Open Access Article
Open Access ArticleRetracted Article: CircRNA PVT1 modulates cell metastasis via the miR-181a-5p/NEK7 axis and cisplatin chemoresistance through miR-181a-5p-mediated autophagy in non-small cell lung cancer†
Limin Yan‡
a,
Minghe Bai‡a,
Jinheng Xua,
Xuemei Lia,
Chenpeng Wua,
Yuntao Zhoub,
Jidong Yanc and
Zhiyong Zhang *a
*a
aDepartment of Pathology, Tangshan Gongren Hospital, No. 27, Wenhua Road, Lubei District, Tangshan 063000, Hebei, China. E-mail: hfppry@163.com; Tel: +86-315-3722242
bDepartment of Center Labotarary, Tangshan Gongren Hospital, Tangshan 063000, Hebei, China
cDepartment of Thoracic Surgery, Tangshan Gongren Hospital, Tangshan 063000, Hebei, China
First published on 20th December 2019
Abstract
In the initiation and evolution of human cancers, circular RNAs (circRNAs) act as crucial regulators. The aim of this report was to ascertain the functional mechanisms of circRNA plasmacytoma variant translocation 1 (circPVT1) in the metastasis and chemoresistance of non-small cell lung cancer (NSCLC). The levels of circPVT1, microRNA-181a-5p (miR-181a-5p) and non-inherited maternal antigens-related kinase 7 (NEK7) were examined via quantitative real-time polymerase chain reaction (qRT-PCR). The levels of the associated proteins were determined through western blot. Cell counting kit-8 (CCK-8) and flow cytometry were used to assess the half inhibitory concentration (IC50) of cisplatin and cell apoptosis, respectively. Cell invasion was detected by transwell assay. A dual-luciferase reporter assay and RNA immunoprecipitation (RIP) were used to confirm the target relation. The impact of circPVT1 on cisplatin chemoresistance in vivo was investigated using xenograft experiments. CircPVT1 and NEK7 were up-regulated and miR-181a-5p was down-regulated in NSCLC. CircPVT1 knockdown refrained the cisplatin chemoresistance and metastasis of NSCLC cells. MiR-181a-5p was a target of circPVT1 and circPVT1 inhibition alleviated the effects of a miR-181a-5p inhibitor on NSCLC cells. The decrease of circPVT1 accentuated the si-NEK7-inhibited metastasis by the miR-181a-5p/NEK7 axis and relieved the 3-methyladenine (3-MA)-promoted cisplatin chemoresistance by miR-181a-5p-mediated autophagy. Down-regulation of circPVT1 facilitated the cisplatin sensitivity of NSCLC cells in vivo. Due to the modulation of cell metastasis via the miR-181a-5p/NEK7 axis and cisplatin chemoresistance by miR-181a-5p-mediated autophagy in NSCLC, circPVT1 might act as an appreciable therapeutic marker for NSCLC.
Introduction
As one of the frequent fatal malignancies, lung cancer is exceedingly aggressive, with smoking and exposure to carcinogens as high-risk factors.1 Astoundingly, non-small cell lung cancer (NSCLC) accounts for about 90% of lung cancer and has high metastatic threat.2 There are increasing modern treatments for NSCLC, but the conventional cisplatin-based chemotherapy retains the dominant position in the therapies for NSCLC patients, especially in advanced stages.3,4 Interestingly, cell autophagy is considered to regulate the drug chemoresistance in cancers, including for cisplatin.5–7 Therefore, a better understanding of the molecular mechanisms is required, not only for the metastasis of NSCLC, but also for acquired chemoresistance, to promote the therapeutic strategies and the recovery prognosis for NSCLC.Generated by back-splicing of precursor mRNAs (pre-mRNAs), circular RNAs (circRNAs) are ubiquitous in mammals as novel non-coding RNAs (ncRNAs) and competitive endogenous RNAs (ceRNAs). Different from the linear RNAs, circRNAs are more stable and conservative due to the presence of covalent closed-loop structures.8,9 Gradually, circRNAs are being regarded as pivotal regulators and biomarkers in the development of cancers.10,11 Yao et al. declared that circ_100876 had great prognostic value for NSCLC patients with a high expression level.12 Chen et al. reported that circ_100146 regulated proliferation, invasion and apoptosis via targeting miR-615-5p and miR-361-3p in NSCLC.13 And circ-prolyl 4-hydroxylase subunit beta (P4HB) was shown to expedite the epithelial–mesenchymal transition (EMT) and metastasis as a sponge of miR-133a-5p in NSCLC cells.14 Besides, circ-plasmacytoma variant translocation 1 (circPVT1) was up-regulated in NSCLC specimens and cells15 and enhanced the doxorubicin and cisplatin chemoresistance of osteosarcoma cells.16
It is becoming increasingly clear that circRNAs function as miRNA “sponges” to affect downstream gene expression and cancer regulation.17–19 The down-regulation of miR-181a-5p20 and the allusive up-regulation of non-inherited maternal antigens-related kinase 7 (NEK7, a mitotic kinase)21 in NSCLC were shown in previous studies. Taking into consideration the dysregulation of circPVT1, miR-181a-5p and NEK7 in NSCLC, but the potential relation among them remaining unknown, the current report focuses on the regulatory mechanisms among circPVT1, miR-181a-5p and NEK7 regarding the progression and chemoresistance of NSCLC.
