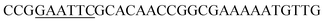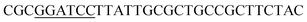Open Access Article
Open Access ArticleEnhanced extracellular recombinant keratinase activity in Bacillus subtilis SCK6 through signal peptide optimization and site-directed mutagenesis†
Jiewei Tian,
Xiufeng Long,
Yongqiang Tian * and
Bi Shi
* and
Bi Shi
Key Laboratory of Leather Chemistry and Engineering (Sichuan University), Department of Biomass and Leather Engineering, Ministry of Education, College of Biomass Science and Engineering (Sichuan University), Sichuan University, Chengdu 610065, PR China. E-mail: yqtian@scu.edu.cn
First published on 17th October 2019
Abstract
Keratinase has a great commercial value owing to its applications in the enzymatic dehairing of goatskins. In this study, we adopted a combined strategy to enhance the extracellular recombinant keratinase activity in Bacillus subtilis SCK6. First, nine signal peptides were screened to enhance the expression of extracellular keratinase. The recombinant strain with SPLipA exhibited the highest extracellular keratinase activity of 739.03 U per mL, which was two-fold higher activity of the wild type. Second, based on the multiple sequence alignment with the bacterial alkaline proteases, the mutant (M123L/V149I/A242N) was introduced into the keratinase. Comparing with the wild type of keratinase, the mutant M123L/V149I/A242N showed an increase in the extracellular keratinase activity, which was about 1.2-fold higher activity of the wild type. Finally, the keratinase expression vector with SPLipA and mutant M123L/V149I/A242N was constructed, and the extracellular keratinase activity reported at 830.91 U per mL was a 2.2-fold activity of the wild type. Then, the mutant keratinase was purified and characterized. The mutant exhibited properties similar to those of the wild type at an optimal temperature of 60 °C and pH 10.0. Conclusively, the extracellular expression of keratinase was enhanced via a combined strategy, and the mutant keratinase demonstrated properties similar to that of the wild type of keratinase.
Introduction
Keratinolytic proteases are a new generation of proteolytic enzymes with an ability to degrade the recalcitrant keratin proteins, such as feathers, horns, hooves, nails, and wool.1 These enzymes have been gaining importance in the last few years for several potential applications such as in dehairing of hides, textiles and keratin waste management, and association with the hydrolysis of keratinous substrates.2 Keratinolytic proteases are one of the excellent biocatalysts to hydrolyze the disulfide bond-rich proteins of hair and inflict little damage to leather. Biological treatment with keratinolytic proteases could largely reduce the quantity and toxicity of wastewater effluent from the leather industry.3Bacillus subtilis and Escherichia coli are the two major hosts for cloning and overexpression of alkaline proteases.4 B. subtilis is among the most widely used hosts for protein production in biotechnology owing to its high efficiency of secretion into the cell medium, high safety, clear inherited backgrounds and mature fermentation technology.5,6 The most commonly used method to enhance the extracellular expression of proteins is the optimization of the signal peptide. Yao et al.7 reported that the extracellular α-amylase activity with an optimal signal peptide (SPYojL) was 3.5-fold greater than that of the control after screening the 173 signal peptides of B. subtilis. Similarly, the α-amylase-producing strain with the best-performing signal peptide (SPpel) yielded a maximum of 1487.85 U per mL amylase after a 48 h cultivation, and it was about 68.4% higher than that of the strain with the native signal peptide.8 Similar results were found by Degering et al. when the signal peptides with the 173 signal peptides of Bacillus subtilis and the 220 signal peptides of Bacillus licheniformis were screened, and the subtilisin BPN-producing strain B. licheniformis H402 with the SPdBli00338 resulted in a 9-fold increase in activity in the medium supernatant when compared with the wild-type SP.9 Song et al. reported that the extracellular β-mannanase activity with the optimal signal peptide (SPLipA) and 72 h of fermentation was 533 ± 32 U per mL after the screening of four signal peptides of Sec pathway and two signal peptides of the Tat pathway.10 In addition, the optimization of the signal peptide was also done to enhance the extracellular production of proteins, such as L-asparaginase,11 β-galactosidase12 and xylanase.13
Although enhancing the extracellular production of protein by signal peptide optimization was feasible, it was difficult to predict the secretion efficiency of different proteins with the same signal peptide and the specific signal peptide for the secretion efficiency of specific proteins. Moreover, there were only a few reports to describe the change in the mature protein to enhance the extracellular expression of enzyme. Directed evolution and site-directed mutagenesis were the most commonly used methods for the modification. Yao et al. described that a recombinant B. subtilis containing an α-amylase with double mutation K82E/S405R showed an α-amylase activity, which was 2.1-fold greater than that of the wild type.7 Feng et al. reported that after the deletion of the N-terminal 25-residues, the activity of L-asparaginase with the signal peptide ASN was 100% higher than that of the intact.11 However, most of the mutants were constructed by improving the catalytic properties, such as activity,14 thermostability,15,16 cold adaptation17,18 and substrate specificity,19 with regard to the modification of the mature enzyme.
