Open Access Article
Open Access ArticleThe hydroxy-analogue of selenomethionine alleviated lipopolysaccharide-induced inflammatory responses is associated with recover expression of several selenoprotein encoding genes in the spleens of Kunming mice†
Jia-Yong Tang‡
ab,
Long-Qiong Wang‡ab,
Gang Jiaab,
Guang-Mang Liu ab,
Xiao-Ling Chenab,
Gang Tianab,
Jing-Yi Caiab,
Hai-Ying Shangab and
Hua Zhao
ab,
Xiao-Ling Chenab,
Gang Tianab,
Jing-Yi Caiab,
Hai-Ying Shangab and
Hua Zhao *ab
*ab
aAnimal Nutrition Institute, Sichuan Agricultural University, No. 211 Huimin Road, Chengdu, Sichuan 611130, China. E-mail: zhua666@126.com; Fax: +86-28-8629-0922; Tel: +86-28-8629-0922
bKey Laboratory of Animal Disease-resistant Nutrition, Sichuan Province, China
First published on 6th December 2019
Abstract
This study aimed to determine whether hydroxy-analogue of selenomethionine (HMSeBA) supplementation could alleviate LPS-induced immunological stress in mice. A total of 90 Kunming mice were randomly assigned into 5 groups. The CON-LPS and CON+LPS groups were fed basal diet (BD), the others were fed BD with different levels of HMSeBA (0.15, 0.30 and 0.45 mg Se per kg) for 4 weeks. Mice were injected with LPS (3 mg per kg BW) or the corresponding physiological saline at 14 d and 28 d. Plasma and spleens were collected at 28 d. The results showed that: (1) LPS injection decreased ADG of mice at the 3rd week, and increased the concentration of IL-6 and TNF-α in plasma and the spleen index; (2) LPS injection induced immunological stress, up-regulated 8 inflammation-related genes and 3 selenoprotein encoding genes, and down-regulated 16 selenoprotein encoding genes in spleens; (3) compared with the CON+LPS group, HMSeBA supplementation increased ADG of mice at 3 weeks and GSH-Px activity in plasma and spleens, decreased spleen index and plasma IL-6 and TNF-α levels, down-regulated mRNA levels of COX-2, ICAM-1, TNF-α, IL-6, and MCP-1, and up-regulated IL-10 and iNOS in spleens. 0.30 mg Se per kg of HMSeBA exhibited the optimal protective effect; (4) HMSeBA supplementation modestly recovered the expression of 8 selenoprotein encoding genes in the spleens of the stressed mice. The results indicated that HMSeBA supplementation alleviated LPS-induced immunological stress accompanied up-regulation of a subset of selenoprotein encoding genes in spleens of mice.
Introduction
Inflammation is a response of an organism to injury caused by physical or chemical noxious stimuli or microbiological toxins,1 such as heavy metal-caused inflammation in chickens.2,3 Inflammatory response is a defensive reaction that helps the body to inactivate or destroy invading organisms, remove irritants, and set the stage for tissue repair.4 However, excessive inflammation with the production of many inflammatory cytokines depresses the growth performance of an animal.5 Decrease in the release or expression of pro-inflammatory mediators can alleviate inflammatory responses in animals or cells. Lipopolysaccharide (LPS) is a potent inducer of inflammation,6,7 which stimulates host cells to produce various pro-inflammatory cytokines such as tumor necrosis factor alpha (TNF-α), interleukin (IL)-6 and IL-1β.8,9 LPS has been commonly used for the evaluation of anti-inflammatory agents in immune cells or animal models.10–13Selenium (Se) is an essential micronutrient that plays an important role in the regulation of inflammation and immunity in neutrophils, NK cells, B lymphocytes and T cells.14–16 Supplementation of Se alleviates the LPS-induced immunological stress in MMECs and RAW264.7 cells through inhibiting expression of pro-inflammatory cytokines.11,17 Se exerts its biological function as selenoproteins in the form of amino acid selenocysteine (Sec).18,19 Many selenoproteins have been involved in innate and adaptive immune responses.14 Previous studies have revealed that the expression of selenoprotein encoding genes are affected by immunological stress and Se pretreatment alleviates LPS-induced immunological stress in murine macrophages through restoring expressions of a small subset of selenoprotein encoding genes.11,15,20 However, the role of dietary Se in the regulation of selenogenome transcription in the immune tissues in response to inflammation remains unclear.
It is well known that organic Se is better bioactive than the inorganic one in improving the growth, survival, and Se content in animals.21,22 Hydroxy-analogue of selenomethionine (HMSeBA) is a new stable organic Se-containing compound available. Various experiments have demonstrated that HMSeBA exhibited a more efficiency in Se deposition in pigs and laying hens, compared with selenite and Se-yeast.23–25 However, whether HMSeBA has the anti-inflammatory effect in LPS-stimulated immunological stress remains unclear. Therefore, Kunming mice were used in this study to determine (1) the protective effect of dietary supplementation of HMSeBA on alleviating LPS-induced inflammatory responses in mice; (2) the possible mechanism linked the protective effect of HMSeBA to selenoproteins expression profiles in immune organ.
