Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Marine unsaturated fatty acids: structures, bioactivities, biosynthesis and benefits
Yingfang Lua,
Yinning Chenb,
Yulin Wua,
Huili Haoa,
Wenjing Liangc,
Jun Liu*d and
Riming Huang *a
*a
aGuangdong Provincial Key Laboratory of Food Quality and Safety, College of Food Science, South China Agricultural University, Guangzhou 510642, China. E-mail: luyingfang@stu.scau.edu.cn; DanielWu@stu.scau.edu.cn; hhlhaohuili@stu.scau.edu.cn; huangriming@scau.edu.cn; Tel: +86 20 8528 3448
bGuangdong Polytechnic College, 526100, Zhaoqing, China. E-mail: Yinning_Chen@163.com
cLonggang No. 2 Vocational School, Shenzhen, 518104, China. E-mail: 345589207@qq.com
dLaboratory of Pathogenic Biology, Guangdong Medical University, Zhanjiang 524023, China. E-mail: lj2388240@gdmu.edu.cn; Tel: +86 7592388240
First published on 31st October 2019
Abstract
Unsaturated fatty acids (UFAs) are an important category of monounsaturated and polyunsaturated fatty acids with nutritional properties. These secondary metabolites have been obtained from multitudinous natural resources, including marine organisms. Because of the increasing numerous biological importance of these marine derived molecules, this review covers 147 marine originated UFAs reported from 1978 to 2018. The review will focus on the structural characterizations, biological properties, proposed biosynthetic processes, and healthy benefits mediated by gut microbiota of these marine naturally originated UFAs.
1 Introduction
Fatty acids other than saturated fatty acids (fatty acids that do not contain double bonds are called saturated fatty acids, and all animal oils, except fish oils, contain saturated fatty acids) are unsaturated fatty acids. Unsaturated fatty acids are a kind of fatty acid that makes up body fat. Unsaturated fatty acids (UFAs) consist of a long-chain hydrocarbon with the presence of at least one double covalent bond and ending in a carboxyl group (–COOH), and are distinguished into monounsaturated fatty acids and polyunsaturated fatty acids, both of which have numerous beneficial properties to human health.1,2 These secondary metabolites have previously been obtained from a variety of natural resources, including marine fish oils that are a good natural source of these UFAs.3,4 In previous decades, marine derived UFAs have attracted a great deal of interest because of their structural diversity and potential biological and nutritional functions.5 In particular, research interest in omega-3 fatty acids,6 eicosapentaenoic acid (EPA) and docosahexaenoic acid (DHA) from marine organisms, has dramatically increased as they are excellent sources of nutrients. These UFAs also can be described as cis fatty acids versus trans fatty acids, which is a description of the geometry of their double bonds. These characteristics in UFAs not only enable them to show a broad range of biological activities, but also allow the development of the nutrient-like physicochemical properties. However, most of marine derived UFAs belong to a relatively unexplored category that may hold a great promise for the potential nutritional application in the future. The structures and potential nutritional applications of UFAs, particularly these with the interesting biological activities have previously been reviewed,7,8 but there is still lack of a comprehensive review about marine derived UFAs. Thus, this review aims to summarize 147 marine organisms-derived UFAs published from 1978 to 2018. The review will focus on the structural characterizations, biological properties, proposed biosynthetic processes, and benefits mediated by gut microbiota of these marine UFAs. In addition, the origin of the isolation of these UFAs is also taxonomically presented.2 Monounsaturated fatty acids
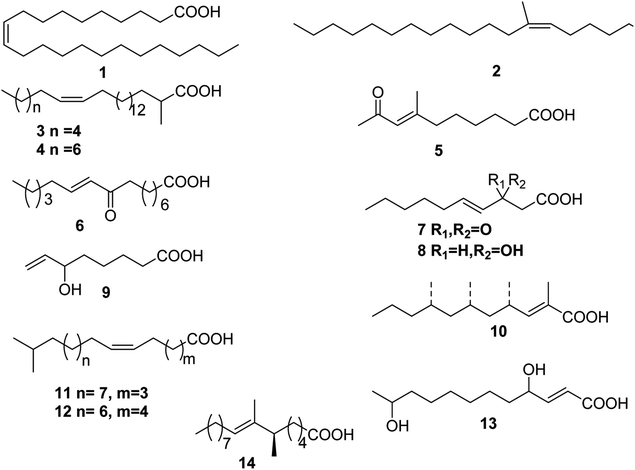
Up to date, there are 14 of total monounsaturated fatty acids obtained from marine organisms, linear and branched monounsaturated fatty acids 1–14 (Table 1 and Fig. 1).| Number | Names | Bioactivities | Sources | Reference(s) |
|---|---|---|---|---|
| 1 | 10-Tricosenoic acid | — | Calyx podatypa | 9 |
| 2 | (6Z)-7-Methyloctadec-6-enoic acid A | — | Holothuria mexicana | 10 |
| 3 | Not given | — | Halichondria panicea | 11 |
| 4 | Not given | — | H. panicea | 11 |
| 5 | Not given | — | Ircinia sp. | 12 |
| 6 | Not given | Antiinflammatory properties | Gracilaria verrucosa | 13 |
| 7 | Not given | — | Ulva fasciata | 14 |
| 8 | Not given | — | U. fasciata | 14 |
| 9 | Not given | — | U. fasciata | 14 |
| 10 | (2E,4S,6S,8S)-2,4,6,8-Tetramethyl-2-undecenoic acid | — | Siphonaria capensis | 15 |
| 11 | Not given | — | S. denticulata | 16 |
| 12 | Not given | — | S. denticulata | 16 |
