Open Access Article
Open Access ArticleRetracted Article: LncRNA OIP5-AS1 contributes to ox-LDL-induced inflammation and oxidative stress through regulating the miR-128-3p/CDKN2A axis in macrophages
Xiaojuan Li *a,
Quansheng Caob,
Yanyu Wangc and
Yongsheng Wangb
*a,
Quansheng Caob,
Yanyu Wangc and
Yongsheng Wangb
aDepartment of Central Sterile Supply, The First Affiliated Hospital of Henan University of Science and Technology, No. 24, Jinghua Road, Luoyang 471003, China. E-mail: erehzf@163.com; Tel: +86-0379-64830544
bDepartment of Cardiovascular Surgery, The First Affiliated Hospital of Henan University of Science and Technology, Luoyang, China
cDepartment of Cardiovascular Medicine, The First Affiliated Hospital of Henan University of Science and Technology, Luoyang, China
First published on 17th December 2019
Abstract
Long non-coding RNA OIP5-AS1 (lncRNA OIP5-AS1) and microRNA-128-3p (miRNA-128-3p) have been reported to play significant roles in human diseases. However, their role in atherosclerosis (AS) has been less studied. The aim of this research was to reveal the roles and functional mechanisms of OIP5-AS1 and miRNA-128-3p in AS development. Quantitative real-time polymerase chain reaction (qRT-PCR) and western blot assays were performed to detect gene expression. Cell proliferation and apoptosis were assessed using a 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay and flow cytometry analysis, respectively. In addition, ELISA was employed to determine the levels of interleukin (IL)-6, IL-1β, and tumor necrosis factor (TNF)-α. Oxidative stress was examined using a relevant kit. Furthermore, the interaction between miR-128-3p and OIP5-AS1 or cyclin-dependent kinase inhibitor 2A (CDKN2A) was predicted using StarBase, and then confirmed using the dual-luciferase reporter assay or the RNA immunoprecipitation (RIP) assay. We found that OIP5-AS1 and CDKN2A levels were upregulated and the miR-128-3p level was downregulated in oxidized low-density lipoprotein (ox-LDL)-induced THP1 cells. OIP5-AS1 knockdown weakened the regulatory effect of ox-LDL on cell progression. Interestingly, OIP5-AS1 directly interacted with miR-128-3p and miR-128-3p-targeted CDKN2A. Furthermore, OIP5-AS1 regulated ox-LDL-induced cell progression through modulating miR-128-3p expression, and miR-128-3p exerted its influence by modulating the CDKN2A level. Finally, we confirmed that OIP5-AS1 suppressed miR-128-3p expression to increase the level of CDKN2A. In conclusion, our findings demonstrate that OIP5-AS1 knockdown repressed the effect of ox-LDL on cell progression through regulating the miR-128-3p/CDKN2A axis, providing a potential target for the treatment of AS.
1. Introduction
Atherosclerosis (AS) is a chronic inflammatory disease and is the major cause of coronary heart and cerebral infarction vascular diseases.1,2 The accumulation of foam cells containing cholesterol in blood vessels is the result of macrophage transformation and is considered to be a sign of AS development.3,4 Present evidence suggests that oxidized low-density lipoprotein (ox-LDL) plays a crucial role in the progression of macrophage activation and foam cell development.5 Therefore, it is urgent to reveal the functional mechanism of ox-LDL in the growth of macrophages.Long non-coding RNAs (lncRNAs), with an estimated 200 nucleotides, are a group of conserved RNAs that act as regulators in many human diseases, such as immune-related diseases, cancers, and metabolic diseases.6–8 Furthermore, lncRNAs have been reported to play a role in the ox-LDL-induced cell processes associated with human diseases.9,10 Also, lncRNA OIP5-AS1 promotes the apoptosis of ox-LDL-induced vascular endothelial cells by regulating GSK-3β,11 meaning that OIP5-AS1 is related to the functional mechanism of ox-LDL. However, the role of OIP5-AS1 in ox-LDL-induced macrophages is unclear.
MicroRNAs (miRNAs), a type of small non-coding RNAs, have about 20 nucleotides and play pivotal roles in many cell processes, including proliferation, mobility, apoptosis, and autophagy.12–14 As a miRNA, miR-128-3p has been reported to be involved in the development of a variety of diseases. For example, Zhao et al. revealed that miR-128-3p modulates the injury of doxorubicin-treated liver cells through the regulation of sirtuin-1.15 Chen et al. reported that miR-128-3p repressed tumor growth in anaplastic thyroid cancer.16 Liu et al. confirmed that miR-128-3p affected chemo-resistance by modulating Bmi1 and MRP5 in colorectal cancer cells.17 These data suggest that miR-128-3p plays an important role in human diseases. However, the analysis of the role of miR-128-3p in AS is rare.
Cyclin-dependent kinase inhibitor 2A (CDKN2A) has been identified as a cell cycle regulator and is related to the modulation of platelets.18,19 The location of CDKN2A was found to be close to chromosome 9p21 (chr9p21). Motterle et al. indicated that chr9p21 variation affected the level of CDKN2A through regulating lncRNA ANRIL expression, which modulates the proliferation of vascular smooth muscle cells and enhances AS.18,20 Therefore, CDKN2A is clearly related to the development of AS. However, whether CDKN2A mediates ox-LDL-induced cell progression in AS is unknown.
The aim of our present study was to analyze the mechanism of OIP5-AS1 in ox-LDL-induced cells. Firstly, we investigated the levels of OIP5-AS1 and explored the function OIP5-AS1 through its downregulation. Furthermore, the downstream genes of OIP5-AS1 were predicted, and then confirmed. Finally, the roles of miR-128-3p and CDKN2A in OIP5-AS1-regulated cell progression were analyzed.