Materials and methods
Clinical specimens
After obtaining informed consent from 25 patients scheduled to undergo a pneumonectomy at Tangshan Gongren Hospital, paired NSCLC tissues and normal contiguous tissues were collected and preserved at −80 °C with rapid speed. Ethically, the present research received approval from the Ethics Committee of Tangshan Gongren Hospital.Cell culture and treatment
A normal human bronchial epithelial cell line (16HBE) was purchased from Millipore (Bedford, MA, USA) and two NSCLC cell lines (A549 and H1299) were bought from American Type Culture Collection (ATCC, Manassas, VA, USA). In a 37 °C humid incubator with 5% CO2, 16HBE and H1299 cells were cultured in Roswell Park Memorial Institute-1640 medium (RPMI-1640; Gibco, Carlsbad, CA, USA) and A549 cells were maintained in F-12K medium (Gibco), both supplemented with 10% fetal bovine serum (FBS; Gibco) and antibiotics (100 U mL−1 penicillin and 100 μg mL−1 streptomycin; Gibco). A549 and H1299 cells were treated with cisplatin (Sigma-Aldrich, St. Louis, MO, USA) and 3-methyladenine (3-MA; Millipore) according to the requirements of the assays.Cell transfection
Small interfering RNA (siRNA) against circPVT1 and NEK7 (si-circPVT1 and si-NEK7), short hairpin RNA (shRNA) against circPVT1 (sh-circPVT1), miR-181a-5p mimic (miR-181a-5p), miR-181a-5p inhibitor and respective negative controls (si-NC, sh-NC, miR-NC and miR-NC inhibitor) were constructed by GenePharma (Shanghai, China). The sequence of circPVT1 was cloned into the pcDNA basic vector (Invitrogen, Carlsbad, CA, USA) to obtain the overexpression vector pcDNA-circPVT1, and its negative control was marked as pcDNA-NC. These oligonucleotides or vectors were mixed with Lipofectamine 3000 reagent (Invitrogen) and transfected into NSCLC cells with 60% confluence. Harvested cells at different time points were prepared for further experiments.Quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA was isolated from tissues and cells using Trizol reagent (Invitrogen). Then, SuperScript III Reverse Transcriptase (Thermo Fisher Scientific, Waltham, MA, USA) was applied to transcribe the total RNA into complementary DNA (cDNA), which was used as a template to perform the qRT-PCR reaction with SYBR Green Master Mix (Applied Biosystems, Foster City, CA, USA) through an ABI Prism 7500 sequence detection system (Applied Biosystems) as per the user manuals. The 2−ΔΔCt approach was used to analyze the relative expression with glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as the internal control for circPVT1 or NEK7 and U6 for miR-181a-5p. The primers obtained from Sangon Biotech (Shanghai, China) are listed below: circPVT1 (forward: 5′-ATCGGTGCCTCAGCGTTCGG-3′, reverse: 5′-CTGTCCTCGCCGTCACACCG-3′); miR-181a-5p (forward: 5′-CCGCGAACATTCAACGCTGTCG-3′, reverse: 5′-ATCCAGTGCAGGGTCCGAGG-3′); NEK7 (forward: 5′-CCACTGGGATGGTAAAACTTG-3′, reverse: 5′-AAGGACTTTGTAATGCAGCCAT-3′); GAPDH (forward: 5′-TTGCCCTCAACGACCACTTT-3′, reverse: 5′-TGGTCCAGGGGTCTTACTCC-3′); U6 (forward: 5′-CTCGCTTCGGCAGCACA-3′, reverse: 5′-AACGCTTCACGAATTTGCGT-3′).Western blot
After the exaction of total proteins by radioimmunoprecipitation assay (RIPA) buffer with proteinase inhibitor (Beyotime, Shanghai, China), sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was implemented for segregating 40 μg of proteins, which were then transferred onto polyvinylidene fluoride (PVDF) membranes (Millipore), followed by blocking with 5% skim milk (Sangon Biotech) for 4 h. In a refrigerator at 4 °C, the primary antibodies were incubated overnight, including anti-NEK7 (Abcam, Cambridge, MA, USA, ab133514, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000), anti-B-cell lymphoma-2 (anti-Bcl-2; Abcam, ab32124, 1
1000), anti-B-cell lymphoma-2 (anti-Bcl-2; Abcam, ab32124, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000), anti-Bcl-2-Associated X (anti-Bax; Abcam, ab32503, 1
1000), anti-Bcl-2-Associated X (anti-Bax; Abcam, ab32503, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000), anti-E-cadherin (Abcam, ab40772, 1
1000), anti-E-cadherin (Abcam, ab40772, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000), anti-Vimentin (Abcam, ab92547, 1
1000), anti-Vimentin (Abcam, ab92547, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000), anti-N-cadherin (Abcam, ab76011, 1
1000), anti-N-cadherin (Abcam, ab76011, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000) or anti-beta Actin (anti-β-Actin, Abcam, ab8227, 1
1000) or anti-beta Actin (anti-β-Actin, Abcam, ab8227, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3000). Subsequently, the cells were incubated with secondary antibody (Abcam, ab205718, 1
3000). Subsequently, the cells were incubated with secondary antibody (Abcam, ab205718, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5000) for 45 min at indoor temperature. Ultimately, the specific blots were visualized through ImageJ software (NIH, Bethesda, MD, USA) using an enhanced chemiluminescence kit (Millipore).
5000) for 45 min at indoor temperature. Ultimately, the specific blots were visualized through ImageJ software (NIH, Bethesda, MD, USA) using an enhanced chemiluminescence kit (Millipore).