| 13 | Seco-patulolide | — | unidentified fungal strain | 17 |
| 14 | Not given | — | Sinularia sp. | 18 |
2.1 Linear monounsaturated fatty acids
2.2 Branched monounsaturated fatty acids
3 Polyunsaturated fatty acids
3.1 Linear chain polyunsaturated fatty acids
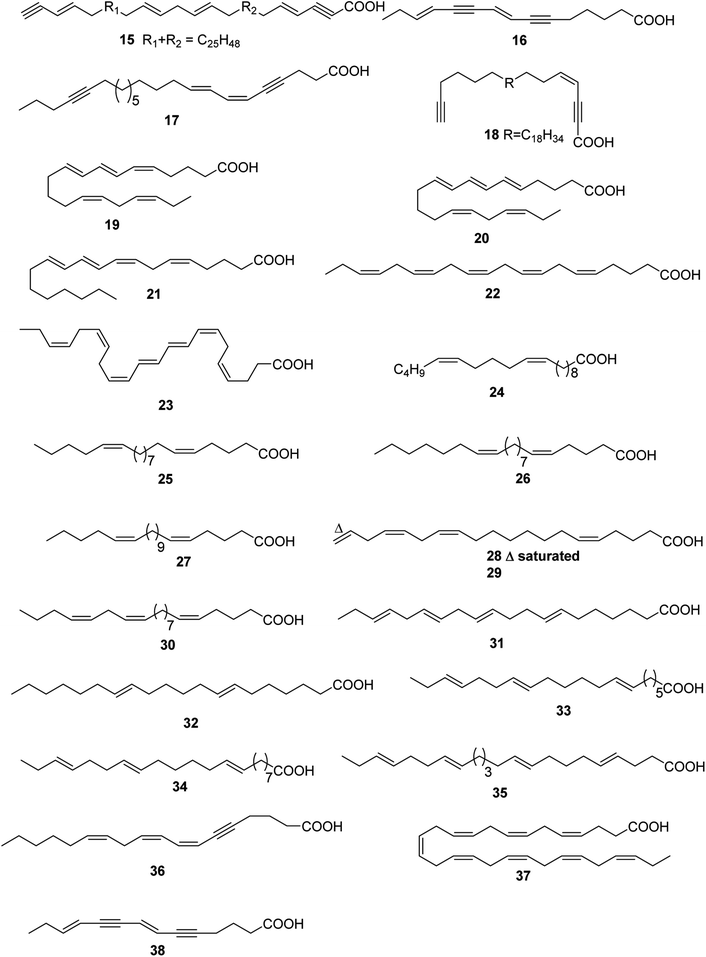
Up to date, there are 24 of total linear chain polyunsaturated fatty acids 15–38 obtained from marine organisms (Table 2 and Fig. 2).| Number | Names | Bioactivities | Sources | Reference(s) |
|---|---|---|---|---|
| 15 | Not given | — | Petrosia ficiformis | 19 |
| 16 | Not given | Antimicrobial | Oceanapia sp. | 20 |
| 17 | Carduusyne A | — | Phakellia carduus | 21 and 22 |
| 18 | Petroformynic acid | — | P. ficiformis | 23 |
| 19 | (5Z,7E,9E,14Z,17Z)-Icosa-5,7,9,14,17-pentaenoic acid | — | Ptilota jilicina | 24 |
| 20 | (5E,7E,9E,14Z,17Z)-Icosa-5,7,9,14,17-pentaenoicacid | — | P. jilicina | 24 |
| 21 | 5(Z),8(Z),10(E),12(E),14(Z)-Eicosapentaenoic acid | — | Bossiella orbigniana | 25 |
| 22 | (5Z,8Z,11Z,14Z,17Z)-Eicosapentaenoic acid | Inhibiting growth of the green alga Monostroma oxyspermum | Neodilsea yendoana | 26 |
| 23 | (4Z,7Z,9E,11E,13Z,16Z,19Z)-Docosaheptaenoic acid | — | Anadyomene stellata | 27 |
| 24 | 10,15-Eicosadienoic acid | — | Haminaea templadoi | 28 and 29 |
| 25 | (5Z,15Z)-5,15-Eicosadienoic acid | — | Calyptogena phaseoliformis | 30 |
| 26 | (5Z,14Z)-5,14-Heneicosadienoic acid | — | C. phaseoliformis | 30 |
| 27 | (5Z,16Z)-5,16-Heneicosadienoic acid | — | C. phaseoliformis | 30 |
| 28 | (5Z,13Z,16Z)-5,13,16-Eicosatrienoic acid | — | C. phaseoliformis | 30 |
| 29 | (5Z,13Z,16Z)-5,13,16,19-Eicosatetraenoic acid | — | C. phaseoliformis | 30 |
| 30 | (5Z,14Z,17Z)-5,14,17-Heneicosatrienoic acid | — | C. phaseoliformis | 30 |
| 31 | 7,11,14,17-Eicosatetraenoic acid | Anti-inflammatory | Perna canaliculus | 31 |
| 32 | 7,13-Eicosadienoic acid | — | Ophiura sarsi | 32 |
| 33 | 7,13,17-Eicosatrienoic acid | — | O. sarsi | 32 |
| 34 | 9,15,19-Docosatrienoic acid | — | O. sarsi | 32 |
| 35 | 4,9,15,19-Docosatetraenoic acid | — | O. sarsi | 32 |
| 36 | (7Z,9Z,12Z)-Octadeca-7,9,12-trien-5-ynoic acid | — | Liagora farinosa | 33 |
| 37 | 4,7,10,13,16,19,22,25-Octacosaoctaenoic acid | — | Marine dinoflagellate species | 33 |
| 38 | 7,11-Tetradecadiene-5,9-diynoic acid | — | Marine dinoflagellate species | 33 |
3.2 Branched chain polyunsaturated fatty acids
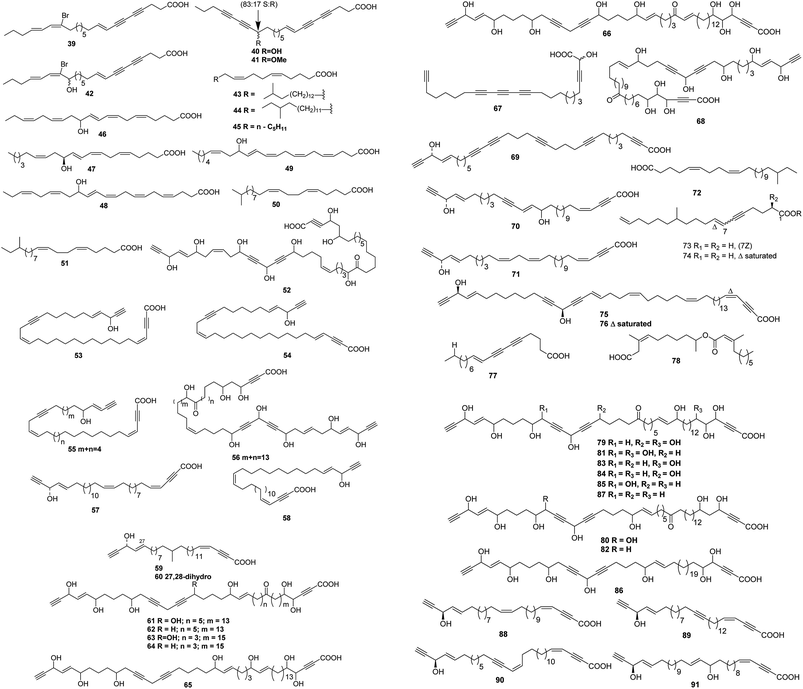
Up to date, there are 109 of total linear chain polyunsaturated fatty acids 39–147 obtained from marine organisms (Tables 3–5 and Fig. 3–5).| Number | Names | Bioactivities | Sources | Reference(s) |
|---|---|---|---|---|
| 39 | Not given | — | P. carduus | 21 |
| 40 | Not given | — | P. carduus | 21 |
| 41 | Not given | — | P. carduus | 21 |
| 42 | Not given | — | P. carduus | 21 |
| 43 | (Z,Z)-25-Methyl-5,9-hexacosadienoic acid | — | Jaspis stellifera | 35 |
| 44 | (Z,Z)-24-Methyl-5,9-hexacosadienoic acid | — | J. stellifera | 35 |
| 45 | (5Z,9Z)-Hexadeca-5,9-dienoic acid | — | Chondrilla nucula | 36 |
| 46 | 5,8,10,14,17-Eicosapentaenoic acid | — | Echinochalina mollis | 37 |
| 47 | Not given | — | E. mollis | 37 |
| 48 | 4,7,10,12,16,19-Docosahexaenoic acid | — | E. mollis | 37 |
| 49 | Not given | — | E. mollis | 37 |
| 50 | 5,9-Eicosadienoic acid | — | Erylus forrnosus | 38 and 39 |
| 51 | 5,9-Eicosadienoic acid | — | E. forrnosus | 38 and 39 |
| 52 | Petrosolic acid | Inhibited HIV reverse transcriptase | Petrosia sp. | 40 |
| 53 | Corticatic acid A | Antifungal | Petrosia corticata | 41 |
| 54 | Corticatic acid B | Antifungal | P. corticata | 41 |
| 55 | Corticatic acid C | Antifungal | P. corticata | 41 |
| 56 | Nepheliosyne A | — | Xestospon | 42 |
| 57 | Triangulynic acid | Against leukemia and colon tumour lines | Pellina triangulata | 43 |
| 58 | Pellynic acid | Inhibited inosine monophosphate dehydrogenase in vitro | P. triangulata | 44 |
| 59 | Aztequynol A | — | Petrosia sp. | 45 |
| 60 | Aztequynol B | — | Petrosia sp. | 45 |
| 61 | Osirisyne A | — | Haliclona osiris | 46 |
| 62 | Osirisyne B | — | H. osiris | 46 |
| 63 | Osirisyne C | — | H. osiris | 46 |
| 64 | Osirisyne D | — | H. osiris | 46 |
| 65 | Osirisyne E | — | H. osiris | 46 |
| 66 | Osirisyne F | — | H. osiris | 46 |
| 67 | Aikupikanyne F | — | Callyspongia sp. | 20 |
| 68 | Haliclonyne | — | Haliclona sp. | 47 |
| 69 | Callyspongynic acid | α-glucosidase inhibitor | P. corticata | 41, 48 and 49 |
| 70 | Corticatic acid D | Geranylgeranyltransferase type I inhibitor | P. corticata | 41, 48 and 49 |
| 71 | Corticatic acid E | P. corticata | 41, 48 and 49 | |
| 72 | (5Z,9Z)-22-Methyl-5,9-tetracosadienoic acid | Cytotoxic activity against mouse Ehrlich carcinoma cells and a hemolytic effect on mouse erythrocytes | Stelletta sp. | 50 |