2. Materials and methods
2.1. Cell culture
Tohoku Hospital Pediatrics-1 (THP1) and Human Embryonic Kidney 293T (HEK293T) cells were provided by the American Type Culture Collection (ATCC, Manassas, VA, USA) and grown in DMEM (Dulbecco's modified Eagle medium) containing 10% fetal bovine serum (FBS; Gibco, Gran Island, NY, USA) and 100 U mL−1 penicillin/streptomycin (Invitrogen; Carlsbad, CA, USA) at 37 °C with 5% CO2. 0, 15, 25, or 50 μg mL−1 ox-LDL (Shanghai Yeasen BioTechnologies Co., Ltd., Shanghai, China) was applied to treat THP1 cells. THP1 cells were incubated with 25 μg mL−1 ox-LDL for 0 h, 12 h, 24 h, or 48 h.2.2. RNA extraction and quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA was obtained from THP-1 cells by the use of TRIzol reagent (Invitrogen). The reverse transcription reaction was carried out for the synthesis of cDNA using the SuperScript First-Stand Synthesis system (Invitrogen) based on the recommended instructions. Next, qRT-PCR was performed using the SYBR Green I detection kit (Donghuan Biotech, Shanghai, China). U6 and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) were selected for normalization. The 2−ΔΔCt method was applied for the analysis of gene expression. The primers against OIP5-AS1, miR-128-3p, CDKN2A, U6, and GAPDH were: OIP5-AS1 (forward (F), 5′-GGTCGTGAAACACCGTCG-3′; reverse (R), 5′-GTGGGGCATCCAGGGT-3′), miR-128-3p (F, 5′-GACTGCCGAGCGAGCG-3′; R, 5′-GACGCCGAGGCACTCTCTCCT-3′), CDKN2A (F, 5′-TGGTCACTGTGAGGATTCAGC-3′; R, 5′-TCGCACGAACTTCACCAAGA-3′), U6 (F, 5′-TGCGGGTGCTCGCTTCGGCAGC-3′; R, 5′-CCAGTGCAGGGTCCGAGGT-3′), and GAPDH (F, 5′-ATTCCATGGCACCGTCAAGGCTGA-3′; R, 5′-TTCTCCATGGTGGTGAAGACGCCA-3′).2.3. Plasmid and transfection
Si-OIP5-AS1, miR-128-3p mimics, miR-128-3p inhibitor, and negative controls were obtained from Ribobio (Shanghai, China). The sequences of MALAT1 and TRIM65 were cloned into the pcDNA3.1 plasmid (Genepharma, Shanghai, China) to generate MALAT1 and TRIM65 overexpression vectors. Lipofectamine 2000 (Invitrogen) was employed to perform the transfection assay in line with the user's manual.2.4. Cell proliferation assay
The 3-(4,5)-dimethylthiahiazo(-z-y1)-3,5-diphenytetrazoliumromide (MTT) kit was employed to examine cell proliferation according to the user's manual (Promega, Madison, WI, USA). Briefly, THP1 cells were treated with ox-LDL and transfected with the construct used in this assay, and then cultured for 24, 48, or 72 h. Subsequently, 20 μL of MTT solution (5 mg mL−1) was added to the cells (1 × 104). After 4 h of incubation, 200 μL dimethyl sulfoxide was applied to dissolve the formazan product. Finally, the absorbance was examined using an ELISA plate reader (Bio-Rad, Hercules, CA, USA) at 570 nm.2.5. Cell apoptosis assay
Cell apoptosis was measured using an Annexin V-FITC/propidium iodide (PI) Apoptosis Detection Kit (Vazyme Biotech, Nanjing, China) based on the manufacturer's protocol. In brief, THP1 cells (1 × 105) were harvested, washed with PBS, and stained using Annexin V-FITC and PI. Finally, the analysis of cell apoptosis was carried out using a flow cytometer (BD Biosciences, San Jose, CA, USA).2.6. Western blot assay
Briefly, proteins were obtained from THP1 cells treated with ox-LDL or transfected with the construct used in this research using RIPA buffer. Then, the same amount of protein was subjected to 10% dodecyl sulfate, sodium salt-polyacrylamide gel electrophoresis (SDS-PAGE), transferred onto a polyvinyl difluoride membrane (PVDF), and blocked by phosphate buffered solution (PBST) containing 5% non-fat skimmed milk. Next, the membranes were incubated with primary antibodies against CDKN2A, B-cell lymphoma 2 (Bcl-2), BCL2 associated X (Bax), cleaved caspase3 (C-caspase3), cleaved poly ADP-ribose polymerase (C-PARP), or GAPDH (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000; Abcam, Cambridge, MA, USA), and were then incubated with relative secondary antibodies (1
1000; Abcam, Cambridge, MA, USA), and were then incubated with relative secondary antibodies (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 400; Abcam). Finally, a chemiluminescence reagent (Beyotime, Haimen, China) was applied to visualize the protein bands.
400; Abcam). Finally, a chemiluminescence reagent (Beyotime, Haimen, China) was applied to visualize the protein bands.