3-(4,5-Dimethylthiazol-2-y1)-2,5-diphenyl tetrazolium bromide (MTT) assay
The half inhibitory concentration (IC50) of cisplatin was evaluated from cell viability through the MTT assay. After treatment with different concentrations of cisplatin, transfected NSCLC cells were digested by 0.25% trypsin (Gibco) and made into a cell suspension with complete medium, followed by seeding into 96-well plates with 100 μL of suspension per well. Then cells were incubated with 20 μL of MTT for 4 h and 200 μL of dimethyl sulfoxide (DMSO; Sigma-Aldrich) for 10 min. Through a microplate reader (Thermo Fisher Scientific), the optical density (OD) value of the lysate was examined at 490 nm and IC50 was calculated.Flow cytometry
Firstly, NSCLC cells were seeded into a 6-well plate with 2 × 104 cells per well. After different treatments (cisplatin, transfection or 3-MA), the cells were harvested with 0.25% trypsin (Gibco) and washed using cold phosphate buffer solution (PBS; Hyclone, Logan, UT, USA). Next, the washed cells were dyed using an Annexin V-fluorescein isothiocyanate (Annexin V-FITC)/propidium iodide (PI) detection kit (BD Biosciences, San Diego, CA, USA) by following the producer's instructions. Eventually, the apoptotic cells were discerned by means of a flow cytometer (BD Biosciences).Transwell assay
A Transwell 24-well chamber (Corning Life Sciences, Corning, NY, USA) was implemented for the invasion assay. Before seeding cells, the lower side of the upper chamber was coated with Matrigel (BD Bioscience). Then, a cell suspension made in serum-free medium was supplemented into the upper chamber. Simultaneously, 600 μL of relative medium containing 10% FBS was added into the lower chamber. After incubation for 24 h, the liquid of the upper chamber was blotted up and moved into 800 μL of methanol (Supelco, Bellefonte, PA, USA) to fix the cells, followed by staining through crystal violet (Supelco) for 20 min. Finally, uninvaded cells were erased by wet cotton swab and the membrane was bottomed up to count the invaded cells using a microscope.Dual-luciferase reporter assay
The putative common points between circPVT1 and miR-181a-5p or miR-181a-5p and NEK7 were predicted using Starbase2.0. The wild-type (WT) and mutant (MUT) sequences of circPVT1 and 3′UTR NEK7 were cloned into the pGL3 basic plasmid (Promega, Madison, WI, USA) to construct the reporter plasmids of the wild-types (WT circPVT1 and WT NEK7) and mutants (MUT circPVT1 and MUT NEK7). With the assistance of Lipofectamine 3000 (Invitrogen), the co-transfection of four luciferase reporters with miR-181a-5p or miR-NC was executed. Following transfection for 48 h, the cells were collected using 1× passive lysis buffer (PLB; Promega), and the activity of firefly luciferase was quantified through the Dual-Luciferase Reporter Assay System (Promega) with renilla activity as the internal reference.RNA immunoprecipitation (RIP) assay
The RIP assay was administrated by the Magna RNA immunoprecipitation kit (Millipore). Briefly, NSCLC cells were lysed using RIP buffer, followed by the incubation of magnetic beads enveloped with anti-Argonaute2 (anti-Ago2; Abcam, ab32381) and the internal control of anti-Immunoglobulin G (anti-IgG; Abcam, ab205718). After the washing of the magnetic beads, RNA was purified applying the phenol-chloroform-isoamyl alcohol reagent (Sangon Biotech). Finally, the RNA enrichment was analyzed by the detection of qRT-PCR.Xenograft tumor assay
Six-week-old BALB/c nude mice were gained from Shanghai SIPPR-BK Laboratory Animal Co. Ltd. (Shanghai, China) and subcutaneously injected with A549 cells transfected with sh-circPVT1 or sh-NC (10 mice per group). Then two groups of mice were intraperitoneally injected with 3 mg kg−1 cisplatin or PBS once every week, respectively. Tumor volume (length × width2 × 0.5) was calculated weekly. After 4 weeks, euthanasia was executed for all mice and the excised tumor tissues were weighed. Then the expression of circPVT1 was examined by qRT-PCR with the extracted RNA as the amplified template. This animal experiment was carried out following the stipulations of National Animal Care and approved by the Animal Ethics Committee of Tangshan Gongren Hospital.Statistical analysis
All assays were performed with three biological replications. Data were presented as mean ± standard deviation (SD) and processed through SPSS 19.0 and GraphPad Prism 7. Spearman's correlation coefficient was applied for analyzing the linear correlation in the NSCLC tissues. The comparisons between two groups were analyzed by Student's t-test and the difference analysis of multiple groups was conducted via a one-way analysis of variance with Tukey's test. At the biostatistical level, a P value less than 0.05 was considered as significant.Results