| 73 | Stellettic acid C | Exhibited marginal to moderate toxicity to five human tumour cell lines | Stelletta sp. | 51 |
| 74 | Not given | Cytotoxic to human leukemia cells | Stelletta sp. | 52 |
| 75 | Petroformynic acid B | Cytotoxic | Petrosia | 53 |
| 76 | Petroformynic acid C | Petrosia | 53 | |
| 77 | Heterofibrin A1 | Inhibited lipid droplet formation | Spongia sp. | 54 |
| 78 | Officinoic acid B | — | Spongia officinalis | 55 |
| 79 | Fulvyne A | Against a chloramphenicol-resistant strain of Bacillus subtilis | Haliclona fulva | 56 |
| 80 | Fulvyne B | H. fulva | 56 | |
| 81 | Fulvyne C | H. fulva | 56 | |
| 82 | Fulvyne D | H. fulva | 56 | |
| 83 | Fulvyne E | H. fulva | 56 | |
| 84 | Fulvyne F | H. fulva | 56 | |
| 85 | Fulvyne G | H. fulva | 56 | |
| 86 | Fulvyne H | H. fulva | 56 | |
| 87 | Fulvyne I | H. fulva | 56 | |
| 88 | Petrosynic acid A | — | Petrosia sp. | 57 |
| 89 | Petrosynic acid B | — | Petrosia sp. | 57 |
| 90 | Petrosynic acid C | — | Petrosia sp. | 57 |
| 91 | Petrosynic acid D | — | Petrosia sp. | 57 |
| Number | Names | Bioactivities | Sources | Reference(s) |
|---|---|---|---|---|
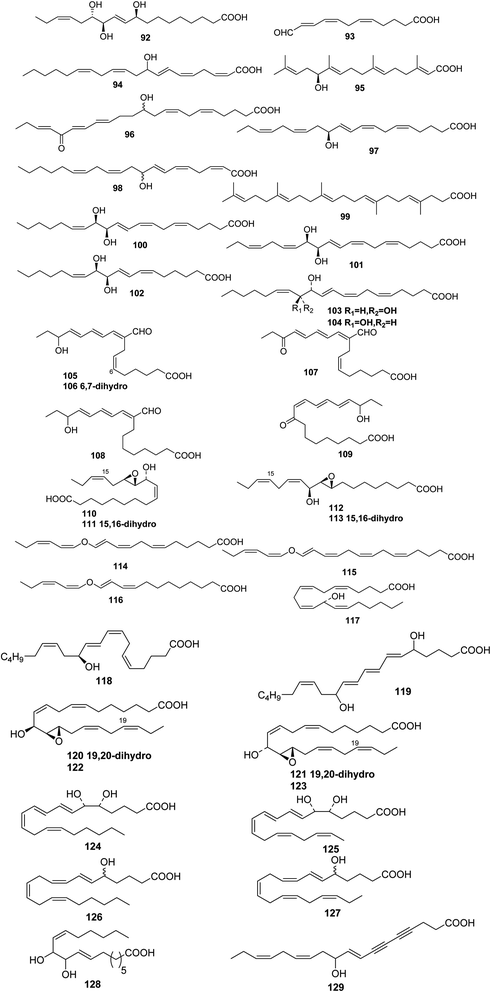
| 92 | (10E,15Z)-(9S,12R,13S)-9,12,13-Trihydroxyoctadeca-10,14-dienoicacid | — | Lyngbya majuscula | 58 |
| 93 | (5Z,8E,10E)-11-Fomylundeca-5,8,10-trienoic acid | Antimicrobial | Laurencia hybrida | 59 |
| 94 | (2Z,5Z,7E,11Z,14Z)-9-Hydroxyeicosa-2,5,7,11,14-pentaenoic acid | Antimicrobial | L. hybrida | 59 |
| 95 | Acyclicditerpene | — | Bifurcaria bifurcate | 60 |
| 96 | Ptilodene | Inhibited both 5-lipoxygenase and Na+/K+A TPase | Ptilota filicina | 61 |
| 97 | 12-(S)-Hydroxyeicosapentaenoic acid | Inhibitor of platelet aggregation | Murrayella periclados | 62 |
| 98 | 9-Hydroxypentaenoic acid | — | Laurencia hybrid | 63 |
| 99 | Turbinaric acid | Cytotoxic | Turbinaria ornata | 64 |
| 100 | (12R,13R)-Dihydroxyeicosa-5(Z),8(Z),10(E),14(Z)-tetraeonic acid | Modulated fMLP-induced superoxide anion generation in human neutrophils; inhibited the conversion of arachidonic acid to lipoxygenase products by human neutrophils; inhibited the functioning of the dog kidney Na+/K+ ATPase | Farlowia mollis | 65 |
| 101 | (12R,13R)-Dihydroxyeicosa-5(Z),8(E),10(E),14(Z),17(Z)-pentaenoic acid | F. mollis | 65 | |
| 102 | (10R,11R)-Dihydroxyoctadeca-6(Z),8(E),12(Z)-trienoic acid | F. mollis | 65 | |
| 103 | (5Z,8Z,10E,12R,13R,14Z)-12,13-Dihydroxyeicosa,5,8,10,14-tetraenoic acid | — | F. mollis | 65 |
| 104 | (5Z,8Z,10E,12R,13S,14Z)-12,13-dihydroxyeicosa-5,8,10,14-tetraenoic acid | — | F. mollis | 66 |
| 105 | (6Z,9E,11E,13E)-9-Formyl-15-hydroxyheptadeca-6,9,11,13-tetraenoic acid | — | Acrosiphonia coalita | 67 |
| 106 | (9E,11E,13E)-9-Formyl-15-hydroxyheptadeca-9,11,13-trienoic acid | — | A. coalita | 67 |
| 107 | (6Z,9E,11E,13E)-9-formyl- 15-oxoheptadeca-6,9,11,13-tetraenoic acid | — | A. coalita | 67 |
| 108 | (10E,12Z,14E)-16-Hydroxy-9-oxooctadeca-10,12,14-trienoic acid | — | A. coalita | 67 |
| 109 | (10E,12E,14E)-16-hydroxy-9-oxooctadeca-10,12,14-trienoic acid | — | A. coalita | 67 |
| 110 | (9Z,11R,12S,13S,152)-12,13-Epoxy-11-hydroxyoctadeca-9,15-dienoic acid | — | A. coalita | 67 |
| 111 | (9Z,11R,12S,13S)-12,13-Epoxy-11-hydroxyoctadeca-9-enoic acid | — | A. coalita | 67 |
| 112 | (9R,10R,11S,12Z,152)-9,10-Epoxy-11-hydroxyoctadeca-12,15-dienoic acid | — | A. coalita | 67 |
| 113 | (9R,10R,11S,122)-9,10-Epoxy-11-hydroxyoctadeca- 12-enoic acid | — | A. coalita | 67 |
| 114 | Not given | — | Laminaria sincluirii | 68 |
| 115 | Not given | — | L. sincluirii | 68 |
| 116 | 9,11-Dodecadienoic acid | — | L. sincluirii | 68 |
| 117 | (13R)-13-hydroxyarachidonic acid | — | Lithothamnion coralloides | 69 and 70 |
| 118 | (12S)-12-Hydroxyeicosatetraenoic acid | — | M. periclados | 71 |
| 119 | (6E)-Leukotriene B4 | — | M. periclados | 71 |
| 120 | Hepoxilin B3 | — | M. periclados | 71 |
| 121 | Hepoxilin B3 | — | M. periclados | 71 |
| 122 | Hepoxilin B4 | — | M. periclados | 71 |
| 123 | Hepoxilin B4 | — | M. periclados | 71 |
| 124 | (5R,6S,7E,9E,11Z,14Z)-5,6-Dihydroxyicosa-7,9,11,14-tetraenoic acid | — | Rhodymenia pertusa | 72 |
| 125 | (5R*,6S*,7E,9E,11Z,14Z,17Z)-5,6-Dihydroxyicosa-7,9,11,14,17-pentaenoic acid | — | R. pertusa | 72 |
| 126 | (6E,8Z,11Z,14Z)-5-Hydroxyicosa-6,8,11,14-tetraenoic acid | — | R. pertusa | 72 |
| 127 | (6E,8Z,11Z,14Z,17Z)-5-Hydroxyicosa-6,8,11,14,17-Pentaenoic acid | — | R. pertusa | 72 |
| 128 | 8,12-Octadecadienoic acid | — | Corallina officinalis | 73 |
| 129 | (8E,12Z,15Z)-10-Hydroxy-8,12,15-trien-4,6-diynoic acid | — | Caulerpa racemosa | 74 |
| Number | Names | Bioactivities | Sources | Reference(s) |
|---|---|---|---|---|