2.7. Enzyme-linked immunosorbent assay (ELISA)
Interleukin (IL)-6, IL-1β, and tumor necrosis factor (TNF)-α levels were examined using ELISA kits (R&D Systems, Minneapolis, MN, USA) according to the recommended protocol.2.8. ROS, SOD, MDA measurements
The contents of reactive oxygen species (ROS), superoxide dismutase (SOD), and malonic dialdehyde (MDA) were detected using ROS Assay Kit (Beyotime), SOD Activity Assay Kit (Beyotime), and Lipid Peroxidation MDA Assay Kit (Beyotime), respectively. Each assay was performed based on the user's manual.2.9. The dual-luciferase reporter assay
Firstly, OIP5-AS1 wide-type (WT)/mutant (MUT) and CDKN2A 3′untranslated region (UTR) WT/MUT were constructed by cloning their sequences into the pGL3 vector (Promega). Then, each of them was co-transfected into HEK293T cells with miR-128-3p mimics or NC mimics. After 48 h of incubation, the dual-luciferase reporter assay kit (Donghuan Biotech) was used to determine luciferase activity.2.10. RNA immunoprecipitation (RIP) assay
The Magna RNA immunoprecipitation kit (Millipore, Billerica, MA, USA) was applied to perform the RIP assay in line with the user's protocol. Briefly, THP1 cells were lysed with RIP buffer, and then incubated with magnetic beads coated with anti-Argonaute2 (AGO2) or anti-Immunoglobulin G (IgG) antibodies. Subsequently, immunoprecipitated RNAs were analyzed using the qRT-PCR assay.2.11. Statistical analysis
All data were presented as mean ± standard deviation (SD) from at least three independent measurements. The student's t-test was used for data analysis. P < 0.05 was considered to be a significant difference.3. Results
3.1. OIP5-AS1 level is upregulated by the treatment of ox-LDL
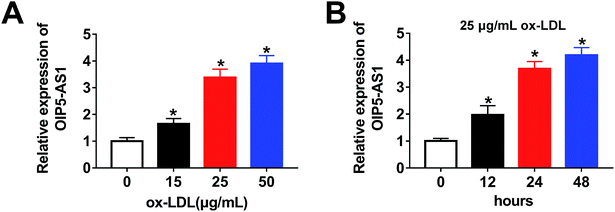
Firstly, to investigate the expression level of OIP5-AS1 in ox-LDL-induced cells, a qRT-PCR assay was performed. As shown in Fig. 1A, OIP5-AS1 expression was significantly induced by ox-LDL treatment in a dose-dependent manner in THP1 cells. Moreover, we observed that the OIP5-AS1 level increased in a time-dependent manner in ox-LDL-induced cells (Fig. 1B). These results indicate that OIP5-AS1 is related to ox-LDL-induced cell development.3.2. OIP5-AS1 knockdown reverses the effect of ox-LDL on cell progression
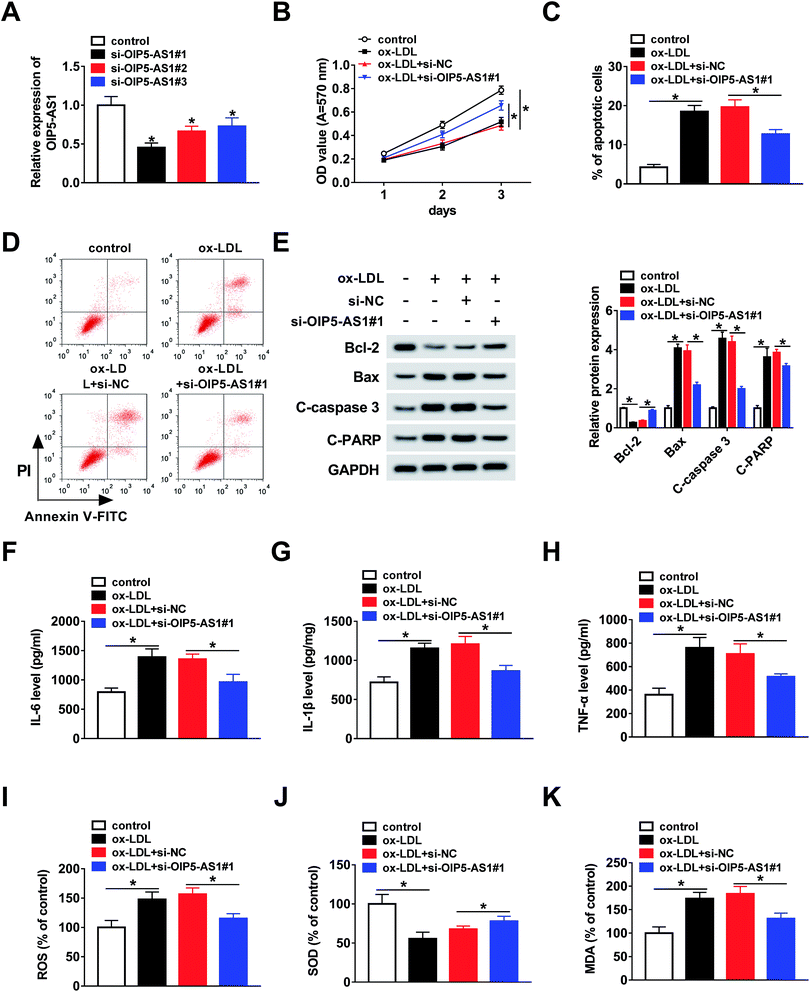
To further explore whether OIP5-AS1 regulates the growth of ox-LDL-induced cells, THP1 cells were transfected with si-OIP5-AS1 (#1, #2, and #3). The QRT-PCR assay suggested that OIP5-AS1 expression was dramatically downregulated by the transfection with si-OIP5-AS1 (#1, #2, and #3), especially si-OIP5-AS1#1 (Fig. 2A). Then, the MTT assay was employed to assess cell proliferation ability. The results confirmed that OIP5-AS1 knockdown remarkably promoted cell proliferation suppressed by ox-LDL (Fig. 2B). Moreover, we found that ox-LDL treatment induced cell apoptosis of THP1 cells, whereas this action was weakened by the downregulation of OIP5-AS1 (Fig. 2C and