CircPVT1 and NEK7 were up-regulated while miR-181a-5p was down-regulated in NSCLC tissues and cells
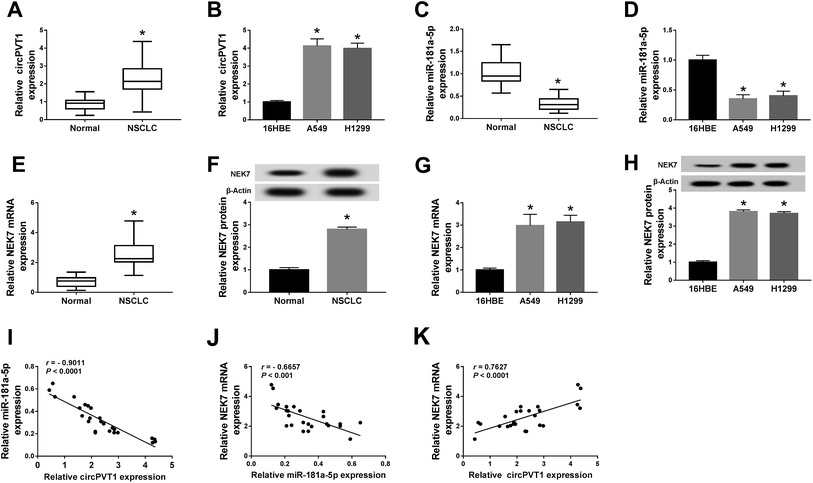
At the beginning, the expression of circPVT1, miR-181a-5p and NEK7 was assayed by qRT-PCR and western blot. For circPVT1, up-regulation was found in the NSCLC tissues (Fig. 1A) and cells (A549 and H1299) (Fig. 1B) in comparison to the normal tissues and 16HBE cells. On the contrary, the miR-181a-5p level was much lower in both NSCLC tissues and cells than in normal tissues and cells (Fig. 1C and D). Interestingly, the mRNA and protein expression of NEK7 was obviously increased in the NSCLC tissues (Fig. 1E and F) and cells (Fig. 1G and H). After the analysis of Spearman's correlation coefficient, a noteworthy negative correlation was discovered not only between circPVT1 and miR-181a-5p (r = −0.9011, P < 0.0001) but also between miR-181a-5p and NEK7 (r = −0.6657, P < 0.001) in NSCLC tissues (Fig. 1I and J), while the relation between circPVT1 and NEK7 was positive (r = 0.7627, P < 0.0001) (Fig. 1K). The aberrant expression of circPVT1, miR-181a-5p and NEK7 might be relevant to the regulation of NSCLC.Knockdown of circPVT1 suppressed the chemoresistance of cisplatin and metastasis in NSCLC cells
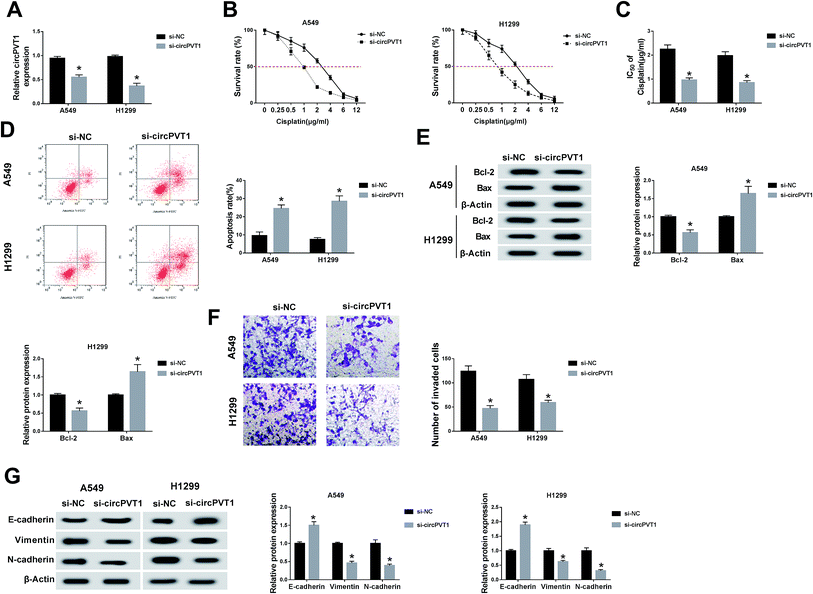
For the sake of investigating the function of circPVT1 in NSCLC, we first disturbed the expression of circPVT1 in NSCLC cells via transfection with si-circPVT1 using si-NC as the negative control. As shown in Fig. 2A, si-circPVT1 transfection inhibited circPVT1 expression in both A549 and H1299 cells by comparison with the si-NC transfection group. Thereafter, the impact of circPVT1 on NSCLC was analyzed. With the addition of cisplatin, the cell survival rate was significantly decreased in the si-circPVT1 group compared with the si-NC group (Fig. 2B), so the calculated lower IC50 of cisplatin (Fig. 2C) implied the depression of cisplatin resistance by knockdown of circPVT1. After treatment with 2 μg mL−1 cisplatin, circPVT1 knockdown resulted in the enhancement of the apoptosis percentage in A549 and H1299 cells, as shown in flow cytometry (Fig. 2D). Simultaneously, the expression of Bcl-2 (anti-apoptosis marker) was decreased, but the Bax (pro-apoptosis marker) level was increased by transfecting with si-circPVT1 relative to si-NC (Fig. 2E). Moreover, the transwell assay indicated that inhibition of the invasion capacity was evoked via introducing si-circPVT1, in contrast to si-NC introduction (Fig. 2F). Also, the elevation of E-cadherin (EMT-inhibited marker) and the reduction of Vimentin and N-cadherin (EMT-promoted markers) were notable in NSCLC cells by knockdown of circPVT1 (Fig. 2G). The suppressive role of si-circPVT1 in the cisplatin resistance and metastasis of NSCLC cells was preliminarily confirmed.CircPVT1 induced targeted regulation of the level of miR-181a-5p
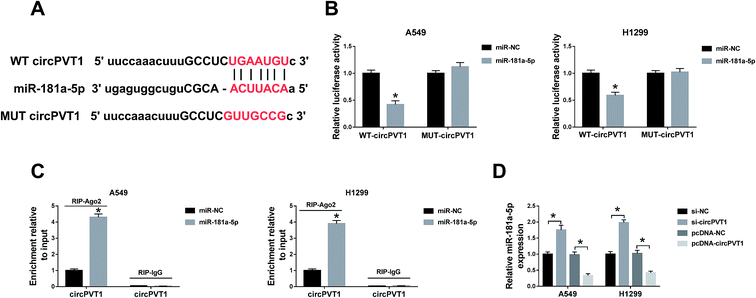
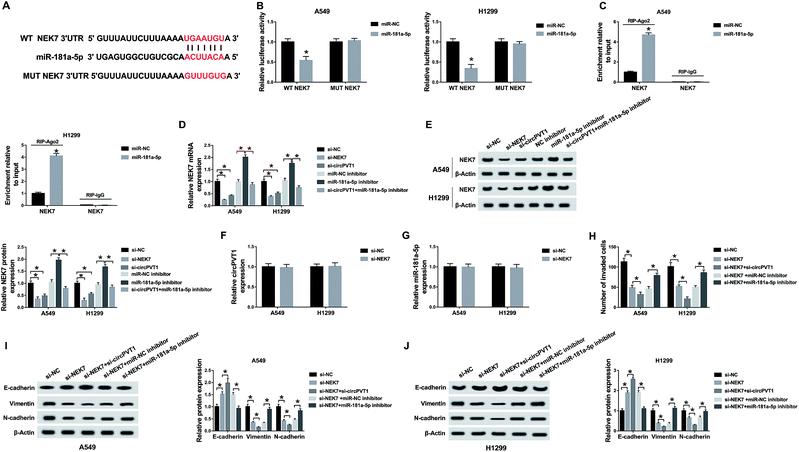
Through the analysis of Starbase2.0, the latent conjugation sites between circPVT1 and miR-181a-5p were found (Fig. 3A). Subsequently, a dual-luciferase reporter assay and an RIP assay were administrated to confirm the predication. Noticeably, the luciferase activities of A549 and H1299 cells co-transfected with WT circPVT1 and miR-181a-5p were repressed, while there was no apparent change after co-transfection with MUT circPVT1 and miR-181a-5p (Fig. 3B). Furthermore, both circPVT1 and miR-181a-5p were enriched in the Ago particle, in contrast to the IgG group and miR-NC group (Fig. 3C). In addition, transfection of si-circPVT1 evidently facilitated the expression of miR-181a-5p, while the introduction of pcDNA-circPVT1 generated the opposite effect on the level of miR-181a-5p in A549 and H1299 cells (Fig. 3D). The results unraveled that circPVT1 serves as a sponge of miR-181a-5p.Down-regulation of circPVT1 ameliorated the effects of miR-181a-5p inhibitor on NSCLC cells
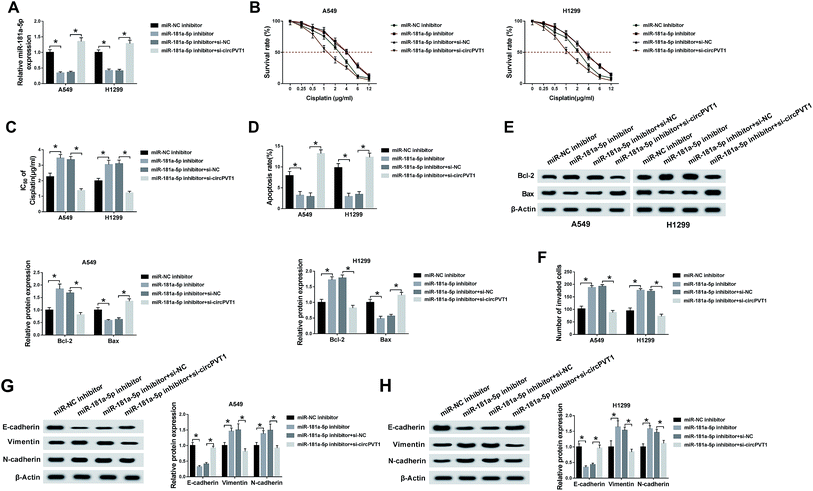