| 130 | Leiopathic acid | — | Leiopathes sp. | 75 |
| 131 | 5,9,11,14,17-Eicosapentaenoic acid | — | Leiopathes sp. | 75 |
| 132 | 5,9,11,14,17-Eicosapentaenoic acid | — | Leiopathes sp. | 75 |
| 133 | (11R)-Hydroxyeicosatetraenoic acid | — | Plexaurella dichotoma | 76 |
| 134 | (5Z,9Z)-14-methylpentadeca-5,9-dienoic acid | Inhibited the growth of Gram positive bacteria | Eunicea succinea | 77 |
| 135 | 6,9,12,16,18-Tetracosapentaenoic acid | Inhibited tube-formation in a human endothelial cell line model of angiogenesis | Sinularia numerosa | 78 |
| 136 | Dendryphiellic acid A | — | Dendryphiella salina | 79 and 80 |
| 137 | Dendryphiellic acid B | — | D. salina | 79 and 80 |
| 138 | Curvulalic acid | — | Curvularia sp. | 81 |
| 139 | 2,4-Decadienoic acid | — | Xylaria sp. | 82 |
| 140 | (5Z,8R,9E,11Z,14Z,17Z)-8-hydroxyeicosa-5,9,11,14,17-pentaenoic acid | — | Balanus balanoides, Eliminus modestus | 83 |
| 141 | 8,13-Dihydroxyeicosapentaenoic acid | A muscle stimulatory factor in the barnacle Balanus balanus | Balanus balanus | 84 |
| 142 | (9Z,12Z)-7-hydroxyoctadeca-9,12-dien-5-ynoic acid | Ichthyotoxic | L. farinosa | 33 |
| 143 | Macrolactic acid | — | Unidentified Gram-positive bacterium | 85 |
| 144 | Isomacrolactic acid | — | Unidentified Gram-positive bacterium | 85 |
| 145 | Ieodomycin C | Antimicrobial | Bacillus sp. | 86 |
| 146 | Ieodomycin D | Bacillus sp. | 86 | |
| 147 | Linieodolide B | Antibacterial; antifungal | Bacillus sp. | 87 and 88 |
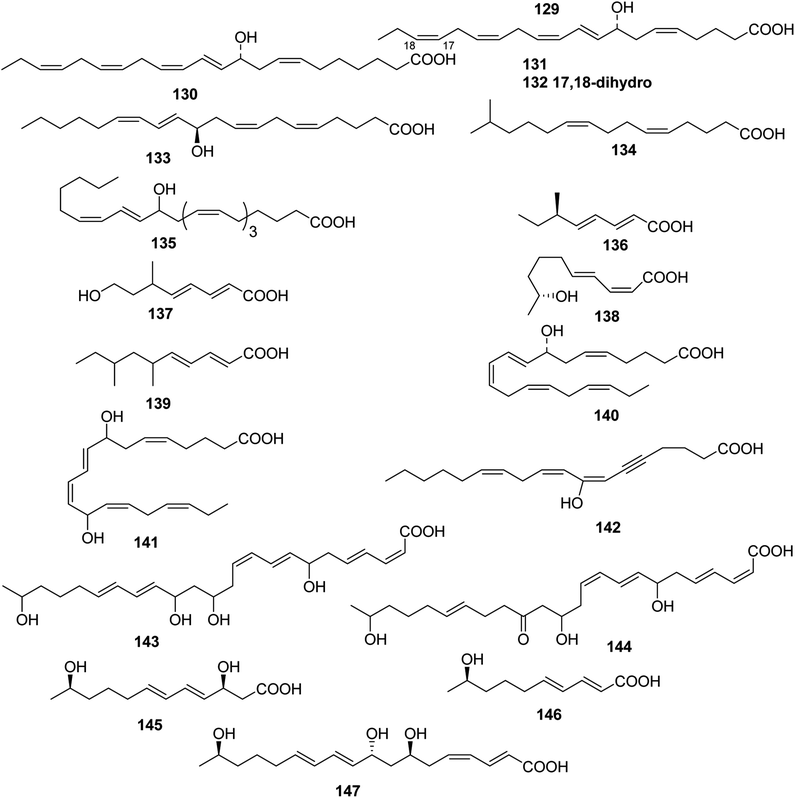 | ||
| Fig. 5 Structures of branched chain polyunsaturated fatty acids from Coelenterate, Marine fungus, Arthropoda, Bacterium. | ||
A cytotoxic fatty acid, (5Z,9Z)-22-methyl-5,9-tetracosadienoic acid 72 was isolated from Geodinella robusta collected from the Sea of Okhotsk, Russia.50 An undescribed Korean species of Stelletta was found to contain a cytotoxic acetylenic acid: stellettic acid C 73 that exhibited marginal to moderate toxicity to five human tumour cell lines.51 From a seemingly identical Stelletta species, collected at a different Korean location, a desmethoxy analogue 74, was isolated; it was mildly cytotoxic to human leukemia cells.52 The cytotoxic petroformynic acids B 75 and C 76 were obtained from a Petrosia species (Katsuo-jim Is., Wakayama Pref., Japan).53 One acetylenic compound heterofibrin A1 77 was isolated from a Spongia (Heterofibria) sp. collected by dredging in the Great Australian Bight. Heterofibrin A1 inhibited lipid droplet formation at 10 mM yet was not cytotoxic at similar concentrations.54 Officinoic acid B 78 is linear diterpene from Spongia officinalis (off Mazara del Vallo, Sicily).55 An extract of Haliclona fulva (Procida Is., Gulf of Naples, Italy) contained the nine acetylenes fulvyne A–I 79–87.56 Petrosynic acids A–D 88–91 (Petrosia sp., Tutuila, American Samoa) all displayed similar activity versus various HTCLs and non-proliferative human fibroblasts and hence no therapeutic window is available.57
The green alga Acrosiphonia coalita contains the oxylipins coalital, which may be an artefact caused by photoisomerization of the natural product, racemic (6Z,9E,11E,13E)-9-formyl-15-hydroxyheptadeca-6,9,11,13-tetraenoic acid 105, (9E,11E,13E)-9-formyl-15-hydroxyheptadeca-9,11,13-trienoic acid 106, (6Z,9E,11E,13E)-9-formyl-15-oxoheptadeca-6,9,11,13-tetraenoic acid 107, (10E,12Z,14E)-16-hydroxy-9-oxooctadeca-10,12,14-trienoic acid 108, (10E,12E,14E)-16-hydroxy-9-oxooctadeca-10,12,14-trienoic acid 109, (9Z,11R,12S,13S,152)-12,13-epoxy-11-hydroxyoctadeca-9,15-dienoic acid 110, (9Z,11R,12S,13S)-12,13-epoxy-11-hydroxyoctadeca-9-enoic acid 111, (9R,10R,11S,12Z,152)-9,10-epoxy-11-hydroxyoctadeca-12,15-dienoic acid 112, and (9R,10R,11S,122)-9,10-epoxy-11-hydroxyoctadeca-12-enoic acid 113, the acids all being isolated as the corresponding methyl esters.67 Three divinyl ethers, 114–116, were isolated along with a number of hydroxylated fatty acids from the Oregon brown alga Laminaria sincluirii and were identified by interpretation of spectral evidence.68 The absolute stereochemistry of (13R)-13-hydroxyarachidonic acid 117, which is a known eicosanoid from Lithothamnion coralloides,69 was determined by degradation and its biosynthesis from arachidonic acid was studied.70
The Caribbean alga Murrayella periclados contains a number of eicosanoids that include (12S)-12-hydroxyeicosatetraenoic acid 118, (6E)-leukotriene B4, 119 and erythro and threo isomers of hepoxilins B3, 120/121 and B4, 122/123.71 Four oxylipins (5R,6S,7E,9E,11Z,14Z)-5,6-dihydroxyicosa-7,9,11,14-tetraenoic acid 124, (5R*,6S*,7E,9E,11Z,14Z,17Z)-5,6-dihydroxyicosa-7,9,11,14,17-pentaenoic acid 125, (6E,8Z,11Z,14Z)-5-hydroxyicosa-6,8,11,14-tetraenoic acid 126, and (6E,8Z,11Z,14Z,17Z)-5-hydroxyicosa-6,8,11,14,17-pentaenoic acid 127 were isolated from Rhodymenia pertusa.72 An oxylipin 128 was obtained from Aspergillus flavus, (red alga Corallina officinalis, Yantai, China).73 Studies on a Caulerpa racemosa (Zhanjiang coastline, China) led to the isolation of the acetylenic fatty acid (8E,12Z,15Z)-10-hydroxy-8,12,15-trien-4,6-diynoic acid 129.74