D). Meanwhile, the levels of cell apoptosis-related proteins, including Bcl-2, Bax, C-caspase3, and C-PARP, were measured using western blot assay. As indicated in Fig. 2E, OIP5-AS1 depletion reversed the effect of ox-LDL on these protein levels. Subsequently, ELISA was carried out to determine the levels of IL-6, IL-1β, and TNF-α. The data suggested that their levels were upregulated by ox-LDL treatment, and then partly recovered by OIP5-AS1 knockdown (Fig. 2F–H). Furthermore, the negative effect of OIP5-AS1 depletion on the amounts of ox-LDL-regulated ROS, SOD, and MDA was observed in THP1 cells (Fig. 2I–K). These results indicate that OIP5-AS1 plays a crucial role in ox-LDL-regulated cell progression.3.3. MiR-128-3p is a target of OIP5-AS1
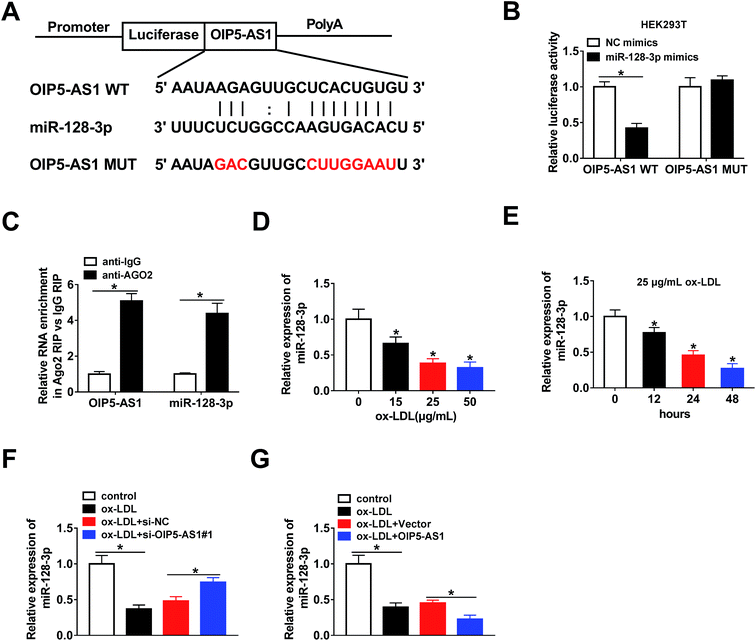
The bioinformatics tool StarBase predicted that miR-128-3p is a potential target gene of OIP5-AS1 (Fig. 3A). Then, OIP5-AS1 WT or OIP5-AS1 MUT and miR-128-3p mimics or NC mimics were co-transfected into HEK293T cells to perform the dual-luciferase reporter assay. The analysis of luciferase activity demonstrated that the miR-128-3p mimics significantly reduced the reporter activity of OIP5-AS1 WT, whereas they did not affect the reporter activity of OIP5-AS1 MUT (Fig. 3B). Thus, OIP5-AS1 interacted with miR-128-3p. Moreover, this interaction was confirmed by the RIP assay (Fig. 3C). Subsequently, whether ox-LDL regulates miR-128-3p expression was determined in THP1 cells. As shown in Fig. 3D and E, ox-LDL dramatically downregulated the expression level of miR-128-3p in a dose-dependent manner and a time-dependent manner. Finally, si-OIP5-AS1 or OIP5-AS1 was transfected into THP1 cells to investigate the effect of OIP5-AS1 on miR-128-3p expression. As expected, the miR-128-3p level inhibited by ox-LDL was upregulated by OIP5-AS1 knockdown and downregulated by OIP5-AS1 overexpression (Fig. 3F and G). These data confirmed that OIP5-AS1 negatively modulates miR-128-3p expression through interaction.3.4. OIP5-AS1 regulates ox-LDL-induced cell progression through modulating miR-128-3p expression
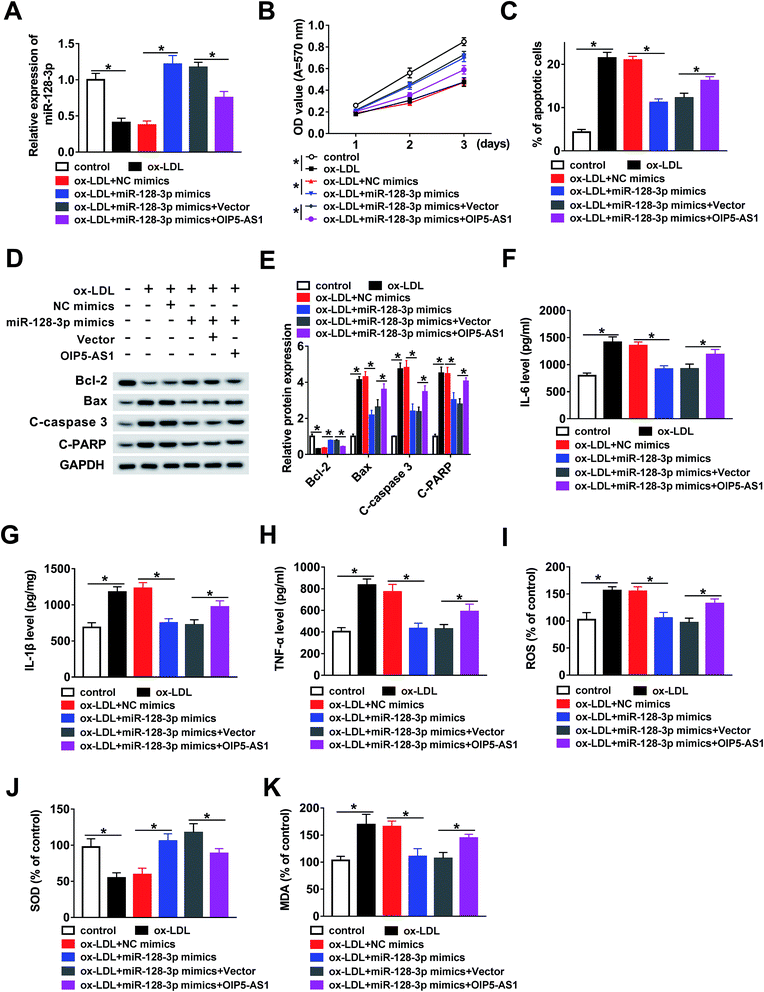
In this study, we hypothesized that OIP5-AS1 mediates miR-128-3p expression to regulate the growth of THP1 cells treated with ox-LDL. To verify this hypothesis, ox-LDL-induced THP1 cells were transfected with NC mimics, miR-128-3p mimics, miR-128-3p mimics + vector, or miR-128-3p mimics + OIP5-AS1. Then, the qRT-PCR assay was employed to determine miR-128-3p expression. As demonstrated in Fig. 4A, the ox-LDL-downregulated miR-128-3p level was upregulated due to the overexpression of miR-128-3p, and then this change was weakened by OIP5-AS1 upregulation. Subsequently, the MTT assay was conducted to examine cell proliferation ability. The results suggest that cell proliferation repressed by ox-LDL treatment was significantly promoted by miR-128-3p overexpression, and then suppressed by the upregulation of OIP5-AS1 (Fig. 4B). Flow cytometry