Previously, a keratinase from Bacillus sp. LCB12, heterologously expressed by B. subtilis SCK6, was purified and characterized. The characterization of the recombinant keratinase revealed that this enzyme was a serine protease with an optimal temperature of 60 °C and pH 10.0. The keratinase was used for the enzymatic dehairing of goatskins and exhibited attractive properties that might develop an efficient and eco-friendly enzymatic dehairing of animal skins and/or hides method in the leather processing industry.20 However, the extracellular expression of keratinase was low. In this study, the signal peptide optimization and site-directed mutagenesis were used for improving the extracellular recombinant keratinase activity.
Materials and methods
Bacterial strains, plasmids and media
All of the strains and plasmids used in this study are summarized in Table 1. For cloning, plasmid pMA0911 and Escherichia coli DH5α were used. Bacillus subtilis SCK6 was used as the heterologous expression host.21 Bacillus sp. LCB12 was previously isolated from alkaline soil.22 An LB-milk solid medium (tryptone 10 g L−1, yeast extract 5 g L−1, NaCl 10 g L−1, non-fat powdered milk 20 g L−1, agar 20 g L−1, pH 7.2–7.4) was used for the functional analysis of recombinant B. subtilis SCK6. The recombinant B. subtilis SCK6 was incubated in a Luria–Bertani medium (tryptone 10 g L−1, yeast extract 5 g L−1, NaCl 10 g L−1, agar 20 g L−1, pH 7.2–7.4) at 37 °C and 200 rpm for the production of heterologous keratinase. When applicable, antibiotics, such as 100 μg mL−1 ampicillin, 50 μg mL−1 kanamycin and 1 μg mL−1 erythromycin, were used.| Strains/plasmids | Properties | Reference |
|---|---|---|
| Strains | ||
| E. coli DH5α | F−, φ80, lacZ△M15, △(lacZYA-argF) U169 endA1, recA1, hsdR17(rk−, mk+) supE44, λ−, thi−1, gyrA96, relA1, phoA | Tiangen |
| B. subtilis SCK6 | ErmR, his, nprR2, nprE18, ΔaprA3, ΔeglS102, ΔbglT/bglSRV, lacA::PxylA-comK | BGSC 1A976 (ref. 21) |
| Bacillus sp. LCB12 | Wild type | 20 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||
| Plasmids | ||
| pMA0911-SPYwb N-keratinase | Kan+ (B. subtilis), Amp+ (E. coli), PhapII | This study |
| pMA0911-SPLip A-keratinase | Kan+ (B. subtilis), Amp+ (E. coli), PhapII | This study |
| pMA0911-SPAmy X-keratinase | Kan+ (B. subtilis), Amp+ (E. coli), PhapII | This study |
| pMA0911-SPWap A-keratinase | Kan+ (B. subtilis), Amp+ (E. coli), PhapII | This study |
| pMA0911-SPYnc M-keratinase | Kan+ (B. subtilis), Amp+ (E. coli), PhapII | This study |
| pMA0911-SPNpr E-keratinase | Kan+ (B. subtilis), Amp+ (E. coli), PhapII | This study |
| pMA0911-SPVpr-keratinase | Kan+ (B. subtilis), Amp+ (E. coli), PhapII | This study |
| pMA0911-SPYvg O-keratinase | Kan+ (B. subtilis), Amp+ (E. coli), PhapII | This study |
| pMA0911-SPYwe A-keratinase | Kan+ (B. subtilis), Amp+ (E. coli), PhapII | This study |
| pMA0911-keratinase | Kan+ (B. subtilis), Amp+ (E. coli), PhapII | This study |
Plasmid construction and transformation
The expression vector pMA0911-keratinase was previously constructed.20 The keratinase gene without the signal peptide was amplified from pMA0911-keratinase using the primers KF/KR (Table 2), and the PCR products were digested using restriction enzymes EcoR I and BamH I. Then, the expression vector pMA0911 with different signal peptides (SPYnc M, SPYwe A, SPNpr E, SPVpr, SPYvg O, SPYwb N, SPLip A, SPAmy X and SPWap A) was digested using the restriction enzymes EcoR I and BamH I. The purified keratinase gene fragment was inserted into the corresponding expression vector pMA0911 by using the Takara's DNA ligation kit (Takara, Dalian, China). The ligation products were transformed into E. coli DH5α cells. The positive clones were selected and amplified. Then, the plasmids were extracted and sequenced by Sangon Biotech (Shanghai, China). The recombinant plasmids were transformed into B. subtilis SCK6 according to the previous description.20 Then, the competent cells were spread on the LB-milk solid medium with kanamycin (50 μg mL−1) for the functional analysis of the recombinant B. subtilis SCK6.Site-directed mutagenesis