4 Biosynthetic pathways
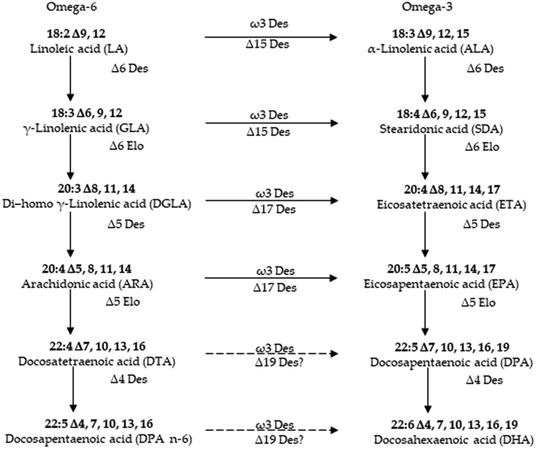
PUFAs are gaining importance due to their innumerable health benefits. The most common source of PUFAs is of marine origin. Hence, understanding their biosynthesis in marine origin has attained prominence in recent years.89,90 Rabbitfish Siganus canaliculatus was the first marine teleost demonstrated to have the ability to biosynthesize C20–22 long-chain polyunsaturated fatty acid (LC-PUFA) from C18 PUFA precursors, which is generally absent or low in marine teleosts.91 The marine diatom Phaeodactylum tricornutum accumulates eicosapentaenoic acid (EPA, 20:5n-3) as its major component of fatty acids. To improve the EPA production, delta 5 desaturase, which plays a role in EPA biosynthetic pathway, was characterized in marine diatom Phaeodactylum tricornutum.90 There is currently considerable interest in understanding how the biosynthetic pathways of highly unsaturated fatty acids (HUFA) are regulated in fish. The aim is to know if it is possible to replace fish oils (FO), rich in HUFA, by vegetable oils (VO), poor in HUFA and rich in their 18 carbon fatty acid precursors, in the feed of cultured fish species of commercial importance.92 Although many better insights into the synthesis of eicosapentaenoic acid (EPA) and docosahexaenoic acid (DHA) in marine microalgae,93 there are still a little known about biosynthetic processes of most isolated UFAs of marine resources.70,94 Thus, more investigation should be carried out for these marine derived UFAs in the coming researches (Fig. 6).5 Beneficial application
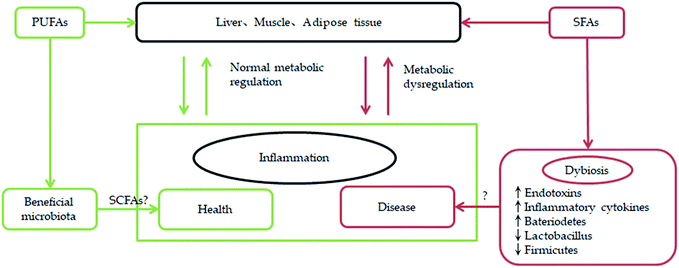
It is well-known that polyunsaturated fatty acids n-3 (PUFAn-3) are very important for human health and nutrition.1 As an example, highly unsaturated long-chain omega-3 fatty acids, derived from the liver of white lean fish, flesh of fatty fish, and blubber of marine mammals, exhibit important biological activities.95 They also serve as the building block fatty acids in the brain, retina, and other organs with electrical activity. Hence, inclusion of oils containing docosahexaenoic acid (DHA) in the diet of pregnant and lactating women as well as infants is encouraged.95In addition, some polyunsaturated fatty acids from marine microalgae are found to modulate lipid metabolism disorders and gut microbiota.96 According to the survey results, high saturated fatty acid and high monounsaturated fatty acid diets have an adverse effect on the gut microbiota and high saturated fatty acids are associated with unhealthy metabolic status, while polyunsaturated fatty acid does not have a negative impact on gut microbiota.97 Through previous studies we find that connecting with gut microbiota, PUFAs can be more beneficial for human health. For example, increasing anti-obesogenic microbial species in the gut microbiota population by appropriate n-3 PUFAs can be an effective way to control or prevent metabolic diseases.98 Furthermore, a link has been established between n-3 PUFAs and gut microbiota especially with respect to inflammation (Fig. 7). A few related researchs show that after omega-3 PUFA supplementation, Faecalibacterium, often associated with an increase in the Bacteroidetes and butyrate-producing bacteria belonging to the Lachnospiraceae family, has decreased. Omega-3 PUFAs perform a positive action on diseases by reverting the microbiota composition and increasing the production of anti-inflammatory compounds like short-chain fatty acids.99 According to the link between n-3 PUFAs and gut microbiota, which is associated with inflammation, some scholars proposing that an optimal level of LC-PUFAs nurtures the suitable gut microbiota that will prevent dysbiosis. The synergy between optimal LC-PUFAs and gut microbiota helps the immune system overcome the immunosuppressive tumour microenvironment.100
Although many scholars have devoted themselves to the study of polyunsaturated fatty acids, they are limited to the more famous unsaturated fatty acids. There is still lack of investigation of the beneficial application of these polyunsaturated fatty acid derivatives with similar structural characteristics. Thus, more investigation should focus on fatty acid physiological roles and applications in human health and disease and the interaction with gut microbiota.101
6 Conclusions
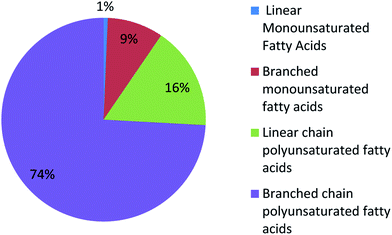