analysis indicated that ox-LDL-induced cell apoptosis was inhibited by miR-128-3p upregulation, and then partly recovered due to OIP5-AS1 overexpression (Fig. 4C). Also, a similar trend for cell apoptosis-related protein levels was observed in THP1 cells (Fig. 4D and E). Furthermore, we measured the levels of IL-6, IL-1β, and TNF-α, and found that OIP5-AS1 overexpression reversed the effect of miR-128-3p upregulation on these levels (Fig. 4F–H). Furthermore, the negative effect of OIP5-AS1 upregulation on miR-128-3p overexpression regulated the amounts of ROS, SOD, and MDA in ox-LDL-induced THP1 cells (Fig. 4I–K). Taken together, we can conclude that OIP5-AS1 mediates ox-LDL-induced THP1 cell progression by regulating miR-128-3p expression.3.5. MiR-128-3p downregulates CDKN2A expression via interaction
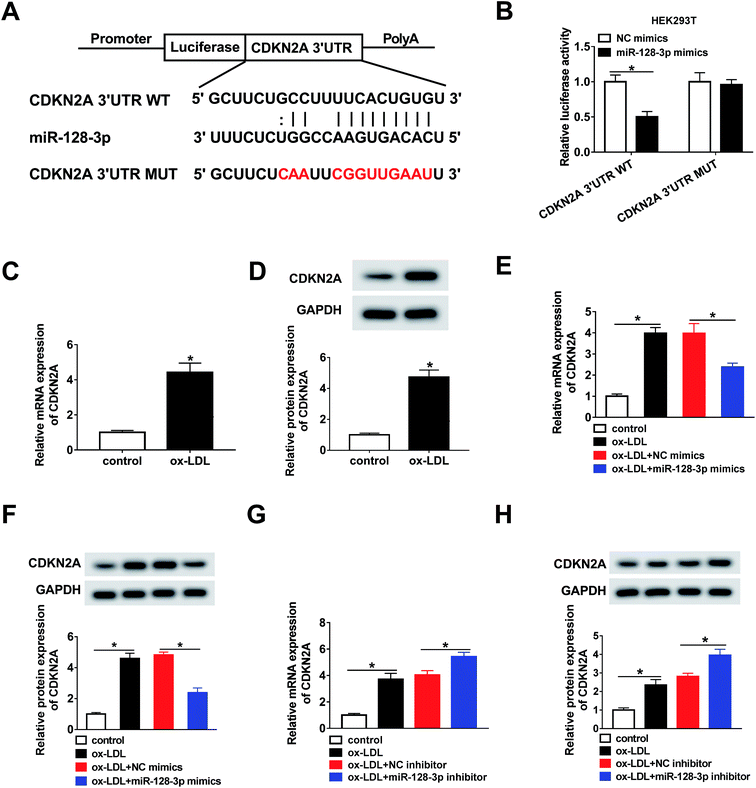
Next, the targets of miR-128-3p were predicted using the bioinformatics tool StarBase. The results demonstrated that CDKN2A possesses a complementary sequence to that of miR-128-3p (Fig. 5A). Then, the dual-luciferase reporter assay was carried out to confirm the interaction between miR-128-3p and CDKN2A. As shown in Fig. 5B, the luciferase activity of CDKN2A 3′UTR WT, but not CDKN2A 3′UTR MUT, was reduced by the miR-128-3p mimics, revealing that miR-128-3p interacted with CDKN2A. Furthermore, we explored the effect of ox-LDL on CDKN2A expression, and found that the CDKN2A level was strongly upregulated by ox-LDL treatment (Fig. 5C and D). Furthermore, we investigated whether miR-128-3p modulated CDKN2A expression. From Fig. 5E–H, we found that CDKN2A expression promoted by ox-LDL was remarkably downregulated by miR-128-3p overexpression and upregulated by miR-128-3p knockdown. Thus, miR-128-3p reduces CDKN2A levels through targeting 3′UTR of CDKN2A.3.6. MiR-128-3p mediates CDKN2A level to affect cell progression
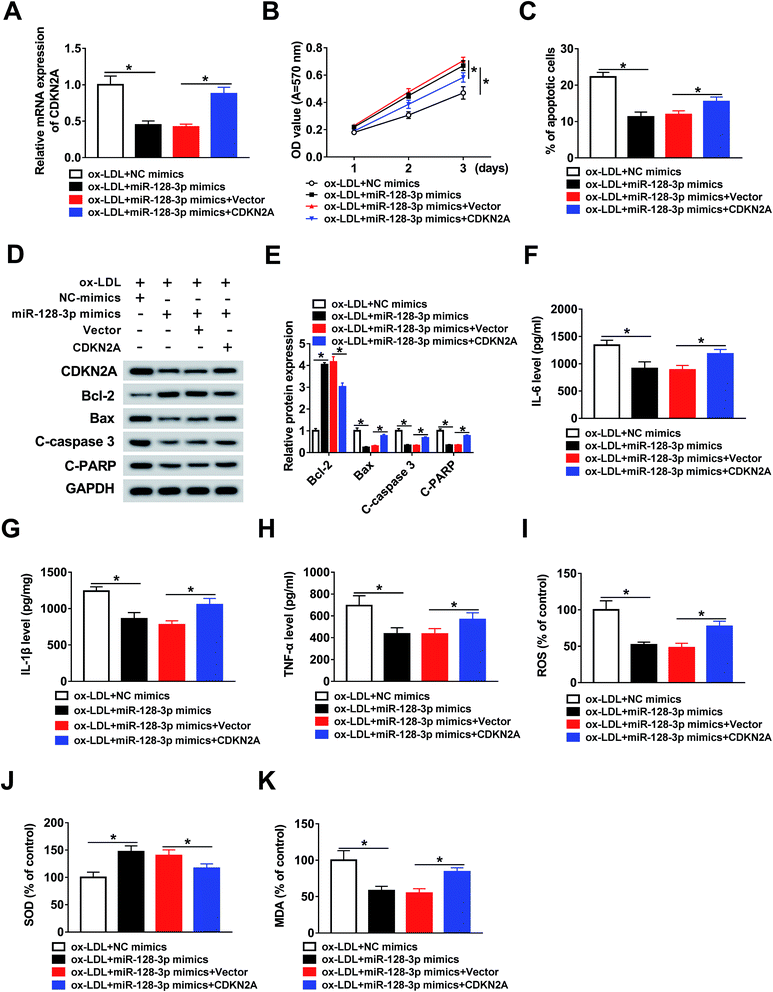
To investigate the function of CDKN2A in miR-128-3p-regulated cell progression. THP1 cells were treated with ox-LDL and transfected with NC mimics, miR-128-3p mimics, miR-128-3p mimics + vector, or miR-128-3p mimics + CDKN2A. Then, the qRT-PCR assay was performed to determine the expression of CDKN2A. As indicated in Fig. 6A, CDKN2A expression was downregulated by miR-128-3p overexpression, and then upregulated by CDKN2A overexpression. Subsequently, the MTT assay was carried out to assess cell proliferation ability. The results suggest that miR-128-3p overexpression significantly promoted the proliferation of ox-LDL-induced THP1 cells, whereas this action was reversed by CDKN2A upregulation (Fig. 6B). Next, the cell apoptosis rate was examined using flow cytometry analysis. From Fig. 6C, we concluded that cell apoptosis was inhibited due to the increased miR-128-3p level, and was subsequently partly restored by CDKN2A overexpression. Meanwhile, the negative effect of CDKN2A upregulation on the change of miR-128-3p overexpression-regulated