The site-directed mutagenesis of keratinase was performed as described by Jakob et al.23 The primers used in the site-directed mutagenesis are shown in Table 2. PCR was performed in a total volume of 50 μL PCR buffer containing 10 μL 5× EVO buffer, 10 μL 5× PCR enhancer, 1 μL dNTPs (10 mM each), 2 μL 100 μM each primer, 1 μL DNA template (pMA0911-keratinase ∼80 ng), 23 μL dd H2O, 1 μL Phanta EVO HS super-fidelity DNA polymerase. The amplification procedure was as follows: initial denaturation at 98 °C for 3 min, 20 cycles of denaturation at 98 °C for 15 s, annealing at 60 °C for 15 s, and extension at 72 °C for 4.5 min. The PCR products were examined on a regular 1% agarose gel and digested with Dpn I at 37 °C for 1 h. The enzyme digestion products were transformed into E. coli DH5α cells. The positive clones were selected and amplified. Then, the plasmids were extracted and sequenced by Sangon Biotech (Shanghai, China). The recombinant plasmids were transformed into B. subtilis SCK6.Production and purification of keratinase and its mutants
For the production of keratinase, the recombinant B. subtilis SCK6 was inoculated into the LB solid medium for 12 h at 37 °C. The single colony was picked up and cultured in the LB liquid medium for 12 h at 37 °C as seed cells. Then, the seed cells were inoculated (2%, v/v) into the LB liquid medium for 48 h at 37 °C.| Signal peptide | Amino acid sequence | Charge N-regionb | Hydrophobic amino acidc |
|---|---|---|---|
| a The cleavage sites of signal peptides were underlined.b The net charge of the N-region was calculated with the amino acids aspartate and glutamate defined as −1; arginine and lysine defined as +1 and any other amino acid defined as 0.c The hydrophobic amino acids of each signal sequence was calculated with amino acids G, A, V, L, I, M, F, W and P defined as hydrophobic; any other amino acid being characterized as hydrophilic. | |||
| Ywb N | 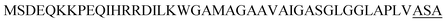 |
2 | 29 |
| Ync M | 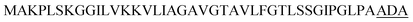 |
4 | 32 |
| Lip A | 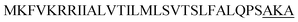 |
4 | 19 |
| Ywe A | 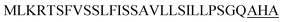 |
2 | 18 |
| Amy X | 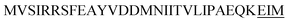 |
1 | 16 |
| Npr E | 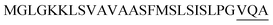 |
2 | 19 |
| Vpr | 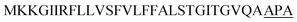 |
3 | 23 |
| Yvg O | 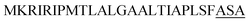 |
3 | 19 |
| Wap A | 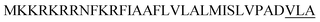 |
8 | 21 |
The fermentation broths were centrifuged at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm at 4 °C for 30 min to obtain a supernate. The supernate was precipitated by the gradient ammonium sulphate fractionation. The supernate was initially precipitated by ammonium sulfate at 30% saturation (4 °C) to remove other proteins. Then, the supernate was precipitated by ammonium sulfate at 50% saturation (4 °C) to obtain the precipitated crude enzyme. The buffer A (50 mM HAc–NaAc, 20 mM NaCl, pH 5.0) was used to resuspend the crude enzyme, and the insoluble substances were removed via centrifugation. The supernatant was loaded on an SP-sepharose fast flow column that was pre-equilibrated with buffer A. The proteins were separated by a linear gradient of NaCl 0–1 M. The active fractions were pooled and stored at −20 °C for further analysis.