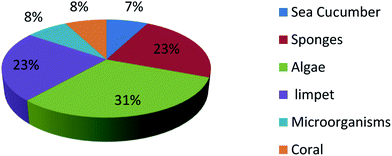
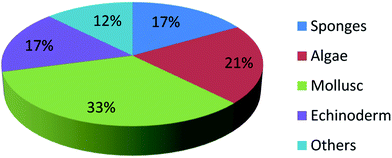
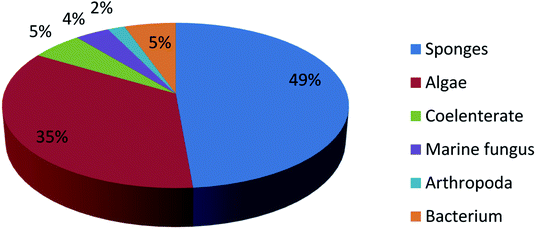
UFAs are ubiquitous in many marine organisms.3,102,103 Although these UFA secondary metabolites have been obtained since the early 20th century, they only recently draw significant interests because of the diverse range of their biological and nutritional properties.104 However, there is still lack of a comprehensive review about the structural characterizations, biological and nutritional properties, proposed biosynthetic processes, and beneficial application of marine derived UFAs. 1978 to 2018, the main structural types of UFAs obtained from marine organisms is branched chain PUFAs, accounting for 74% of the total (Fig. 8), the main natural source of branched monounsaturated fatty acids isolated from marine organisms is coral, accounting for 31% (Fig. 9), while linear chain polyunsaturated fatty acids obtained from marine organisms is mollusc, accounting for 33% (Fig. 10), the preponderant natural marine source of PUPAs is arthropoda, accounting for 49% (Fig. 11). Although omega-3 fatty acid,6 eicosapentaenoic acid (EPA) and docosahexaenoic acid (DHA) from marine organisms, have dramatically increased as excellent sources of nutrients, it is indicated that the biological activities of most of the UPAs are not investigated (Tables 1–3), and the little known about the biosynthetic pathways of these isolated UPAs. In addition, there is no report about new UFAs isolated from marine resources during 2016 to 2018. Thus, the further investigation of marine derived PUPAs should focus on their and beneficial application mediated by gut microbiota.Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
This research was funded by Science and Technology Planning Project of Guangdong Province, Guangzhou Planned Program in Science and Technology, Program of Department of Ocean and Fisheries of Guangdong Province, Natural Science Foundation of Guangdong, Guangdong Provincial Key Laboratory of Applied Botany, South China Botanical Garden, Chinese Academy of Sciences, Finance Special Project of Zhanjiang City, Natural Science Foundation of Guangdong, grant number 2017A020217002, 201803020003, GDME2018C014, 2016A030313151, AB2018004, 2018A01044 and 2018A0303070018.Notes and references
- E. B. Rimm, L. J. Appel, S. E. Chiuve, L. Djousse, M. B. Engler, P. M. Kris-Etherton, D. Mozaffarian, D. S. Siscovick, A. H. Lichtenstein, C. L. C. Hlth, C. E. Prevention, C. C. D. Young, C. C. S. Nursing and C. C. Cardiology, Circulation, 2018, 138, E35–E47 CrossRef CAS.
- M. E. Riveros and M. A. Retamal, Front. Physiol., 2018, 9, 693 CrossRef.
- A. Tsoupras, R. Lordan, K. Shiels, S. K. Saha, C. Nasopoulou and I. Zabetakis, Mar. Drugs, 2019, 17 CrossRef CAS PubMed.
- E. Alexandri, A. Raheel, H. Siddiqui, M. I. Choudhary, C. G. Tsiafoulis and I. P. Gerothanassis, Molecules, 2017, 22, 1633–1671 CrossRef.
- T. Gluck and P. Alter, Vasc. Pharmacol., 2016, 82, 11–19 CrossRef.
- D. S. Im, Eur. J. Pharmacol., 2016, 785, 36–43 CrossRef CAS.
- P. Kuppusamy, I. Soundharrajan, S. Srigopalram, M. M. Yusoff, G. P. Maniam, N. Govindan and K. C. Cho, Indian J. Geo-Mar. Sci., 2017, 46, 663–667 Search PubMed.
- M. Masson, T. Loftsson and G. G. Haraldsson, Pharmazie, 2000, 55, 172–177 CAS.
- N. M. Carballeira, M. Pagan and A. D. Rodriguez, J. Nat. Prod., 1998, 61, 1049–1052 CrossRef CAS PubMed.
- N. M. C. Carballeira, C. Clarisa and A. Sostre, J. Nat. Prod., 1996, 59, 1076–1078 CrossRef CAS.
- M. Perpelescu, M. Tsuda, M. Suzuki, S. Yoshida and J. Kobayashi, Nat. Med., 2004, 58, 86 CAS.
- I. I. Tatli, F. Kong, X. Feng, G. Carter, K. V. Rao and M. T. Hamann, J. Chem. Res., 2008, 50–51, DOI:10.3184/030823408x287131.
- H. T. Dang, H. J. Lee, E. S. Yoo, P. B. Shinde, Y. M. Lee, J. Hong, D. K. Kim and J. H. Jung, J. Nat. Prod., 2008, 71, 232–240 CrossRef CAS.
- G. S. E. Abou-ElWafa, M. Shaaban, K. A. Shaaban, M. E. E. El-Naggar and H. Laatsch, Z. Naturforsch., B: J. Chem. Sci., 2009, 64, 1199–1207 CAS.
- D. R. D.-C. Beukes and T. Michael, Tetrahedron, 1999, 55, 4051–4056 CrossRef CAS.
- N. M. Carballeira, H. Cruz, C. A. Hill, J. J. De Voss and M. Garson, J. Nat. Prod., 2001, 64, 1426–1429 CrossRef CAS.
- C. J. Smith, D. Abbanat, V. S. Bernan, W. M. Maiese, M. Greenstein, J. Jompa, A. Tahir and C. M. Ireland, J. Nat. Prod., 2000, 63, 142–145 CrossRef CAS.
- K. Watanabe, R. Makino, H. Takahashi, K. Iguchi, H. Ohrui and K. Akasaka, Chem. Pharm. Bull., 2008, 56, 861–863 CrossRef CAS.
- G. Cimino, A. De Giulio, S. De Rosa, S. De Stefano and G. Sodano, J. Nat. Prod., 1985, 48, 22–27 CrossRef CAS.
- S. Matsunaga, Y. Okada, N. Fusetani and R. W. M. Van Soest, J. Nat. Prod., 2000, 63, 690–691 CrossRef CAS.