cell apoptosis-related protein levels was discovered in ox-LDL-treated THP1 cells (Fig. 6D and E). Subsequently, ELISA was employed to measure the levels of IL-6, IL-1β, and TNF-α. The results revealed that the overexpression of CDKN2A increased the levels of these compounds, which were downregulated by miR-128-3p upregulation (Fig. 6F–H). Furthermore, we found that the increase of CDKN2A weakened the effect of miR-128-3p overexpression on the amounts of ROS, SOD, and MDA in ox-LDL-treated THP1 cells (Fig. 6I–K). Taken together, we can conclude that miR-128-3p mediates the growth of ox-LDL-induced cells by modulating CDKN2A expression.3.7. OIP5-AS1 inhibits miR-128-3p expression to increase CDKN2A level
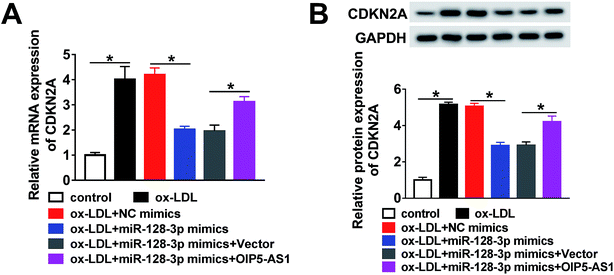
Based on the above results, it was speculated that OIP5-AS1 repressed miR-128-3p expression to upregulate the CDKN2A level in ox-LDL-induced cells. To test this hypothesis, ox-LDL-induced THP1 cells were transfected with NC mimics, miR-128-3p mimics, miR-128-3p mimics + vector, or miR-128-3p mimics + OIP5-AS1. Subsequently, the qRT-PCR assay and western blot assay were carried out to detect the expression levels of CDKN2A. The results revealed that CDKN2A expression was inhibited by miR-128-3p overexpression, and then partly recovered by OIP5-AS1 upregulation (Fig. 7A and B). Therefore, OIP5-AS1 positively regulates the CDKN2A level by inhibiting miR-128-3p expression.4. Discussion
In recent years, lncRNAs have been reported to be involved in the development of AS.21,22 For instance, Yao et al. reported that lncRNA ENST00113 positively regulated cell growth and survival by mediating the PI3K/Akt/mTOR axis in AS.23 Zhang et al. confirmed that lncRNA TUG1 depletion promotes AS by regulation of the miR-133a level.24 Pan demonstrated that lncRNA H19 enhances AS development through modulating the MAPK pathway.25 These studies suggest that lncRNAs play essential roles in AS development. Therefore, it is of urgency to explore the functional mechanisms of lncRNAs for the treatment of AS.In the present study, we found that OIP5-AS1 expression was upregulated by the treatment of ox-LDL in a dose-dependent manner and time-dependent manner. Furthermore, OIP5-AS1 knockdown reversed the effect of ox-LDL on THP1 cell proliferation, apoptosis, inflammation, and oxidative stress, confirming that ox-LDL regulates AS development through regulating OIP5-AS1. Previous evidence suggests that OIP5-AS1 expression is increased in ox-LDL-induced cells and OIP5-AS1 acts as a positive regulator to modulate cell growth in ox-LDL-regulated cell progression.11 These data are consistent with our results. This evidence confirms that OIP5-AS1 plays a positive role in the progression of AS, also OIP5-AS1 has been reported to promote the development of many other human diseases, such as hemangioma,26 melanoma,27 and osteosarcoma.28 Importantly, our work shows that ox-LDL activates OIP5-AS1 expression to regulate cell progression.
