Open Access Article
Open Access ArticleDetection of carboxylesterase by a novel hydrosoluble near-infrared fluorescence probe†
Mengyao Li ac,
Chen Zhai*a,
Shuya Wanga,
Weixia Huanga,
Yunguo Liuc and
Zhao Li
ac,
Chen Zhai*a,
Shuya Wanga,
Weixia Huanga,
Yunguo Liuc and
Zhao Li *b
*b
aNutrition & Health Research Institute, COFCO Corporation, Beijing Key Laboratory of Nutrition & Health and Food Safety, Beijing 102209, China. E-mail: zhaichen@cofco.com
bShaanxi Engineering Laboratory for Food Green Processing and Safety Control, College of Food Engineering and Nutritional Science, Shaanxi Normal University, Xi'an 710062, China
cCollege of Life Science and Technology, Xinjiang University, Urumqi 830002, China
First published on 9th December 2019
Abstract
A novel hydrosoluble near-infrared fluorescence off–on probe has been developed for detecting carboxylesterase activity. The probe was designed by introducing (4-acetoxybenzyl)oxy as a quenching and recognizing moiety to the decomposed product of IR-783, which exhibits excellent near-infrared fluorescence features and good water solubility. The responding mechanism of novel probe 1 to carboxylesterase was investigated. It would lead to the cleavage of the carboxylic ester bond by carboxylesterase catalyze the spontaneous hydrolysis of the probe, resulting in the release of a near-infrared fluorophore. This behaviour leads to the development of a simple and sensitive fluorescent method for assaying carboxylesterase activity, with a detection limit of 3.4 × 10−3 U mL−1. Moreover, the probe displays excellent selectivity toward carboxylesterase over other substances. Notably, the imaging experimental results showed that the probe 1 is cell membrane permeable, and its applicability has been demonstrated for monitoring carboxylesterase activity in HeLa cells.
Introduction
Carboxylesterases are a family of hydrolases, which can catalyze the hydrolysis of carboxylic esters to generate acids and alcohols.1–4 They are widely found in flora and fauna. The enzymes play a crucial role in drug targets and prodrug activators,5–10 and have received much attention in recent decades because of their human health importance.11–14 Therefore, detection of carboxylesterases in living biosystems is of great significance for better understanding their biological functions as well as the evaluation of therapeutic drugs.In recent years, an assortment of methods including immunology, chromatography, chemiluminescence, mass spectrometry and fluorescence, have been reported for carboxylesterase detection.15–22 Among them, fluorescent probes for detecting carboxylesterase have become more and more attractive because of their advantages such as high sensitivity, selectivity and high temporal-spatial resolution.23–26 For example, Zhou et al. reported a new lysosome-targeted fluorescence probe with emission 575 nm for fluorescence sensing of carboxylesterase in live cells, sera and tissues.27 As for in vivo imaging studies, near-infrared (NIR) fluorescent probes are more desired because they have excellent tissue penetration, low biological and autofluorescence damage.28–32 As far as we know, most of the current probes have a severe limitation on application for imaging in living systems, since they have a short excitation (usually <500 nm) and emission wavelength (usually <630 nm) and there are limited reports on NIR fluorescent probes for carboxylesterase detection in recent years. For instance, Yang et al. designed a NIR fluorescent probe which has been used to detect carboxylesterases in vitro and in vivo.33 Li et al. prepared a NIR fluorescence off–on probe for detection and imaging of carboxylesterase in living HepG-2 cells and zebrafish pretreated with pesticides.34 To the best of our knowledge, only two NIR fluorescent probe for carboxylesterase have been reported so far. Hence, NIR fluorescent probes are still necessary for carboxylesterase assay. In particular, the development of readily accessible and water-soluble fluorescent probes with improved detection selectivity and sensitivity remains challenging and desirable for imaging in vivo.