To explore the potential regulatory mechanism between circPVT1 and miR-181a-5p in NSCLC cells, A549 and H1299 cells were transfected with miR-181a-5p inhibitor, miR-181a-5p inhibitor + si-circPVT1 or matched controls (miR-NC inhibitor and miR-181a-5p inhibitor + si-NC). QRT-PCR demonstrated that the down-regulation of miR-181a-5p expression induced by the miR-181a-5p inhibitor was attenuated by the knockdown of circPVT1 in A549 and H1299 cells (Fig. 4A). After treatment with cisplatin of different concentrations, the miR-181a-5p inhibitor improved the cell viability of A549 and H1299 cells compared to the miR-NC inhibitor group, whereas this impact was alleviated following transfection of si-circPVT1 (Fig. 4B), indicating the weakening of the miR-181a-5p inhibitor-induced IC50 by circPVT1 knockdown (Fig. 4C). After A549 and H1299 cells were exposed to 2 μg mL−1 cisplatin, flow cytometry indicated that the apoptosis rate of the miR-181a-5p inhibitor group was lower than that of the miR-NC inhibitor group, while this inhibition was reduced through repressing circPVT1 (Fig. 4D). Subsequently, miR-181a-5p inhibitor-induced up-regulation of Bcl-2 and the decrease of Bax were both mitigated via si-circPVT1 transfection in a western blot (Fig. 4E). For the metastasis capacity, transfection of si-circPVT1 markedly reduced the miR-181a-5p inhibitor-induced stimulative effects on cell invasion (Fig. 4F) and the levels of Vimentin and N-cadherin, as well as the suppressive effect on the expression of E-cadherin (Fig. 4G and H). Briefly, circPVT1 down-regulation abated the promoted effects of the miR-181a-5p inhibitor on cisplatin resistance and cell metastasis by promoting miR-181a-5p.Inhibition of circPVT1 aggravated the si-NEK7-inhibited metastasis via reducing NEK7 by promoting miR-181a-5p
Through bioinformatics analysis by Starbase2.0, the presumptive combining sites of miR-181a-5p acting on the 3′-UTR of NEK7 are displayed in Fig. 5A. Furthermore, miR-181a-5p transfection obviously decreased the luciferase activity of the WT NEK7 group in A549 and H1299 cells, while this decrease was abrogated in the MUT NEK7 group (Fig. 5B). Furthermore, the RIP assay proved that the enrichment of miR-181a-5p and NEK7 in the Ago pellet was much higher than that in the IgG pellet and miR-NC group (Fig. 5C). Then, qRT-PCR and western blot revealed that the knockdown effect of si-NEK7 was conspicuous, and there was a decrease of NEK7 in the si-circPVT1 group compared to that in the si-NC group and an elevation by the miR-181a-5p inhibitor in contrast to the miR-NC inhibitor group; moreover, the miR-181a-5p inhibitor relieved the reduction of NEK7 caused by si-circPVT1 (Fig. 5D and E). There was no obvious change in the expression of circPVT1 and miR-181a-5p after knockdown of NEK7, implicating that NEK7 was indeed a downstream target of miR-181a-5p (Fig. 5F and G). In transwell assay, knockdown of NEK7 led to the decline of invaded cells, which was exacerbated by transfection with si-circPVT1 but was eased by miR-181a-5p inhibitor (Fig. 5H). Similarly, circPVT1 knockdown accentuated the up-regulation of E-cadherin and repression of Vimentin and N-cadherin induced by si-NEK7, but miR-181a-5p inhibitor generated the inverse effects (Fig. 5I and J). Thus, the decline of circPVT1 inhibited NEK7 expression by up-regulating miR-181a-5p to aggravate the si-NEK7-induced restrained effect on the metastasis of NSCLC cells.Knockdown of circPVT1 relieved the 3-MA-promoted chemoresistance of cisplatin via elevating miR-181a-5p-mediated autophagy
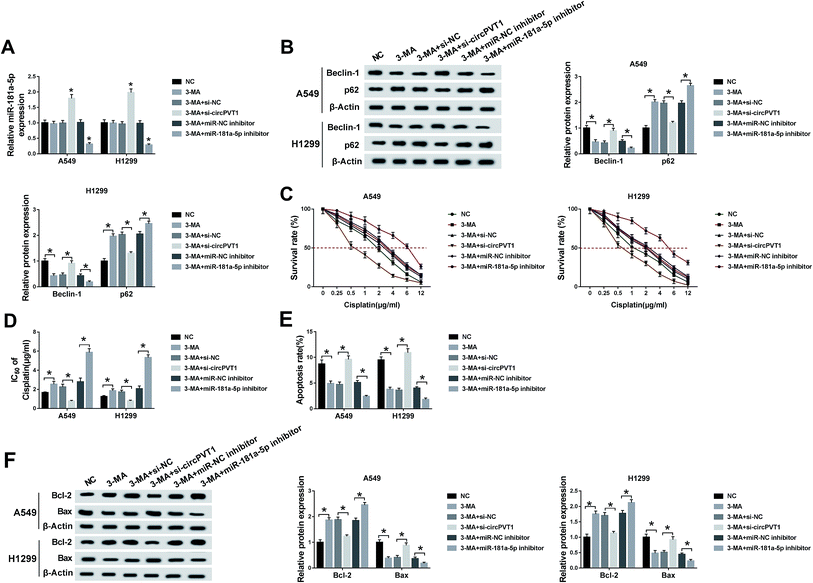
When A549 and H1299 cells were treated with 5 mM 3-MA (an autophagy inhibitor) for 24 h and transfected with si-circPVT1, miR-181a-5p or the corresponding controls (si-NC and miR-NC inhibitor), qRT-PCR indicated that 3-MA treatment had no influence on miR-181a-5p expression compared to the NC group, but miR-181a-5p was up-regulated in the 3-MA + si-circPVT1 treatment group and down-regulated in the 3-MA + miR-181a-5p inhibitor group in contrast to their control groups (Fig. 6A). A western blot was first performed to examine the levels of autophagy-associated proteins. As depicted in Fig. 6B, treatment of 3-MA overtly reduced the expression of Beclin1 (pro-autophagy marker) but enhanced the level of p62 (anti-autophagy marker), which was weakened following transfection of si-circPVT1 and aggravated by the miR-181a-5p inhibitor in comparison to the si-NC and miR-NC inhibitor groups, indicating that circPVT1 knockdown mitigated 3-MA-inhibited autophagy via increasing miR-181a-5p. Under the conditions of different concentrations of cisplatin, the introduction of si-circPVT1 reduced the elevation of cell viability induced by 3-MA, while the miR-181a-5p inhibitor had the opposite effect in contrast to their controls (Fig. 6C). Likewise, the same results were found for the IC50 of cisplatin after calculation (Fig. 6D). Flow cytometry and western blot were used to evaluate the cell apoptosis of A549 and H1299 cells treated with 2 μg mL−1 cisplatin and transfected with the above groups. The results indicated that 3-MA brought about the repression of the apoptosis rate and Bax level but an increase in Bcl-2 expression, whereas down-regulation of circPVT1 abolished these effects and the inhibition of miR-181a-5p reversely caused the exacerbation of these effects compared with the transfection of si-NC or miR-NC inhibitor, respectively (Fig. 6E and F). Taken together, down-regulation of circPVT1 relieved the chemoresistance of cisplatin promoted by 3-MA through facilitating autophagy via elevating miR-181a-5p.The decrease of circPVT1 heightened the cisplatin sensitivity of NSCLC cells in vivo