000 rpm at 4 °C for 30 min to obtain a supernate. The supernate was precipitated by the gradient ammonium sulphate fractionation. The supernate was initially precipitated by ammonium sulfate at 30% saturation (4 °C) to remove other proteins. Then, the supernate was precipitated by ammonium sulfate at 50% saturation (4 °C) to obtain the precipitated crude enzyme. The buffer A (50 mM HAc–NaAc, 20 mM NaCl, pH 5.0) was used to resuspend the crude enzyme, and the insoluble substances were removed via centrifugation. The supernatant was loaded on an SP-sepharose fast flow column that was pre-equilibrated with buffer A. The proteins were separated by a linear gradient of NaCl 0–1 M. The active fractions were pooled and stored at −20 °C for further analysis.
SDS-PAGE analysis
The fermented centrifugal supernatant sample was precipitated by 0.4 M trichloroacetic acid, and then washed using precooled ethyl alcohol absolute. The SDS-PAGE was performed in a 12.5% (v/v) polyacrylamide gel (Baihe technology, Chengdu, China).Enzyme activity assays
The keratinase activity was determined using the keratin obtained from wool as the substrate, and the reactions were proceeded in accordance with the previous description.20 One unit of keratinase activity was defined as the amount of enzyme that caused the release of 1 μg tyrosine per min at 60 °C.Effect of pH and temperature on the enzyme activity and stability
The optimum pH of keratinase and the mutant enzyme was measured using 50 mM glycine–NaOH (pH 8.5–12.0) and by using keratin as the substrate. In order to determine the optimal temperature of keratinase and the mutant enzyme, the samples were used to hydrolyze keratin at different temperatures ranging from 30 to 80 °C in 50 mM glycine–NaOH (pH 10.0).Results and discussion
Host and vector selection
B. subtilis and E. coli are the two major hosts used for cloning and overexpression of alkaline proteases.4 Compared with E. coli, B. subtilis showed higher advantages in producing proteins, such as its high efficiency of secretion into the cell medium (high product yields 20–25 g L−1), well-characterized secretion pathways, high safety (nonpathogenic and free of exotoxins and endotoxins), clear inherited backgrounds and mature fermentation technology.5,6,24,25Therefore, B. subtilis SCK6 (ErmR, his, nprR2, nprE18, ΔaprA3, ΔeglS102, ΔbglT/bglSRV, lacA::PxylA-comK), a protease-deficient strain, was used as the host for keratinase production. The shuttle plasmid pMA0911 was used as the expression plasmid for keratinase production.20 The fragment of the target protein was inserted between the promoter PHpaII and T4 terminator, obtaining the recombinant expression plasmid pMA0911-keratinase. Subsequently, a series of recombinant expression plasmid pMA0911-keratinases with nine different signal peptides were constructed to facilitate the target protein secretion.