In this paper, we successfully designed and synthesized a hydrosoluble, selective, and sensitive NIR fluorescence probe, (E)-2-(2-(6-(4-acetoxybenzyloxy)-2,3-dihydro-1H-xanthen-4-yl)vinyl)-3,3-dimethyl-1-(4-sulfobutyl)-3H-indolium (probe 1), for the detection of carboxylesterase activity. We selected the decomposed product of the unstable precursor of IR-783 as fluorophore, which shows excellent near-infrared spectroscopic feature and good water solubility due to the existence of sulfonic acid group.35 In addition, (4-acetoxybenzyl)oxy is identified as a quenching and recognizing moiety due to the specificity and high sensitivity to carboxylesterase.36 The presence of carboxylesterase will cut off the bonds that connect fluorophore with recognize moiety, resulting in the release of the fluorophore decomposed product of IR-783, which has reached the purpose of detecting carboxylesterase (Scheme 1). The decomposed product of the unstable precursor cyanine dyes not only exhibits high stability but also preserves the characteristics of near-infrared fluorescence excitation and emission of cyanine dyes. Furthermore, the probe 1 indeed exhibits excellent biological properties, and high fluorescence quantum yield, which applies to the detection of endogenous carboxylesterase in living cells. As the starting point of this work, the probe 1 was synthesized through two steps as illustrated in Scheme 1. Detailed synthetic steps and characterization of probe 1 were described in ESI.† The probe 1 was characterized by NMR and ESI-MS (Fig. S1–S3†).
Experimental section
Materials and chemicals
Electrospray ionization mass spectrum (ESI-MS) was recorded in positive mode with a Shimadzu LC-MS 2010A instrument (Kyoto, Japan). 1H NMR and 13C NMR spectra were measured on Brucker DMX-600 spectrometer in CD3OD. Absorption spectra were measured with a TU-1900 spectrophotometer (Beijing, China) in 1 cm quartz cells. Fluorescence spectra were collected on a Hitachi F-4600 spectrofluorimeter with both excitation and emission slit widths of 10 nm. The absorbance for MTT analysis was recorded on a microplate reader (BIO-TEK Synergy HT, USA). Fluorescence imaging was made on a TCS SP5 confocal laser scanning microscope (Leica, Germany) with excitation at 635 nm.IR-783 iodide, resorcinol, 4-(2-aminoethyl)benzenesulfonyl fluoride hydrochloride (AEBSF), 4-(chloromethyl)phenyl acetate, and carboxylesterase were purchased from Sigma-Aldrich. A phosphate buffered saline solution was obtained from Invitrogen Company. Dulbecco's modified eagle media (DMEM), fetal bovine serum, penicillin and streptomycin were obtained from Invitrogen Corporation. 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazoliumb-romide (MTT) and was obtained from Serva Electrophoresis GmbH (Germany). The water used in the experiment is Milli-Q ultrapure water, and all other reagents used are analytical pure. HeLa cells were obtained from the Experimental Animal Center of the Academy of Military Medical Sciences (Beijing, China). All animal procedures were performed in accordance with the Guidelines for Care and Use of Laboratory Animals of Beijing and approved by the Animal Ethics Committee of Beijing.
Synthesis and characterization of probe 1
First of all, the fluorophore 2 is used in the previous procedure.35 Secondly, to a solution of fluorophore 2 (253 mg, 0.5 mmol) in anhydrous DMF (10 mL), K2CO3 (103.5 mg, 0.75 mmol) was added, followed by stirring at 45 °C for 20 min under an Ar atmosphere. Then, a solution of 4-(chloromethyl)phenyl acetate (182 mg, 1.0 mmol) in DMF (5 mL) was added dropwise. The resulting mixture was stirred at 45 °C for 3 h and then diluted with dichloromethane (20 mL). The organic layer was separated, washed with water and brine, and then dried over dry Na2SO4. The solvent was removed by evaporation, and the residue was subjected to silica gel chromatography, affording a blue-green solid (probe 1).1H NMR (600 MHz, CD3OD) δ 8.75 (d, J = 14.8 Hz, 1H), 7.68–7.63 (m, 1H), 7.59 (d, J = 8.0 Hz, 1H), 7.52 (t, J = 8.5 Hz, 3H), 7.49–7.43 (m, 2H), 7.36 (s, 1H), 7.16 (d, J = 8.5 Hz, 2H), 7.08 (d, J = 2.1 Hz, 1H), 7.04 (dd, J = 8.6, 2.3 Hz, 1H), 6.56 (d, J = 14.9 Hz, 1H), 5.27 (s, 2H), 4.39 (t, J = 7.5 Hz, 2H), 2.91 (t, J = 7.2 Hz, 2H), 2.81–2.71 (m, 4H), 2.28 (s, 3H), 2.07 (d, J = 7.0 Hz, 2H), 2.00–1.90 (m, 4H), 1.82 (s, 6H).