- R. A. C. Barrow and J. Robert, Aust. J. Chem., 1994, 47, 1901–1918 CrossRef CAS.
- P. D. Charoenying, D. Huw, D. McKerrecher and R. J. K. Taylor, Tetrahedron Lett., 1996, 37, 1913–1916 CrossRef CAS.
- Y. G. Guo, M. Gavagnin, C. Salierno and G. Cimino, J. Nat. Prod., 1998, 61, 333–337 CrossRef CAS.
- A. G. Lopez and H. William, Lipids, 1987, 22, 190–194 CrossRef CAS.
- J. R. D. l. R. Burgess, I. Roger, R. S. Jacobs and A. Butler, Lipids, 1991, 26(2), 162–165 CrossRef CAS.
- M. Suzuki, I. Wakana, T. Denboh and M. Tatewaki, Phytochemistry, 1996, 43, 63–65 CrossRef CAS.
- M. V. B. Mikhailova, L. Debra, M. L. Wise, W. H. Gerwick, J. N. Norris and R. S. Jacobs, Lipids, 1995, 30, 583–589 CrossRef CAS.
- N. M. Carballeira, E. Anastacio, J. Salva and M. J. Ortega, J. Nat. Prod., 1992, 55, 1783–1786 CrossRef CAS PubMed.
- B. A. Kulkarni, A. Chattopadhyay and V. R. Mamdapur, J. Nat. Prod., 1994, 57, 537–538 CrossRef CAS.
- H. Saito, J. Chromatogr. A, 2007, 1163, 247–259 CrossRef CAS.
- A. P. Treschow, L. D. Hodges, P. F. A. Wright, P. M. Wynne, N. Kalafatis and T. A. Macrides, Comp. Biochem. Physiol., Part B: Biochem. Mol. Biol., 2007, 147, 645–656 CrossRef CAS PubMed.
- D. Sato, Y. Ando, R. Tsujimoto and K. Kawasaki, Lipids, 2001, 36, 1372–1375 CrossRef.
- V. J. Paul and W. Fenical, Tetrahedron Lett., 1980, 21, 3327–3330 CrossRef CAS.
- M. P. V. Mansour, K. John, D. G. Holdsworth, A. E. Jackson and S. I. Blackburn, Phytochemistry, 1999, 50, 541–548 CrossRef CAS.
- N. T. Carballeira, E. Janice, E. Ayanoglu and C. Djerassi, J. Org. Chem., 1986, 52, 2751–2756 CrossRef.
- N. M. Carballeira and L. Maldonado, Lipids, 1986, 21, 470–471 CrossRef CAS.
- A. Guerriero, M. D'Ambrosio, F. Pietra, O. Ribes and D. Duhet, J. Nat. Prod., 1990, 53, 57–61 CrossRef CAS.
- N. M. Carballeira and V. Negron, J. Nat. Prod., 1991, 54, 305–309 CrossRef CAS.
- B. A. Kulkarni, A. Chattopadhyay and V. R. Mamdapur, Nat. Prod. Lett., 1993, 3, 251–255 CrossRef CAS.
- S. Isaacs, Y. Kshman, S. Loya, A. Hizi and Y. Loya, Tetrahedron, 1993, 49, 10435–10438 CrossRef CAS.
- H.-Y. Li, S. Matsunaga and N. Fusteani, J. Nat. Prod., 1994, 57, 1464–1467 CrossRef CAS.
- J. Kobayashi, K. Naitoh, K. Ishida, H. Shigemori and M. Ishibashi, J. Nat. Prod., 1994, 57, 1300–1303 CrossRef CAS.
- J.-R. H. Dai, F. Yali, J. H. Cardellina II, G. N. Gray and M. R. Boyd, J. Nat. Prod., 1996, 59, 860–865 CrossRef CAS.
- X. Fu, S. A. Abbas, F. J. Schmitz, I. Vidavsky, M. L. Gross, M. Laney, R. C. Schatzman and R. D. Cabuslay, Tetrahedron, 1997, 53, 799–814 CrossRef CAS.
- A. Guerriero, C. Debitus, D. Laurent, M. D'Ambrosio and F. Pietra, Tetrahedron Lett., 1998, 39, 6395–6398 CrossRef CAS.
- J. Shin, Y. Seo, K. W. Cho, J. R. Rho and V. J. Paul, Tetrahedron, 1998, 54, 8711–8720 CrossRef CAS.
- L. Chill, A. Miroz and Y. Kashman, J. Nat. Prod., 2000, 63, 523–526 CrossRef CAS.
- Y. Nakao, T. Uehara, S. Matunaga, N. Fusetani and R. W. M. van Soest, J. Nat. Prod., 2002, 65, 922–924 CrossRef CAS.
- S. Nishimura, S. Matsunaga, M. Shibazaki, K. Suzuki, N. Harada, H. Naoki and N. Fusetani, J. Nat. Prod., 2002, 65, 1353–1356 CrossRef CAS.
- T. N. Makarieva, E. A. Santalova, I. A. Gorshkova, A. S. Dmitrenok, A. G. Guzii, V. I. Gorbach, V. I. Svetashev and V. A. Stonik, Lipids, 2002, 37, 75–80 CrossRef CAS.
- Q. C. Zhao, T. A. Mansoor, J. K. Hong, C. O. Lee, K. S. Im, D. S. Lee and J. H. Jung, J. Nat. Prod., 2003, 66, 725–728 CrossRef CAS.
- H. S. Lee, J. R. Rho, C. J. Sim and J. Shin, J. Nat. Prod., 2003, 66, 566–568 CrossRef CAS.
- C. Okamoto, Y. Nakao, T. Fujita, T. Iwashita, W. M. van Soest, N. Fusetani and S. Matsunaga, J. Nat. Prod., 2007, 70, 1816–1819 CrossRef CAS.
- A. A. Salim, J. Rae, F. Fontaine, M. M. Conte, Z. Khalil, S. Martin, R. G. Parton and R. J. Capon, Org. Biomol. Chem., 2010, 8, 3188–3194 RSC.
- E. Manzo, M. L. Ciavatta, G. Villani, M. Varcamonti, S. M. Abu Sayem, R. van Soest and M. Gavagnin, J. Nat. Prod., 2011, 74, 1241–1247 CrossRef CAS.
- G. Nuzzo, M. L. Ciavatta, G. Villani, E. Manzo, A. Zanfardino, M. Varcamonti and M. Gavagnin, Tetrahedron, 2012, 68, 754–760 CrossRef CAS.
- E. J. Mejia, L. B. Magranet, N. J. De Voogd, K. TenDyke, D. Y. Qiu, Y. Y. Shen, Z. R. Zhou and P. Crews, J. Nat. Prod., 2013, 76, 425–432 CrossRef CAS.
- J. H. Cardellina II and R. E. Moore, Tetrahedron, 1980, 36, 993–996 CrossRef.
- M. D. Higgs and L. J. Mulheirn, Tetrahedron, 1981, 37, 4259–4262 CrossRef CAS.
- L. Z. Semmak, Abdelfetta, R. Valls, B. Banaigs, G. Jeanty and C. Francisco, Phytochemistry, 1988, 27, 2347–2349 CrossRef CAS.
- A. G. Lopez and H. William, Tetrahedron Lett., 1988, 29, 1505–1506 CrossRef CAS.
- M. D. Higgs, Tetrahedron, 1981, 37, 4255–4258 CrossRef CAS.
- M. G. Bernart and H. William, Tetrahedron Lett., 1988, 29, 2015–2018 CrossRef CAS.
- F. Asari, T. Kusumi and H. Kakisawa, J. Nat. Prod., 1989, 52, 1167–1169 CrossRef CAS.