Optimization of the signal peptides for enhancing extracellular keratinase production
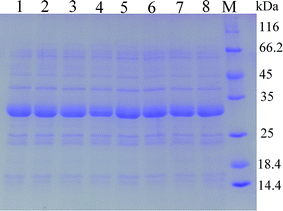
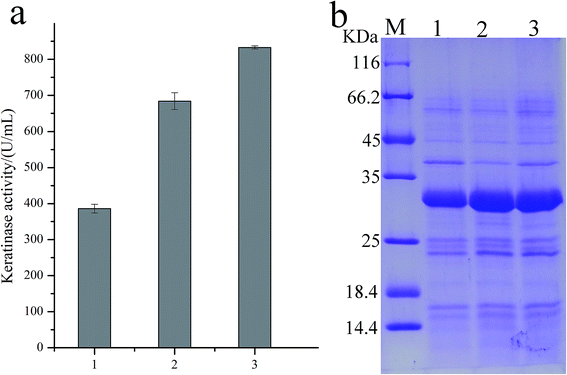
As predicted in a previous report,20 the open reading frame (ORF) of keratinase from Bacillus sp. LCB12 consisted of an N-terminal signal peptide (29 amino acids), the propeptide (76 amino acids) and the mature keratinase (274 amino acids) (Fig. S1†). There are four typical secretion pathways in B. subtilis, such as the general secretory (Sec) pathway, the twin-arginine translocation (Tat) pathway, the pseudopilin export pathway for competence development, and the pathways using the ATP-binding cassette (ABC) transporters, particularly the general secretory (Sec) pathway and the twin-arginine (Tat) translocation pathway. We had selected five signal peptides (SPYnc M, SPYwe A, SPNpr E, SPVpr and SPYvg O) from the Sec pathway and four signal peptides (SPYwb N, SPLip A, SPAmy X, and SPWap A) from the Tat pathway for improving the extracellular expression of keratinase (Table 3). The keratinase gene fragment without itself signal peptide from Bacillus sp. LCB12. Then, the recombinant expression plasmids were transformed into the B. subtilis SCK6, and the recombinant strains were incubated in the LB–milk solid medium for the functional analysis of the recombinant strains. After 12 h of cultivation at 37 °C, five recombinant strains with different signal peptides (SPNpr E, SPVpr, SPYwe A, SPLip A and SPWap A) showed clear hydrolysis ring of milk on the plate (Fig. S2†).Then, the five recombinant strains were incubated into the LB liquid medium. After 48 h of fermentation, the extracellular keratinase activity was determined. SDS-PAGE was used for the analysis of the extracellular production of keratinase, and the prominent bands corresponding to the size of 30 kDa were observed, which were consistent with the theoretical molecular weight of the mature keratinase (Fig. 1). Among the three Sec pathway signal peptides, SPYwe A was the most efficient one with a secretion efficiency of 380.19 U per mL, slightly higher than the wild type, while the other signal peptides (SPNpr E and SPVpr) showed a low secretion efficiency (Fig. 2). Moreover, SPLip A and SPWap A showed high secretion efficiency and SPLip A showed the highest extracellular keratinase activity with 739.03 U per mL, a 1.95-fold higher activity of wild-type (Fig. 3). SDS-PAGE results of the supernatant samples also showed that the two the Tat-dependent signal peptides, SPLip A and SPWap A, could efficiently secrete keratinase into the culture medium (Fig. 1). Thus, we concluded that keratinase tended to be highly secreted under the direction of Tat-pathway signal peptide SPLip A.
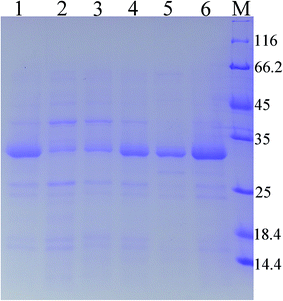 | ||
| Fig. 1 SDS-PAGE analysis of extracellular keratinase of the recombinant strains with different signal peptide. 1: wild-type; 2: SPNpr E; 3: SPVpr; 4: SPYwe A; 5: SPLip A; 6: SPWap A; M: marker. | ||