13C NMR (151 MHz, CD3OD) δ 169.72, 162.30, 161.74, 154.52, 150.79, 145.90, 142.27, 141.73, 134.26, 133.50, 129.57, 128.88, 128.71, 128.43, 127.40, 126.97, 122.34, 121.64, 116.13, 114.63, 113.87, 112.61, 103.66, 101.30, 69.85, 50.63, 50.18, 44.55, 28.91, 28.70, 27.01, 26.14, 23.67, 22.11, 20.26, 19.52.
ESI-MS m/z: calculated for probe 1 (C38H40NO7S+, [M]+), 654.2520; found, 654.2499.
General procedure for carboxylesterase detection
The fluorescence of probe 1 (10 μM) reacting with carboxylesterase was determined in PBS buffer (pH 7.4). The stock solutions (1 mM, 50 μL) of probe 1 were dissolved in 1 mL of PBS, followed by addition of carboxylesterase and then add appropriate PBS up to 2 mL. After incubation at 37 °C for 20 min in a shaker incubator, a 2 mL portion of the reaction solution was transferred to a 1 cm quartz cell to measure the absorbance or fluorescence with λex/em = 670/706 nm and both excitation and emission slit widths of 10 nm.Cytotoxicity assay
Standard MTT assay was performed to determine the cytotoxicity of probe 1 towards HeLa cells.37,38Fluorescence imaging of carboxylesterase in HeLa cells
HeLa cells were cultured in DMEM media, supported with 10% FBS, 100 U mL−1 penicillin, and 1% L-glutamine in a humidified 5% CO2/95% air incubator at 37 °C. Prior to imaging, the medium was removed, the control group is the cells in PBS buffer. The second group is that the cells were incubated with 10 μM probe 1 for 20 min. The third group is that the cells were pretreated with 0.5 mM AEBSF for 30 min and then incubated with 10 μM probe 1 for 20 min. The fourth group is that the cells were pretreated with 1 mM AEBSF for 30 min and then incubated with 10 μM probe 1 for 20 min. After washing with PBS solution, the imaging was performed with confocal laser scanning microscope (Leica, Germany) with 635 nm and then washed three times with the PBS buffer (pH 7.4). The pixel intensity collected from fluorescence image at least 10 cells were measured using Image J software.Results and discussion
Absorption and fluorescence studies
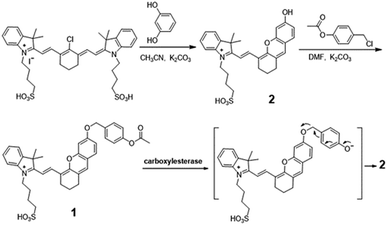
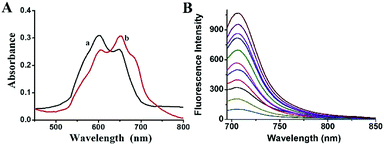
The spectroscopic evaluation of the probe 1 in the absence and presence of carboxylesterase were carried out in phosphate buffered saline (PBS, 10 mM, pH = 7.4) at 37 °C. In the presented UV-vis spectrum (Fig. 1A), the probe 1 exhibited absorption peak at 600 nm in PBS, but after reaction with carboxylesterase, the maximum absorption peak was red-shifted to 670 nm. In addition, the probe 1 had a high sensitivity to carboxylesterase results in the sharp fluorescence off-on response at 706 nm (Fig. 1B), which was attributed to the characteristic emission of fluorophore 2, the decomposed product of IR-783.35 It was worth to note that both the absorption and fluorescence spectra from the reaction system resemble those of fluorophore 2, suggesting that the carboxylesterase-triggered cleavage reaction caused the release of free fluorophore 2 (Scheme 1). The results indicated that probe 1 displayed excellent sensitivity for carboxylesterase detection in abiotic systems. The generation of fluorophore 2 was further proved by electrospray ionization mass spectral analysis (m/z 506.2 [M]+; Fig. S4†).Optimization conditions
The effects of pH, temperature and reaction time on the fluorescence intensity of the reaction system were examined. As anticipated, the changes of both pH from 5 to 9 scarcely activate the fluorescence of probe 1 itself, but do turn on that of the reaction solution of probe 1 with carboxylesterase (Fig. S5†). In addition, the kinetic curves of probe 1 reacting with varied concentrations of carboxylesterase are presented in Fig. S6.† Obviously, with the passing of time, the fluorescence intensity gradually increased and the reaction reached the platform in about 20 min. By contrast, the probe 1 (control) showed no significant fluorescence change during the same period of time, which indicated that probe 1 is very stable in the detection system. These all results clearly indicate that the reaction of probe 1 has the potential to be used to detect carboxylesterase under physiological conditions (pH 7.4 and 37 °C).Selectivity detection