LncRNAs have been considered as a kind of miRNA sponges that mediate downstream gene expression by binding to targets.29 We used the bioinformatics tool StarBase to predict the potential target genes of OIP5-AS1, and found that miR-128-3p likely interacts with OIP5-AS1. Next, this interaction was confirmed by the dual-luciferase reporter assay and RIP assay. Moreover, we analyzed the effect of OIP5-AS1 on miR-128-3p expression. As expected, we found that OIP5-AS1 negatively regulates miR-128-3p expression. Furthermore, a decreased miR-128-3p level was observed in ox-LDL-induced cells. The present evidence suggests that miR-128-3p is a tumor suppressor in human diseases, including glioma,30 esophageal squamous-cell cancer,31 rheumatoid arthritis,32 and hepatocellular carcinoma.33 Moreover, miR-128 knockdown regulated by lncRNA NEAT1 leads to ox-LDL-regulated inflammation and oxidative stress.34 The results in our study are in agreement with the previously reported data. Based on the above results, it was speculated that OIP5-AS1 regulates miR-128-3p expression to modulate ox-LDL-induced cell progression. Subsequently, this hypothesis was verified by our assay, which demonstrated that OIP5-AS1 upregulation weakens the effect of miR-128-3p overexpression on the growth of ox-LDL-induced cells.
Accumulating evidence demonstrates that miRNAs play roles as mRNA sponges to regulate gene expression through targeting 3′UTR of target mRNA.35 Next, the bioinformatics tool StarBase was used to predict the potential targets of miR-128-3p. CDKN2A was found to be a gene that possesses a complementary sequence to that of miR-128-3p. Then, the interaction between miR-128-3p and CDKN2A was confirmed by the dual-luciferase reporter assay. Meanwhile, we confirmed that miR-128-3p downregulates the level of CDKN2A. Furthermore, CDKN2A expression in ox-LDL-induced cells was found to be higher than that in the control cells. These data are in agreement with previous reports.36 Our results also indicate that miR-128-3p regulates the growth of ox-LDL-induced cells through modulation of CDKN2A expression. Finally, the mechanism by which OIP5-AS1 represses miR-128-3p expression to upregulate the CDKN2A level was elucidated in this research.
5. Conclusion
In conclusion, we suggested that OIP5-AS1 knockdown weakens the effect of ox-LDL on cell proliferation, apoptosis, inflammation, and oxidative stress through modulating the miR-128-3p/CDKN2A axis, providing a theoretical basis for the treatment of AS.Conflicts of interest
The authors declare that they have no financial conflicts of interest.References
- J. Li, Y. Cui, A. Huang, Q. Li, W. Jia, K. Liu and X. Qi, Med. Sci. Monit., 2018, 24, 4992–4999 CrossRef CAS PubMed.
- J. H. Jang, E. A. Kim, H. J. Park, E. G. Sung, I. H. Song, J. Y. Kim, C. H. Woo, K. O. Doh, K. H. Kim and T. J. Lee, J. Cell. Mol. Med., 2017, 21, 2720–2731 CrossRef CAS PubMed.
- K. J. Moore and I. Tabas, Cell, 2011, 145, 341–355 CrossRef CAS PubMed.
- R. Ross, Nature, 1993, 362, 801–809 CrossRef CAS PubMed.
- S. Mitra, T. Goyal and J. L. Mehta, Cardiovasc. Drugs Ther., 2011, 25, 419–429 CrossRef CAS PubMed.
- O. M. Rogoyski, J. I. Pueyo, J. P. Couso and S. F. Newbury, Biochem. Soc. Trans., 2017, 45, 895–904 CrossRef CAS PubMed.
- Y. G. Chen, A. T. Satpathy and H. Y. Chang, Nat. Immunol., 2017, 18, 962–972 CrossRef CAS PubMed.
- M. Giroud and M. Scheideler, Int. J. Mol. Sci., 2017, 18 DOI:10.3390/ijms18122578.
- L. Cao, Z. Zhang, Y. Li, P. Zhao and Y. Chen, Int. Immunopharmacol., 2019, 72, 496–503 CrossRef CAS PubMed.
- X. Xu, C. Ma, Z. Duan, Y. Du and C. Liu, Front. Pharmacol., 2019, 10, 397 CrossRef PubMed.