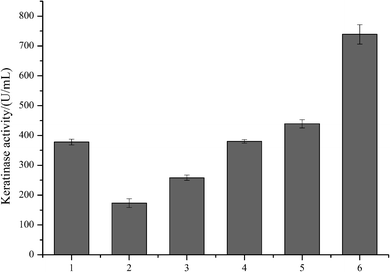 | ||
| Fig. 2 The extracellular keratinase activities of the recombinant strains with different signal peptide. 1: wild-type; 2: SPNpr E; 3: SPVpr; 4: SPYwe A; 5: SPLip A; 6: SPWap A, M: marker. | ||
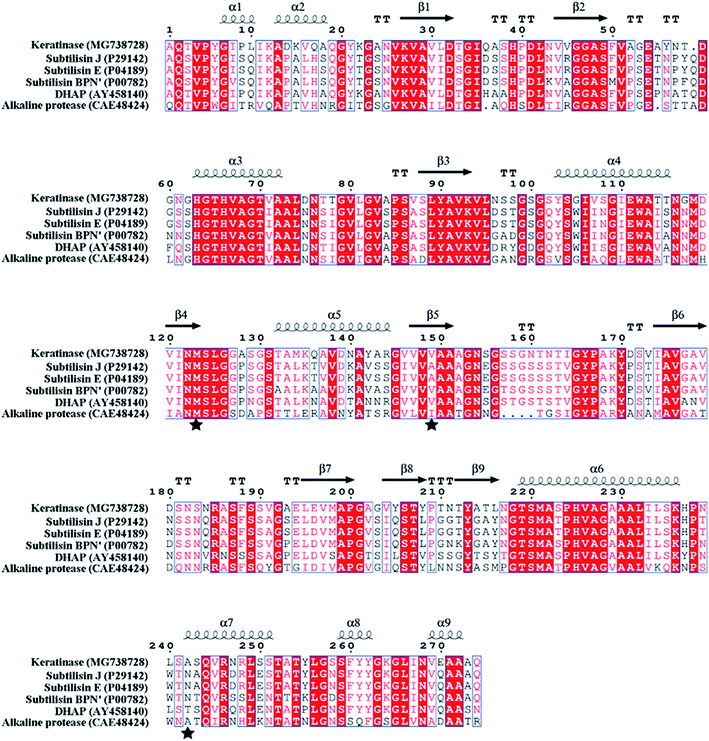 | ||
| Fig. 3 Protein sequence alignment of keratinase and other proteases by ESPript 3.0. “α” represents α helix; “β” represents β sheet; the substitution sites (M123, V149 and A242) are indicated by “★”. | ||
Site-directed mutagenesis for enhancing the extracellular recombinant keratinase activity
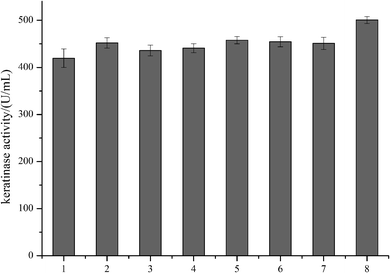
In order to modify the alkaline protease, various methods, such as DNA shuffling, site-directed mutagenesis based on the multiple sequence alignment and the directed evolution, were used.26–28 Based on the multiple sequence alignment of the bacterial alkaline proteases (Fig. 3), we constructed seven mutants (M123L, V149I, A242N, M123L/V149I, M123L/A242N, V149I/A242N, and M123L/V149I/A242N), and the recombinant strains were fermented at 37 °C for 48 h.The extracellular keratinase activity of each enzyme was evaluated at 60 °C. Fig. 4 shows that all of the variants have an increased extracellular keratinase activity compared with that of the wild-type enzyme. The extracellular activity of the double-site variants was higher than that of the single-site variants, and the three-site variants M123L/V149I/A242N showed the highest extracellular activity with 500.58 U per mL, a value ∼1.2-fold greater than that of the wild type. As shown in Fig. 5, there are no significant differences in extracellular expression between the mutants.
Then, the signal peptide optimization and the site-directed mutagenesis were combined to enhance the extracellular expression of keratinase. The pMA0911-keratinase with SPLip A and the three-site variants M123L/V149I/A242N was constructed. After 48 h of fermentation, the extracellular keratinase activity was determined, and SDS-PAGE was used for the analysis of the extracellular production of keratinase. As shown in Fig. 6, the recombinant strain demonstrates a secretion efficiency of 830.91 U per mL, about 2.2-fold greater than that of the wild-type. Besides, the production of keratinase was enhanced from 38.6 mg L−1 to 84.5 mg L−1.
In the previous studies, the mutants (M123L and V149I) showed a higher specific activity than that observed in the wild-type.27,28 M123 was conserved across the bacterial subtilisin and located on the fifth β-sheet, which was part of the substrate-binding pocket. The site V149 was closer to the active site in a 3D structure modeling of subtilisin BPN′ (1GNV), and the mutant V149I might influence the local flexibility of the substrate-binding pocket.17,29 The mutant A242 located at the seventh α-helix (242–251), exposed to the surface of keratinase molecule, and the mutant A242N increased the number of polar amino acid residues of the α-helix improved the flexibility of this region.