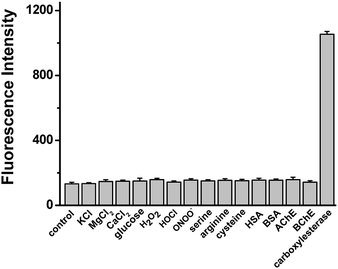
To investigate whether other substances can interfere with the detection of carboxylesterase under the physiological conditions (pH 7.4 and 37 °C), probe 1 was treated with various species commonly found in biological systems, and only carboxylesterase could trigger a fluorescence response. The reaction selectivity was assessed by parallel detection of various possible interfering substances in the same situation, such as metal ions (KCl, MgCl2, CaCl2), glucose, reactive oxygen species (H2O2, HOCl, ONOO−), amino acids (serine, arginine), biothiols (cysteine), human serum albumin (HSA), bovine serum albumin (BSA), acetylcholinesterase (AchE), and butyrylcholinesterase (BchE). The selectivity for over other relative analytes shows that the presence of other substance did not show any significant interference, displaying that probe 1 had outstanding selectivity toward carboxylesterase and these potentially interfering species show insignificant interference in the probing process between carboxylesterase and probe 1. This result indicates the high reliability in a complex biological environment (Fig. 2).Sensitivity detection
Under the optimized reaction conditions (20 min reaction time at 37 °C in pH 7.4 PBS), the fluorescence response of probe 1 to carboxylesterase at varied concentrations was investigated. The fluorescence response of probe 1 presented an excellent linearity in the concentration range of 0.01–0.3 U mL−1 with a regression equation of ΔF = 1985.9 C (U mL−1) + 160.7 (R2 = 0.991), where ΔF is the fluorescence enhancement of probe 1 at 706 nm with and without carboxylesterase. The detection limit (3S m−1, in which S is the standard deviation of blank measurements, n = 11, and m is the slope of the linear equation) is determined to be 3.4 × 10−3 U mL−1 carboxylesterase.Fluorescence imaging in cells
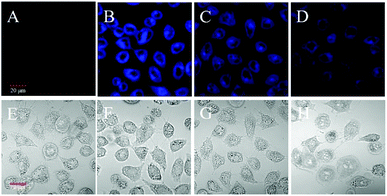
Encouraged by these results as demonstrated above, the practicability of probe 1 to detect carboxylesterase in living cells was investigated. Before fluorescence imaging, since the cytotoxicity is a critical aspect in bio-imaging, a standard MTT assay was performed to evaluate the bio-compatibility of the probe (Fig. S7†). No significant difference changed upon treatment even with 10 μM probe 1 at 37 °C for 24 h, indicating the probe had no marked cytotoxicity and good biocompatibility. There was potential application of probe 1 in live cell systems.In view of the biopermeable and low cytotoxicity of probe 1, these results inspire us to detect the carboxylesterase activity in living cells through the reaction of endogenous carboxylesterase with the probe 1 by fluorescence imaging. HeLa cells were selected to demonstrate this attempt. As shown in Fig. 3, HeLa cells themselves show neglectable intracellular background fluorescence (Fig. 3A), whereas the probe-loaded cells display a strong fluorescence (Fig. 3B). In contrast, when HeLa cells were pre-treated with 0.5 mM of AEBSF (the carboxylesterase inhibitor),39,40 a decreased fluorescence was observed (Fig. 3C). Moreover, a higher concentration of AEBSF (1.0 mM) leads to a weaker fluorescence (Fig. 3D). This clearly indicates that the fluorescence change in HeLa cells arises from the cleavage reaction of probe 1 by endogenous carboxylesterase releasing the free fluorophore 2, the decomposed product of IR-783. In other words, the probe 1 is cell membrane permeable, and can be used to detect the change of intracellular carboxylesterase activity. The relative pixel intensity measurements obtained from the images of HeLa cells were examined. As can be seen from Fig. S8†, the fluorescence intensity from the cells treated with 0.5 and 1.0 mM of AEBSF is deceased to 68% and 39% with respect to that without AEBSF (defined as 1.0), which corresponds to the inhibition of the carboxylesterase activity of 32% and 61%, respectively. The results fully demonstrate that the probe 1 can be applied to image endogenous carboxylesterase in living cells.