- M. L. J. Solem, D. Zhi and W. H. Gerwick, Lipids, 1989, 24, 256–260 CrossRef CAS.
- S. Lumin and J. R. Falck, Tetrahedron Lett., 1990, 31, 2971–2974 CrossRef CAS.
- M. W. Bernart, G. G. Whatley and W. H. Gerwick, Nat. Prod., 1993, 56(2), 245–259 CrossRef CAS.
- P. J. G. Proteau and H. William, Lipids, 1993, 28, 783–787 CrossRef CAS.
- A. Guerriero, M. D'Ambrosio and F. Pietra, Helv. Chim. Acta, 1990, 73, 2183–2189 CrossRef CAS.
- W. H. Gerwick, P. Aasen and M. Hamberg, Phytochemistry, 1993, 34, 1029–1033 CrossRef CAS.
- M. W. G. Bernari and H. William, Phytochemistry, 1994, 36, 1233–1240 CrossRef CAS.
- Z. D. K. Jiang, O. Sharon and W. H. Gerwick, Phytochemistry, 2000, 53, 129–133 CrossRef CAS.
- M. F. Qiao, N. Y. Ji, F. P. Miao and X. L. Yin, Magn. Reson. Chem., 2011, 49, 366–369 CrossRef CAS.
- S. C. Mao, D. Q. Liu, X. Q. Yu and X. P. Lai, Biochem. Syst. Ecol., 2011, 39, 253–257 CrossRef CAS.
- A. D. A. Guerriero, Michele, F. Pietra, O. Ribes and D. Duhet, Helv. Chim. Acta, 1988, 71, 1094–1100 CrossRef CAS.
- V. Di Marzo, M. Ventriglia, E. Mollo, M. Mosca and G. Cimino, Experientia, 1996, 52, 834–838 CrossRef CAS.
- N. M. R. Carballeira, D. Elba, A. Sostre, A. D. Rodriguez, J. L. Rodriguez and F. A. Gonzalez, J. Nat. Prod., 1997, 60, 502–504 CrossRef CAS.
- T. Yamashita, Y. Nakao, S. Matsunaga, T. Oikawa, Y. Imahara and N. Fusetani, Bioorg. Med. Chem., 2009, 17, 2181–2184 CrossRef CAS PubMed.
- A. Guerriero, M. D'Ambrosio, V. Cuomo, F. Vanzanella and F. Pietra, Helv. Chim. Acta, 1989, 72, 438–446 CrossRef CAS.
- A. Guerriero, M. D'Ambrosio, V. Cuomo, F. Vanzanella and F. Pietra, Helv. Chim. Acta, 1988, 71, 57–61 CrossRef CAS.
- K. Trisuwan, V. Rukachaisirikul, S. Phongpaichit, S. Preedanon and J. Sakayaroj, Arch. Pharmacal Res., 2011, 34, 709–714 CrossRef CAS.
- L. Calcul, C. Waterman, W. S. Ma, M. D. Lebar, C. Harter, T. Mutka, L. Morton, P. Maignan, A. Van Olphen, D. E. Kyle, L. Vrijmoed, K. L. Pang, C. Pearce and B. J. Baker, Mar. Drugs, 2013, 11, 5036–5050 CrossRef CAS PubMed.
- T. K. M. Shing, K. H. Gibson, J. R. Wiley and I. F. Watt, Tetrahedron Lett., 1994, 35, 1067–1070 CrossRef CAS.
- B. H. Maskrey, G. W. Taylor and A. F. Rowley, J. Exp. Biol., 2006, 209, 558–566 CrossRef CAS PubMed.
- K. Gustafson, M. Roman and W. Fenical, J. Am. Chem. Soc., 1989, 111, 7519–7524 CrossRef CAS.
- M. A. M. Mondol, J. H. Kim, M. A. Lee, F. S. Tareq, H. S. Lee, Y. J. Lee and H. J. Shin, J. Nat. Prod., 2011, 74, 1606–1612 CrossRef CAS.
- M. A. M. T. Mondol, F. Shahidullah, Ji H. Kim, M. ah Lee, H.-S. Lee, Y.-J. Lee, J. S. Lee and H. J. Shin, J. Nat. Prod., 2011, 74, 2582–2587 CrossRef CAS PubMed.
- M. A. M. Mondol, F. S. Tareq, J. H. Kim, M. A. Lee, H. S. Lee, J. S. Lee, Y. J. Lee and H. J. Shin, J. Antibiot., 2013, 66, 89–95 CrossRef PubMed.
- X. Xie, D. Meesapyodsuk and X. Qiu, Appl. Microbiol. Biotechnol., 2018, 102, 847–856 CrossRef CAS PubMed.
- K. T. Peng, C. N. Zheng, J. Xue, X. Y. Chen, W. D. Yang, J. S. Liu, W. B. Bai and H. Y. Li, J. Agric. Food Chem., 2014, 62, 8773–8776 CrossRef CAS PubMed.
- Q. H. Zhang, C. H. You, F. Liu, W. D. Zhu, S. Q. Wang, D. Z. Xie, O. Monroig, D. R. Tocher and Y. Y. Li, Lipids, 2016, 51, 1051–1063 CrossRef CAS.
- M. Vagner and E. Santigosa, Aquaculture, 2011, 315, 131–143 CrossRef CAS.
- R. Vaezi, J. A. Napier and O. Sayanova, Mar. Drugs, 2013, 11, 5116–5129 CrossRef CAS PubMed.
- N. T. Carballeira, E. Janice, E. Ayanoglu and C. Djerassi, J. Org. Chem., 1986, 51, 2751–2756 CrossRef CAS.
- F. Shahidi, Advances in Seafood Byproducts, 2002 Conference Proceedings, 2003, pp. 247–263 Search PubMed.
- T.-T. T. Li, Ai-Jun, Y.-Y. Liu, Zi-R. Huang, Xu-Z. Wan, Yu-Y. Pan, R.-B. Jia, B. Liu, X.-H. Chen and C. Zhao, Food Chem. Toxicol., 2019, 131 Search PubMed.
- M. Wolters, J. Ahrens, M. Romani-Perez, C. Watkins, Y. Sanz, A. Benitez-Paez, C. Stanton and K. Gunther, Clinical nutrition, Edinburgh, Scotland, 2018, DOI:10.1016/j.clnu.2018.12.024.
- J. Bellenger, S. Bellenger, Q. Escoula, C. Bidu and M. Narce, Biochimie, 2019, 159, 66–71 CrossRef CAS PubMed.
- L. Costantini, R. Molinari, B. Farinon and N. Merendino, Int. J. Mol. Sci., 2017, 18, 18 Search PubMed.
- L. L. Ilag, Medicines, Basel, Switzerland, 2018, vol. 5 Search PubMed.
- E. Tvrzicka, L. S. Kremmyda, B. Stankova and A. Zak, Biomed. Pap., 2011, 155, 117–130 CrossRef CAS PubMed.
- E. Kostetsky, N. Chopenko, M. Barkina, P. Velansky and N. Sanina, Mar. Drugs, 2018, 16 CrossRef CAS PubMed.
- S. H. Jonasdottir, Mar. Drugs, 2019, 17 CrossRef CAS.
- R. B. S. S. Nogueira, A. C. A. Tomaz, D. R. Pessoa, A. L. Xavier, J. C. L. R. Pita, M. V. Sobral, M. L. C. Pontes, H. L. F. Pessoa, M. F. F. M. Diniz, G. E. C. Miranda, M. A. R. Vieira, M. O. M. Marques, M. D. V. Souza and E. V. L. Cunha, Mar. Drugs, 2017, 15 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2019 |