Biochemical characterization of keratinase mutant M123L/V149I/A242N
The wild-type keratinase and its variant with SPLip A and the three-site variants M123L/V149I/A242N were recombinantly produced in B. subtilis SCK6 and purified by ammonium sulfate precipitation and cation exchange chromatography. The optimum pH and the optimal temperature of keratinase and the mutant were determined.Fig. 7a shows the pH dependence of the proteolytic activity profiles of purified wild-type and the mutants (M123L/V149I/A242N) that are determined (pH range 8.5–12; 100 mM glycine–NaOH buffer). The wild type and mutant showed an equal pH dependence with a maximal activity at pH 10.0 and maintained high activity at higher pH (9–12). The effect of temperature on the keratinase activity of wild type and mutant was determined in the range of 30–90 °C. The wild type and mutant also showed a similar temperature dependence with a maximal activity at 60 °C and maintained high activity (>60%) in the range of 50 to 70 °C (Fig. 7b).
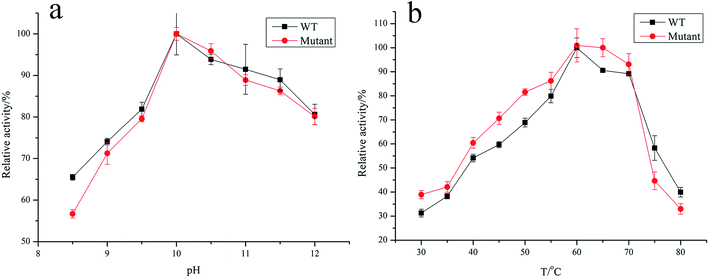 | ||
| Fig. 7 Effect of pH and temperature on the activity of keratinase and mutant. (a) Effect of pH on activity; (b) effect of temperature on activity. | ||
Conclusion
In this study, the signal peptide optimization and the site-directed mutagenesis were adopted to enhance the extracellular keratinase activity in Bacillus subtilis SCK6. In the process of signal peptide optimization, nine signal peptides were screened, among which SPLipA showed the highest extracellular keratinase activity of 739.03 U per mL, nearly two-fold activity of the wild type. Then, based on the multiple sequence alignment with bacterial alkaline proteases, three-site mutations (M123L/V149I/A242N) were introduced into the keratinase, and the mutant keratinase showed an increase in the extracellular keratinase activity, a 1.2-fold activity of the wild type. Finally, the signal peptide optimization and the site-directed mutagenesis were combined to improve the extracellular expression of keratinase. The keratinase expression vector with SPLipA and the mutant (M123L/V149I/A242N) was constructed and the extracellular keratinase of the B. subtilis arrived at 830.91 U per mL, a 2.2-fold activity of the wild type. Moreover, the production of keratinase was enhanced from 38.6 mg L−1 to 84.5 mg L−1. Then, the mutant keratinase was purified and characterized. The mutant keratinase exhibited properties similar to those of the wild type of keratinase.Conflicts of interest
No potential conflict of interest was reported by the authors.Acknowledgements
This study was supported by the National Key Research and Development Program of China (2017YFB0308401) and the Opening Project of Key Laboratory of Leather Chemistry and Engineering (Sichuan University), Ministry of Education (20826041C4159).References
- T. Paul, A. Jana, A. K. Mandal, A. Mandal, P. K. Das Mohpatra and K. C. Mondal, Sustainable Chem. Pharm., 2016, 3, 8–22 CrossRef CAS.
- R. Gupta and P. Ramnani, Appl. Microbiol. Biotechnol., 2006, 70(1), 21–33 CrossRef CAS.
- Z. Fang, Y. C. Yong, J. Zhang, G. C. Du and J. Chen, Appl. Microbiol. Biotechnol., 2017, 101(21), 7771–7779 CrossRef CAS.
- R. Gupta, Q. K. Beg and P. Lorenz, Appl. Microbiol. Biotechnol., 2002, 59(1), 15–32 CrossRef CAS.
- W. Cui, L. Han, F. Suo, Z. Liu, L. Zhou and Z. Zhou, World J. Microbiol. Biotechnol., 2018, 34(10), 145 CrossRef.
- Y. Gu, X. Xu, Y. Wu, T. Niu, Y. Liu, J. Li, G. Du and L. Liu, Metab. Eng., 2018, 50, 109–121 CrossRef CAS.
- D. Yao, L. Su, N. Li and J. Wu, Microb. Cell Fact., 2019, 18, 69 CrossRef.