Conclusions
In summary, we have successfully designed and synthesized a new hydrosoluble NIR spectroscopic off–on probe for visualization of carboxylesterase. The presence of carboxylesterase will cut off the bonds that connect fluorophore with recognize moiety, resulting in the release of the fluorophore, decomposed product of IR-783. The proposed probe demonstrates excellent characteristics in terms of low toxicity, high selectivity, sensitivity, stable photo-stability, enabling the tracking of endogenously carboxylesterase in living HeLa cells. Based on the results of inhibitor AEBSF, this indicates that the fluorescence-on reaction indeed arises from endogenous carboxylesterase in living HeLa cells. All these features of probe 1 suggest that this NIR fluorescent probe holds great potential for further investigation on the biological functions of carboxylesterase in other biosystems.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We are grateful to the financial support from the National Key Research and Development Project (2016YFD0401204) and National Natural Science Foundation of China (21605099).Notes and references
- S. J. Park, Y. J. Kim, J. S. Kang, I. Y. Kim, K. S. Choi and H. M. Kim, Anal. Chem., 2018, 90, 9465–9471 CrossRef CAS.
- S. D. Kodani, M. Barthelemy, S. G. Kamita, B. Hammock and C. Morisseau, Anal. Biochem., 2017, 539, 81–89 CrossRef CAS PubMed.
- S. C. Laizure, V. Herring, Z. Hu, K. Witbrodt and R. B. Parker, Pharmacotherapy, 2013, 33, 210–222 CrossRef CAS PubMed.
- S. Bencharit, C. C. Edwards, C. L. Morton, E. L. Howard-Williams, P. Kuhn, P. M. Potter and M. R. Redinbo, J. Mol. Biol., 2006, 363, 201–214 CrossRef CAS PubMed.
- J. Z. Long and B. F. Cravatt, Chem. Rev., 2011, 111, 6022–6063 CrossRef CAS.
- L. Feng, Z. M. Liu, J. Hou, X. Lv, J. Ning, G. B. Ge, J. N. Cui and L. Yang, Biosens. Bioelectron., 2015, 65, 9–15 CrossRef CAS PubMed.
- D. A. Bachovchin and B. F. Cravatt, Nat. Rev. Drug Discovery, 2012, 11, 52–68 CrossRef CAS PubMed.
- S. Kim, H. Kim, Y. Choi and Y. Kim, Chem. –Eur. J., 2015, 21, 9645–9649 CrossRef CAS PubMed.
- X. J. Wang, H. Liu, J. W. Li, K. G. Ding, Z. L. Lv, Y. G. Yang, H. Chen and X. M. Li, Chem.–Asian J., 2014, 9, 784–789 CrossRef CAS PubMed.
- Z. M. Liu, L. Feng, G. B. Ge, X. Lv, J. Hou, Y. F. Cao, J. N. Cui and L. Yang, Biosens. Bioelectron., 2014, 57, 30–35 CrossRef CAS.
- S. J. Park, H. W. Lee, H. R. Kim, C. Kang and H. M. Kim, Chem. Sci., 2016, 7, 3703–3709 RSC.
- G. Xu, W. Zhang, M. K. Ma and H. L. Mcleod, Clin. Cancer Res., 2002, 8, 2605–2611 CAS.
- M. P. Marrades, P. Gonzalez-Muniesa, J. A. Martinez and M. J. Moreno-Aliaga, Obes. Facts, 2010, 3, 312–318 CrossRef CAS PubMed.
- D. Xiao, D. Shi, D. Yang, B. Barthel, T. H. Koch and B. Yan, Biochem. Pharmacol., 2013, 85, 439–447 CrossRef CAS.
- M. J. Zhao, T. P. Zhang, F. J. Yu, L. X. Guo and B. J. Wu, Biochem. Pharmacol., 2018, 152, 293–301 CrossRef CAS PubMed.
- S. J. Godin, J. A. Crow, E. J. Scollon, M. F. Hughes, M. J. DeVito and M. K. Ross, Drug Metab. Dispos., 2007, 35, 1664–1671 CrossRef CAS.
- S. R. Levine and K. E. Beatty, Chem. Commun., 2016, 52, 1835–1838 RSC.