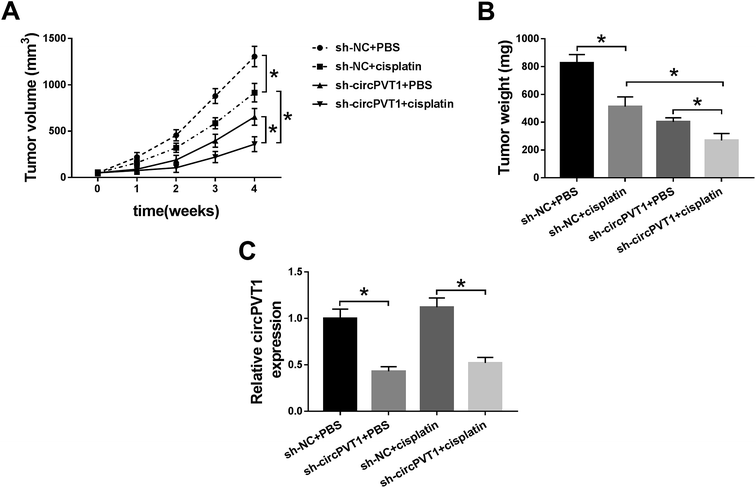
A xenograft model was established to research the impact of circPVT1 on the chemoresistance of cisplatin in vivo. As Fig. 7A and B reveal, the tumor volume and weight were decreased in the sh-NC + cisplatin and sh-circPVT1 + PBS treatment groups compared with the sh-NC + PBS group, which suggested that either cisplatin treatment or circPVT1 knockdown could reduce tumor growth, while this reduction was intensified in the sh-circPVT1 + cisplatin group. Moreover, the lower circPVT1 expression of tumor tissues in the sh-circPVT1 + PBS or sh-circPVT1 + cisplatin groups was shown by qRT-PCR analysis compared to the sh-NC + PBS or sh-NC + cisplatin groups, respectively, but there was no evident difference between the sh-NC + PBS and sh-NC + cisplatin groups (Fig. 7C), suggesting that the exacerbation of the inhibitory effect on tumor growth in the sh-circPVT1 + cisplatin group was achieved by motivating cisplatin sensitivity in NSCLC cells through repressing circPVT1 in vivo.Discussion
Patients that develop NSCLC are at risk of metastasis and acquired chemoresistance after drug treatment.22,23 Due to the dissatisfactory effect of many modern therapeutic strategies, novel molecular indicators might provide hope of preventing the progression of NSCLC. Our report showed not only the abnormal regulation of circPVT1, but also its regulatory mechanisms for metastasis via miR-181a-5p/NEK7 and chemoresistance by autophagy signaling via miR-181a-5p. It is seemingly promising that circPVT1 plays a significant role in the evolution of NSCLC as a therapeutic target.Extensive research studies have been conducted to determine the molecular mechanism of ncRNAs in the progression of human cancers. CircPVT1 has been shown to have great involvement in the regulation of multiple cancers, as a novel ncRNA. For instance, circPVT1 is overexpressed in head and neck squamous cell carcinoma (HNSCC) and regulates the expression of miR-497-5p to affect the proliferation of HNSCC cells, as an oncogene.24 Similarly, Chen et al. exhibited the high expression of circPVT1 in gastric cancer (GC) and its pro-proliferation ability by sponging miR-125 in GC cells.25 Also, knockdown of circPVT1 inhibits the metastasis of colorectal cancer (CRC) cells, partly via the regulation of miR-145,26 and reduces the migration ability through the miR-203/homebox D3 (HOXD3) pathway in human hepatocellular carcinoma.27 Moreover, Qin et al. certified that circPVT1 is expressed at a high level and the down-regulation of circPVT1 restrains the proliferation and motivates the apoptosis of NSCLC cells.28 In agreement with these research studies, the up-regulation of circPVT1 was presented in NSCLC and the depression of circPVT1 inhibited cell metastasis during the current study. In addition, we found that knockdown of circPVT1 played an inhibitory role in the chemoresistance of cisplatin in NSCLC cells, consistent with the effect of promotion of circPVT1 overexpression on cisplatin resistance in osteosarcoma cells in the report of Zhu et al.16
It is well-known that the regulatory role of circRNAs is achieved partly by them acting as miRNA sponges to govern the expression of important genes, implicating the participation of circRNAs in cancer progression via the miRNA-gene network. In this report, miR-181a-5p was identified as a target miRNA for circPVT1 and regulated by circPVT1. MiR-181a-5p was proven to be down-regulated and caused the suppression of the migration29 and invasion of NSCLC cells.30 Additionally, the relevance of dysregulated miR-181a-5p to insulin resistance was confirmed.31 Xue et al. also described the effect of the reduction of miR-181a-5p on the resistance of oxidation in osteoarthritis.32 In line with these studies, we found that low expression of miR-181a-5p in NSCLC and inhibition of miR-181a-5p elevated the cisplatin chemoresistance and cell metastasis. In addition, the alleviation of these effects by knockdown of circPVT1 suggested that the function of circPVT1 in NSCLC was generated by miR-181a-5p at least in part.
