), 89–103.
- Koch C. and Ullman S., (1985), Shifts in selective visual attention: towards the underlying neural circuitry, Hum. Neurobiol., 4, 219–227.
- Kozma R. B. and Russell J., (1997), Multimedia and understanding: expert and novice responses to different representations of chemical phenomena, J. Res. Sci. Teach., 34(9). 949–968.
- Lewalter D., (2003), Cognitive strategies for learning from static and dynamic visuals, Learn. Instr., 13, 177–189.
- Lindgren R. and Schwartz D. L., (2009), Spatial learning and computer simulations in science, Int. J. Sci. Educ., 31(3), 419–438.
- Linenberger, K. J. and Holme, T. A., (2014), Results of a national survey of biochemistry instructors to determine the prevalence and types of representations used during instruction and assessment, J. Chem. Educ., 91, 800–806.
- Lowe R., (1996), Background knowledge and the construction of a situational representation from a diagram, Eur. J. Psychol. Educ., 11(4), 377–397.
- Lowe R., (2003), Animation and learning: selective processing of information in dynamic graphics, Learn. Instr., 13, 157–176.
- Lowe R., (2004), Interrogation of a dynamic visualization during learning, Learn. Instr., 14, 257–274.
- Marton, F. and Booth, S., (1997), Learning and awareness. Mahwah, NJ:Lawrence Erlbaum Associates.
- Mayer R. E., (1993), Illustrations that instruct, in Glaser R. (ed.), Advances in instructional psychology, Hillsdale, NJ: Erlbaum, vol. 4, pp. 253–284.
- Mayer R. E. and Moreno R., (2003), Nine ways to reduce cognitive load in multimedia learning, Educ. Psychol., 38, 43–52.
- Mayer R. E., Bove W., Bryman A., Mars R. and Tapangco L., (1996), Why less is more: meaningful learning from visual and verbal summaries of science textbook lessons, J. Educ. Psychol., 88(1), 64–73.
- Nelson M. M. and Cox D. L., (2000), Lehninger Principles of Biochemistry, 3rd edn, New York, NY: W. H. Freeman.
- Nelson M. M. and Cox, D. L., (2008), Lehninger Principles of Biochemistry, 5th edn, New York, NY: W. H. Freeman.
- Pang M. F. and Marton F., (2003), Beyond “lesson study”: comparing two ways of facilitating the grasp of some economic concepts, Instr. Sci., 31, 175–194.
- Patrick M. D., Carter G. and Wiebe E. N., (2005), Visual representations of DNA replication: middle grades students’ perceptions and interpretations, J. Sci. Educ. Technol., 14(3), 353–365.
- Runesson U., (1999), Teaching as constituting a space of variation, Paper presented at 8th EARLI conference, Göteborg, Sweden.
- Schnotz W. and Lowe R., (2003), External and internal representations in multimedia learning, Learn. Instr., 13, 117–123.
- Schönborn K. J. and Anderson T. R., (2006), The importance of visual literacy in the education of biochemists, Biochem. Mol. Biol. Educ., 34(2), 94–102.
- Schönborn K. J. and Anderson T. R., (2009), A model of factors determining students’ ability to determine external representations in biochemistry, Int. J. Sci. Educ., 31(2), 193–232.
- Schönborn K. J. and Anderson T. R., (2010), Bridging the educational research-teaching practice gap: foundations for assessing and developing biochemistry students’ visual literacy, Biochem. Mol. Biol. Educ., 38(5), 347–354.
- Schönborn K. J., Anderson T. R. and Grayson D. J., (2002), Student difficulties with the interpretation of a textbook diagram of Immunoglobulin G (IgG), Biochem. Mol. Biol. Educ., 30(2), 93–97.
- Schönborn K. J., Anderson T. R. and Mnguni L. E., (2007), Methods to determine the role of external representations in developing understanding in biochemistry, in Lemmermöhle D. et al. (ed.), Professionell lehren—erfolgreich lernen, New York: Waxmann, pp. 291–301.
- Sim, J. H., and Daniel, E. G. S., (2014), Representational competence in chemistry: a comparison between students with different levels of understanding of basic chemical concepts and chemical representations, Cogent. Educ., 1, 1–17.
- Snyder J. L., (2000), An investigation of the knowledge structures of experts, intermediates and novices in physics, Int. J. Sci. Educ., 22(9), 979–992.
- Stanley E. D., (1996), Taking a second look: investigating biology with visual datasets, Bioscene, 22(3), 13–17.
- Stieff M., Hegarty M. and Dixon B., (2010), Alternative strategies for spatial reasoning with diagrams, in Goel A. K., Jamnik M. and Narayanan N. H. (ed.), Diagrammatic representation and interference, Berlin: Springer, pp. 115–127.
- Towns M. H., Raker J. R., Becker N., Harle M. and Sutcliffe J., (2012), The biochemistry tetrahedron and the development of the taxonomy of biochemistry external representations (TOBER), Chem. Educ. Res. Pract., 13(3), 296–306.
- Trumbo J., (1999), Visual literacy and science communication, Sci. Commun., 20(4), 409–425.
- Wellington J. and Osborne J., (2001), Language and Literacy in Science Education, Philadelphia, PA: Open University Press.
| This journal is © The Royal Society of Chemistry 2019 |