- T. Steinkamp, F. Schweppe, B. Krebs and U. Karst, Analyst, 2003, 128, 29–31 RSC.
- Y. Sato, A. Miyashita, T. Iwatsubo and T. Usui, Drug Metab. Dispos., 2012, 40, 1389–1396 CrossRef CAS.
- M. Koitka, J. Hochel, D. Obst, A. Rottmann, H. Gieschen and H. H. Borchert, Anal. Biochem., 2008, 381, 113–122 CrossRef CAS PubMed.
- Y. Y. Zhang, W. Chen, D. Feng, W. Shi, X. H. Li and H. M. Ma, Analyst, 2012, 137, 716–721 RSC.
- S. R. Levine and K. E. Beatty, Chem. Commun., 2016, 52, 1835–1838 RSC.
- W. Chen, A. Pacheco, Y. Takano, J. J. Day, K. Hanaoka and M. Xian, Angew. Chem., Int. Ed., 2016, 55, 9993–9996 CrossRef CAS PubMed.
- H. W. Liu, L. Chen, C. Xu, Z. Li, H. Zhang, X. B. Zhang and W. Tan, Chem. Soc. Rev., 2018, 47, 7140–7180 RSC.
- X. Li, X. Gao, W. Shi and H. Ma, Chem. Rev., 2014, 114, 590–659 CrossRef CAS PubMed.
- M. Zheng, P. Li, C. Yang, H. Zhu, Y. Chen, Y. Tang, Y. Zhou and T. Lu, Analyst, 2012, 137, 1182–1189 RSC.
- H. Zhou, J. Tang, J. Zhang, B. Chen, J. Kan, W. Zhang, J. Zhou and H. Ma, J. Mater. Chem. B, 2019, 7, 2989–2996 RSC.
- X. Tian, Z. Li, Y. Sun, P. Wang and H. Ma, Anal. Chem., 2018, 90, 13759–13766 CrossRef CAS PubMed.
- W. Chen, S. Xu, J. J. Day, D. Wang and M. Xian, Angew. Chem., Int. Ed., 2017, 56, 16611–16615 CrossRef CAS PubMed.
- Z. Li, X. He, Z. Wang, R. Yang, W. Shi and H. Ma, Biosens. Bioelectron., 2015, 63, 112–116 CrossRef CAS PubMed.
- J. Zhang, L. Shi, Z. Li, D. Li, X. Tian and C. Zhang, Analyst, 2019, 144, 3643–3648 RSC.
- B. Wu, Y. Lin, B. Li, C. Zhan, F. Zeng and S. Wu, Anal. Chem., 2018, 90, 9359–9365 CrossRef CAS PubMed.
- Q. Jin, L. Feng, D. D. Wang, J. J. Wu, J. Hou, Z. R. Dai, S. G. Sun, J. Y. Wang, G. B. Ge, J. N. Cui and L. Yang, Biosens. Bioelectron., 2016, 83, 193–199 CrossRef CAS PubMed.
- D. Y. Li, Z. Li, W. Y. Chen and X. B. Yang, J. Agric. Food Chem., 2017, 65, 4209–4215 CrossRef CAS PubMed.
- J. H. Zhang, Z. Li, X. W. Tian and N. Ding, Chem. Commun., 2019, 55, 9463–9466 RSC.
- E. Nagele, M. Schelhaas, N. Kuder and H. Waldmann, J. Am. Chem. Soc., 1998, 120, 6889–6902 CrossRef.
- H. Zhou, J. Tang, L. Lv, N. Sun, J. Zhang, B. Chen, J. Mao, W. Zhang, J. Zhang and J. Zhou, Analyst, 2018, 143, 2390–2396 RSC.
- X. Tian, Z. Li, Y. Pang, D. Li and X. Yang, J. Agric. Food Chem., 2017, 65, 9553–9558 CrossRef CAS PubMed.
- T. Komatsu, Y. Urano, Y. Fujikawa, T. Kobayashi, H. Kojima, T. Terai, K. Hanaokaa and T. Nagano, Chem. Commun., 2009, 45, 7015–7017 RSC.
- H. Siebert, S. Engelke, B. Maruschak and W. Bruck, Brain Res., 2001, 916, 159–164 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Apparatus and reagents, and other supporting data. See DOI: 10.1039/c9ra08150j |
| This journal is © The Royal Society of Chemistry 2019 |