Through online bioinformatics analysis and assay verification, NEK7 was considered as a downstream target of miR-181a-5p. High expression of NEK7 was found in NSCLC tissues and cells and circPVT1 could regulate NEK7 expression by targeting miR-181a-5p. NEK7 was shown to play a potential role in cancer metastasis and tumorigenesis.33 Doubtlessly, the interference of NEK7 expression restrained the metastasis of NSCLC cells, and the data indicated that circPVT1 knockdown decreased cell metastasis by inhibiting NEK7 via promoting miR-181a-5p. The functional mechanism of circPVT1 in NSCLC metastasis was through the miR-181a-5p/NEK7 axis.
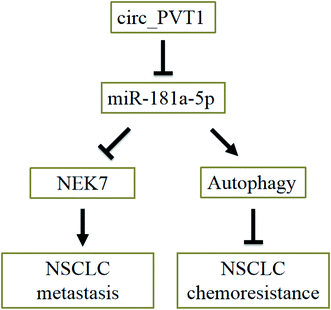
Accumulative evidence demonstrates that cell autophagy has a deep connection with drug resistance. For example, Zhang et al. testified that thioredoxin domain-containing protein 17 (TXNDC17) induced autophagy to facilitate paclitaxel resistance in ovarian cancer;34 Zhang et al. found that p53 and Ras signaling controlled cisplatin resistance via the promotion of autophagy;35 Wang et al. reported that arctigenin increased the cisplatin sensitivity of CRC cells through inducing autophagy.36 In NSCLC cells, we discovered that the repression of autophagy could promote cisplatin resistance. In addition, the reduction of circPVT1 relieved the 3-MA-promoted chemoresistance of cisplatin via activating autophagy, by up-regulating miR-181a-5p. Thus, circPVT1 regulated the cisplatin chemoresistance by miR-181a-5p-mediated autophagy. Furthermore, a decrease in circPVT1 enhanced the cisplatin sensitivity of NSCLC cells in vivo, implying a role of circPVT1 in promoting cisplatin chemoresistance. Therefore, the circ_PVT1/miR-181a-5p axis not only promotes the metastasis of NSCLC by regulating NEK7 expression, but also facilitates the chemoresistance of NSCLC via affecting cell autophagy (Fig. 8).
Conclusion
To summarize, our data indicate the regulatory networks of circPVT1/miR-181a-5p/NEK7 in cell metastasis and circPVT1/miR-181a-5p-mediated autophagy in cisplatin chemoresistance in NSCLC cells. Interestingly, this study provides a forceful explanation for the functional mechanisms of circPVT1 in NSCLC metastasis and chemoresistance, which might be of profound significance in the progression of NSCLC. This research gives a novel perspective and holds great promise in the clinic therapeutic strategies of NSCLC patients with circPVT1 as an advantageous biomarker.Funding
This work was supported by the Science and technology project of Hebei province (Clinicopathological significance of precise detection of gene mutations in non-small cell lung cancer) (No. 162777167), the Medical application tracking project of Hebei province (The clinical and pathological significance of EGFR T790M mutation in advanced non-small cell lung cancer detected by the ARMS method) (No. G2018121) and the Medical science research key project of Hebei province (the expression of c-Met and EGFR protein in lung adenocarcinoma and the significance of EGFR gene mutation) (No. 20171381).Conflicts of interest
The authors declare that they have no financial conflicts of interest.References
- L. A. Torre, F. Bray, R. L. Siegel, J. Ferlay, J. Lortet-Tieulent and A. Jemal, Ca-Cancer J. Clin., 2015, 65, 87–108 CrossRef PubMed.
- A. V. Nascimento, H. Bousbaa, D. Ferreira and B. Sarmento, Curr. Drug Targets, 2015, 16, 1448–1463 CrossRef CAS PubMed.
- T. Reungwetwattana, M. J. Eadens and J. R. Molina, Semin. Resp. Crit. Care Med., 2011, 32, 78–93 CrossRef PubMed.
- D. A. Fennell, Y. Summers, J. Cadranel, T. Benepal, D. C. Christoph, R. Lal, M. Das, F. Maxwell, C. Visseren-Grul and D. Ferry, Cancer Treat. Rev., 2016, 44, 42–50 CrossRef CAS PubMed.
- M. Kim, J. Y. Jung, S. Choi, H. Lee, L. D. Morales, J. T. Koh, S. H. Kim, Y. D. Choi, C. Choi, T. J. Slaga, W. J. Kim and D. J. Kim, Autophagy, 2017, 13, 149–168 CrossRef CAS PubMed.
- H. YiRen, Y. YingCong, Y. Sunwu, L. Keqin, T. Xiaochun, C. Senrui, C. Ende, L. XiZhou and C. Yanfan, Mol. Cancer, 2017, 16, 174 CrossRef PubMed.
- P. P. Naik, S. Mukhopadhyay, P. K. Panda, N. Sinha, C. K. Das, R. Mishra, S. Patil and S. K. Bhutia, Cell Proliferation, 2018, 51, e12411 CrossRef PubMed.
- J. E. Wilusz and P. A. Sharp, Science, 2013, 340, 440–441 CrossRef CAS PubMed.
- S. Memczak, M. Jens, A. Elefsinioti, F. Torti, J. Krueger, A. Rybak, L. Maier, S. D. Mackowiak, L. H. Gregersen, M. Munschauer, A. Loewer, U. Ziebold, M. Landthaler, C. Kocks, F. le Noble and N. Rajewsky, Nature, 2013, 495, 333–338 CrossRef CAS PubMed.
- S. Meng, H. Zhou, Z. Feng, Z. Xu, Y. Tang, P. Li and M. Wu, Mol. Cancer, 2017, 16, 94 CrossRef PubMed.