- M. Wang, Y. Liu, C. Li, Y. Zhang, X. Zhou and C. Lu, Am. J. Transl. Res., 2019, 11, 1827–1834 Search PubMed.
- L. L. Christensen, A. Holm, J. Rantala, O. Kallioniemi, M. H. Rasmussen, M. S. Ostenfeld, F. Dagnaes-Hansen, B. Oster, T. Schepeler, H. Tobiasen, K. Thorsen, O. M. Sieber, P. Gibbs, P. Lamy and T. F. Hansen, et al., PLoS One, 2014, 9, e96767 CrossRef PubMed.
- H. N. Luu, H.-Y. Lin, K. D. Sørensen, O. O. Ogunwobi, N. Kumar, G. Chornokur, C. Phelan, D. Jones, L. Kidd, J. Batra, K. Yamoah, A. Berglund, R. J. Rounbehler, M. Yang and S. H. Lee, et al., BMC Urol., 2017, 17, 18 CrossRef PubMed.
- L. Chen, Y. Zhou, Q. Sun, J. Zhou, H. Pan and X. Sui, Int. Rev. Cell Mol. Biol., 2017, 334, 1–26 CrossRef PubMed.
- X. Zhao, Y. Jin, L. Li, L. Xu, Z. Tang, Y. Qi, L. Yin and J. Peng, Pharmacol. Res., 2019, 146, 104276 CrossRef CAS PubMed.
- J. Chen, D. Zhao and Q. Meng, Biomed. Pharmacother., 2019, 116, 108966 CrossRef CAS PubMed.
- T. Liu, X. Zhang, L. Du, Y. Wang, X. Liu, H. Tian, L. Wang, P. Li, Y. Zhao, W. Duan, Y. Xie, Z. Sun and C. Wang, Mol. Cancer, 2019, 18, 43 CrossRef PubMed.
- S. Zhou, Y. Zhang, L. Wang, Z. Zhang, B. Cai, K. Liu, H. Zhang, M. Dai, L. Sun, X. Xu, H. Cai, X. Liu, G. Lu and G. Xu, J. Transl. Med., 2016, 14, 333 CrossRef PubMed.
- C. Gieger, A. Radhakrishnan, A. Cvejic, W. Tang, E. Porcu, G. Pistis, J. Serbanovic-Canic, U. Elling, A. H. Goodall, Y. Labrune, L. M. Lopez, R. Magi, S. Meacham, Y. Okada and N. Pirastu, et al., Nature, 2011, 480, 201–208 CrossRef CAS PubMed.
- Y. Kong, C. H. Hsieh and L. C. Alonso, Front. Endocrinol., 2018, 9, 405 CrossRef PubMed.
- I. Fernandez-Ruiz, Nat. Rev. Cardiol., 2018, 15, 195 CrossRef PubMed.
- C. H. Wang, H. H. Shi, L. H. Chen, X. L. Li, G. L. Cao and X. F. Hu, Front. Genet., 2019, 10, 123 CrossRef CAS PubMed.
- X. Yao, C. Yan, L. Zhang, Y. Li and Q. Wan, Medicine, 2018, 97, e0473 CrossRef CAS PubMed.
- L. Zhang, H. Cheng, Y. Yue, S. Li, D. Zhang and R. He, Cardiovasc. Pathol., 2018, 33, 6–15 CrossRef CAS PubMed.
- J. X. Pan, Eur. Rev. Med. Pharmacol. Sci., 2017, 21, 322–328 Search PubMed.
- J. Zhang, T. Zhao, L. Tian and Y. Li, Front. Pharmacol., 2019, 10, 449 CrossRef PubMed.
- W. Luan, X. Zhang, H. Ruan, J. Wang and X. Bu, J. Cell. Physiol., 2019 DOI:10.1002/jcp.28335.
- L. Song, Z. Zhou, Y. Gan, P. Li, Y. Xu, Z. Zhang, F. Luo, J. Xu, Q. Zhou and F. Dai, J. Cell. Biochem., 2019, 120, 9656–9666 CrossRef CAS PubMed.
- E. Lopez-Urrutia, L. P. Bustamante Montes, D. Ladron de Guevara Cervantes, C. Perez-Plasencia and A. D. Campos-Parra, Front. Oncol., 2019, 9, 669 CrossRef PubMed.
- L. Huo, B. Wang, M. Zheng, Y. Zhang, J. Xu, G. Yang and Q. Guan, Exp. Ther. Med., 2019, 17, 2921–2930 CAS.
- L. Zhao, R. Li, S. Xu, Y. Li, P. Zhao, W. Dong, Z. Liu, Q. Zhao and B. Tan, Acta Biochim. Biophys. Sin., 2018, 50, 171–180 CrossRef CAS PubMed.
- Z. Xia, F. Meng, Y. Liu, Y. Fang, X. Wu, C. Zhang, D. Liu and G. Li, Biosci. Rep., 2018, 38 DOI:10.1042/BSR20180540.
- C. Y. Huang, X. P. Huang, J. Y. Zhu, Z. G. Chen, X. J. Li, X. H. Zhang, S. Huang, J. B. He, F. Lian, Y. N. Zhao and G. B. Wu, Oncol. Rep., 2015, 33, 2889–2898 CrossRef CAS PubMed.
- D.-D. Chen, L.-L. Hui, X.-C. Zhang and Q. Chang, J. Cell. Biochem., 2019, 120, 2493–2501 CrossRef CAS PubMed.
- J. G. Doench and P. A. Sharp, Genes Dev., 2004, 18, 504–511 CrossRef CAS PubMed.
- L. Yan, Z. Liu, H. Yin, Z. Guo and Q. Luo, Cell Biol. Int., 2019, 43, 409–420 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2019 |