- G. Fu, J. Liu, J. Li, B. Zhu and D. Zhang, J. Agric. Food Chem., 2018, 66, 13141–13151 CrossRef CAS.
- C. Degering, T. Eggert, M. Puls, J. Bongaerts and K. E. Jaeger, Appl. Environ. Microbiol., 2010, 76(19), 6370–6376 CrossRef CAS.
- Y. Song, G. Fu, H. Dong, J. Li, Y. Du and D. Zhang, J. Agric. Food Chem., 2017, 65, 2540–2548 CrossRef CAS.
- Y. Feng, S. Liu, Y. Jiao, H. Gao, M. Wang, G. Du and J. Chen, Appl. Microbiol. Biotechnol., 2017, 101(4), 1509–1520 CrossRef CAS.
- G. Ren, L. Cao, W. Kong, Z. Wang and Y. Liu, J. Agric. Food Chem., 2016, 64, 5708–5716 CrossRef CAS.
- W. Zhang, M. Yang, Y. Yang, J. Zhan, Y. Zhou and X. Zhao, Appl. Microbiol. Biotechnol., 2016, 100(20), 8745–8756 CrossRef CAS.
- J. Kim, J. H. Kim, K. H. Choi, J. H. Kim, Y. S. Song and J. Cha, J. Agric. Food Chem., 2011, 59(16), 8675–8682 CrossRef CAS PubMed.
- Z. Fang, J. Zhang, B. Liu, G. Du and J. Chen, Microb. Biotechnol., 2016, 9(1), 35–46 CrossRef CAS PubMed.
- R. Huang, Q. Yang and H. Feng, Acta Biochim. Biophys. Sin., 2015, 47(2), 98–105 CrossRef CAS.
- C. Zhong, S. Song, N. Fang, X. Liang, H. Zhu, X. Tang and B. Tang, Biotechnol. Bioeng., 2010, 104(5), 862–870 CrossRef.
- H. Zhao and F. Hong, BMC Biotechnol., 2018, 18(1), 34 CrossRef.
- B. Ruan, V. London, K. E. Fisher, D. T. Gallagher and P. N. Bryan, Biochemistry, 2008, 47(25), 6628–6636 CrossRef CAS.
- J. Tian, Z. Xu, X. Long, Y. Tian and B. Shi, High-expression keratinase by Bacillus subtilis SCK6 for enzymatic dehairing of goatskins, Int. J. Biol. Macromol., 2019, 135, 119–126 CrossRef CAS.
- X. Zhang and Y. P. Zhang, Microb. Biotechnol., 2011, 4(1), 98–105 CrossRef.
- J. Tian, Z. Lei, Y. Liu, P. Qiu, L. Wang and Y. Tian, J. Pure Appl. Microbiol., 2013, 7, 2531–2540 CAS.
- F. Jakob, R. Martinez, J. Mandawe, H. Hellmuth, P. Siegert, K. H. Maurer and U. Schwaneberg, Appl. Microbiol. Biotechnol., 2013, 97(15), 6793–6802 CrossRef CAS.
- H. Dong and D. Zhang, Microb. Cell Fact., 2014, 13, 63 CrossRef PubMed.
- Y. Song, J. M. Nikoloff and D. Zhang, J. Microbiol. Biotechnol., 2015, 25(7), 963–977 CrossRef CAS.
- J. E. Ness, M. Welch, L. Giver, M. Bueno, J. R. Cherry, T. V. Borchert, W. P. Stemmer and J. Minshull, Nat. Biotechnol., 1999, 17(9), 893–896 CrossRef CAS.
- H. Y. Zhao, L. Y. Wu, G. Liu and H. Feng, Biosci., Biotechnol., Biochem., 2016, 80(12), 2480–2485 CrossRef CAS.
- R. Martinez, F. Jakob, R. Tu, P. Siegert, K. H. Maurer and U. Schwaneberg, Biotechnol. Bioeng., 2013, 110(3), 711–720 CrossRef CAS.
- O. Almog, D. T. Gallagher, J. E. Ladner, S. Strausberg, P. Alexander, P. Bryan and G. L. Gilliland, J. Biol. Chem., 2002, 277(30), 27553–27558 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9ra07866e |
| This journal is © The Royal Society of Chemistry 2019 |