- S. Qu, Z. Liu, X. Yang, J. Zhou, H. Yu, R. Zhang and H. Li, Cancer Lett., 2018, 414, 301–309 CrossRef CAS PubMed.
- J. T. Yao, S. H. Zhao, Q. P. Liu, M. Q. Lv, D. X. Zhou, Z. J. Liao and K. J. Nan, Pathol., Res. Pract., 2017, 213, 453–456 CrossRef CAS PubMed.
- L. Chen, A. Nan, N. Zhang, Y. Jia, X. Li, Y. Ling, J. Dai, S. Zhang, Q. Yang, Y. Yi and Y. Jiang, Mol. Cancer, 2019, 18, 13 CrossRef PubMed.
- T. Wang, X. Wang, Q. Du, N. Wu, X. Liu, Y. Chen and X. Wang, Biochem. Biophys. Res. Commun., 2019, 513, 904–911 CrossRef CAS PubMed.
- X. Li, Z. Zhang, H. Jiang, Q. Li, R. Wang, H. Pan, Y. Niu, F. Liu, H. Gu, X. Fan and J. Gao, Cell. Physiol. Biochem., 2018, 51, 2324–2340 CrossRef CAS PubMed.
- Z. Kun-Peng, M. Xiao-Long and Z. Chun-Lin, Int. J. Biol. Sci., 2018, 14, 321–330 CrossRef PubMed.
- F. R. Kulcheski, A. P. Christoff and R. Margis, J. Biotechnol., 2016, 238, 42–51 CrossRef CAS PubMed.
- A. C. Panda, Adv. Exp. Med. Biol., 2018, 1087, 67–79 CrossRef CAS PubMed.
- J. Zang, D. Lu and A. Xu, J. Neurosci. Res., 2018, 1–11 CAS.
- Y. Cao, D. Zhao, P. Li, L. Wang, B. Qiao, X. Qin, L. Li and Y. Wang, Cell. Physiol. Biochem., 2017, 42, 346–356 CrossRef CAS PubMed.
- D. Kang, H. S. Cho, G. Toyokawa, M. Kogure, Y. Yamane, Y. Iwai, S. Hayami, T. Tsunoda, H. I. Field, K. Matsuda, D. E. Neal, B. A. Ponder, Y. Maehara, Y. Nakamura and R. Hamamoto, Genes, Chromosomes Cancer, 2013, 52, 126–139 CrossRef CAS PubMed.
- J. A. Mirrielees, J. H. Kapur, L. M. Szalkucki, J. M. Harter, L. R. Salkowski, R. M. Strigel, A. M. Traynor and L. G. Wilke, J. Surg. Res., 2014, 188, 419–431 CrossRef PubMed.
- C. G. Azzoli, L. M. Krug, V. A. Miller, M. G. Kris and R. Mass, Semin. Oncol., 2002, 29, 59–65 CrossRef CAS PubMed.
- L. Verduci, M. Ferraiuolo, A. Sacconi, F. Ganci, J. Vitale, T. Colombo, P. Paci, S. Strano, G. Macino, N. Rajewsky and G. Blandino, Genome Biol., 2017, 18, 237 CrossRef.
- J. Chen, Y. Li, Q. Zheng, C. Bao, J. He, B. Chen, D. Lyu, B. Zheng, Y. Xu, Z. Long, Y. Zhou, H. Zhu, Y. Wang, X. He, Y. Shi and S. Huang, Cancer Lett., 2017, 388, 208–219 CrossRef CAS PubMed.
- Z. Wang, M. Su, B. Xiang, K. Zhao and B. Qin, Biochem. Biophys. Res. Commun., 2019, 512, 716–722 CrossRef CAS PubMed.
- Y. Zhu, Y. Liu, B. Xiao, H. Cai, M. Liu, L. Ma, H. Yin and F. Wang, Biol. Open, 2019, 8, bio043687 CrossRef.
- S. Qin, Y. Zhao, G. Lim, H. Lin, X. Zhang and X. Zhang, Biomed. Pharmacother., 2019, 111, 244–250 CrossRef CAS PubMed.
- Z. Ma, X. Qiu, D. Wang, Y. Li, B. Zhang, T. Yuan, J. Wei, B. Zhao, X. Zhao, J. Lou, Y. Jin and Y. Jin, Acta Biochim. Biophys. Sin., 2015, 47, 630–638 CrossRef CAS PubMed.
- S. Li, J. Yang, Y. Xia, Q. Fan and K. P. Yang, Oncol. Res., 2018, 26, 289–296 CrossRef PubMed.
- J. Lozano-Bartolome, G. Llaurado, M. Portero-Otin, A. Altuna-Coy, G. Rojo-Martinez, J. Vendrell, R. Jorba, E. Rodriguez-Gallego and M. R. Chacon, J. Clin. Endocrinol. Metab., 2018, 103, 1447–1458 CrossRef PubMed.
- J. Xue, Z. Min, Z. Xia, B. Cheng, B. Lan, F. Zhang, Y. Han, K. Wang and J. Sun, BMC Musculoskeletal Disord., 2018, 19, 355 CrossRef CAS PubMed.
- N. H. Eisa, Y. Jilani, K. Kainth, P. Redd, S. Lu, O. Bougrine, H. Abdul Sater, C. A. Patwardhan, A. Shull, H. Shi, K. Liu, N. M. Elsherbiny, L. A. Eissa, M. M. El-Shishtawy, A. Horuzsko, R. Bollag, N. Maihle, J. Roig, H. Korkaya, J. K. Cowell and A. Chadli, J. Biol. Chem., 2019, 294, 5246–5260 CrossRef CAS PubMed.
- S. F. Zhang, X. Y. Wang, Z. Q. Fu, Q. H. Peng, J. Y. Zhang, F. Ye, Y. F. Fu, C. Y. Zhou, W. G. Lu, X. D. Cheng and X. Xie, Autophagy, 2015, 11, 225–238 CrossRef PubMed.
- X. Zhang, Z. Qi, H. Yin and G. Yang, Theranostics, 2019, 9, 1096–1114 CrossRef CAS PubMed.
- Y. Wang, L. Lina, L. Xu, Z. Yang, Z. Qian, J. Zhou and L. Suoni, Biochem. Biophys. Res. Commun., 2019, 520, 20–26 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9ra08872e |
| ‡ These authors contributed equally to the work. |
| This journal is © The Royal Society of Chemistry 2019 |