Materials and methods
Animals, diet and experimental design
The animal protocol was approved by the Animal Care Office of Sichuan Agricultural University, Chengdu, China, and the animal experimental procedure was performed according to the guidelines for the care and use of experimental animal established by the Ministry of Agriculture of People's Republic of China. A total of 90 male SPF Kunming mice (21 days of age, Dashuo Experimental Animal Co., Ltd., Chengdu, China) with body weight (BW) of 9.5–11.0 g were fed on a basal diet (BD, normal commercial feed for mice which supplemented 0.10 mg Se per kg in the form of selenite; Table 1) for 3 days, then the mice were randomly divided into 5 dietary treatments with 6 replicates and 3 mice per replicate. The mice were fed on a basal diet supplemented with 0.00, 0.00, 0.15, 0.30 and 0.45 mg per kg Se (HMSeBA, Adisseo Ltd., France) for 4 weeks. At day 14 and day 28, the mice were intraperitoneal injected with LPS (no. L2880, Sigma; 3 mg per kg BW) and the CON-LPS mice were injected with corresponding physiological saline. Mice were housed at facilities of individually ventilated cages (IVC, Suhang Technology Equipment Co., Ltd., Suzhou, China), in a 12 h light–12 h dark cycle. Room temperature and humidity were controlled (21 ± 2 °C and 50 ± 5%, respectively). During the entire protocol, food and water were available ad libitum. BW was measured weekly and average daily gain (ADG) was calculated.| Ingredients | Content (%) | Nutrient component | Level |
|---|---|---|---|
| a The basal diet was a commercial diet which supplemented 0.1 mg kg−1 selenium in the form of selenite. | |||
| Corn | 25.0 | Water content (%) | 10.0 |
| Wheat | 30.0 | Protein (g/100 g) | 20.5 |
| Soybean | 13.0 | Fat (g kg−1) | 58.0 |
| Soybean meal | 5.0 | Fiber (%) | 2.6 |
| Fish meal | 7.0 | Ash (%) | 6.2 |
| Bran | 6.0 | Ca (%) | 1.34 |
| Others | 14.0 | P (g kg−1) | 0.98 |
Sample collection and preparation
At day 28, mice were fasted for 8 h overnight (0:00 to 8:00 AM) and then challenged with LPS or physiological saline. After 6 hours of LPS challenge, 6 mice of the average body weight per replicate were selected and anaesthetized (isoflurane–N2O–O2) and killed by bleeding (eye extraction) and cervical dislocation. Plasma and spleens were prepared according to methods previously described by our group, and stored at −80 °C until used.26–29 The spleen index was calculated as the ration of spleen weight (mg) to body weight (g).Biochemical assays
Plasma proinflammatory cytokines IL-6 and TNF-α concentration were measured using corresponding assay (ELISA) kits (no. CRE0005 and CRE0003, 4A Biotech Co., Ltd., Beijing, China) according to the manufacturer's instructions. Spleen samples for biochemical analysis were prepared as previously described by our group.28,29 Glutathione peroxidase (GSH-Px), activity of superoxide dismutase (SOD), total antioxidant capability (T-AOC) and concentration of malondialdehyde (MDA) were measured using corresponding assay kits (no. A005-1-2, A001-1-2, A015-1-2, and A003-1-2, Jiancheng Bioengineering, Nanjing, China). Concentrations of protein were determined with the bicinchoninic acid (BCA) method (BCA protein assay kit, no. A045-3-2, Jiancheng Bioengineering, Nanjing, China). For each measurement, the compared samples were run on the same plate and experiments were performed in triplicate.Real-time quantitative PCR (qPCR) analysis of gene expression
Primers for 8 inflammation-related genes, 24 selenoprotein encoding genes, and 2 reference genes (β-ACTIN and GAPDH) were designed with Primer Express 3.0 (Applied Biosystems, USA) and are presented in ESI Table 1.† β-ACTIN was used to normalize target gene transcript levels. Total RNA of spleen was extracted by using Trizol Reagent (no. 15596018, Invitrogen, CA, USA) and cDNA was synthesized using the PrimeScript RT reagent kit (no. RR047A, Takara, Dalian, China). qPCR was performed on QuantStudio 6 Flex system (Applied Biosystems, USA) using SYBR Premix Ex Taq™ II kit (no. RR820A, TaKaRa, Dalian, China) as described previously.11,26–29Statistical analysis
Data are presented as means ± SE. Statistical analysis was performed using SPSS for Windows (Version 13.0, Chicago, IL, USA), values were analyzed using one-way ANOVA followed by Duncan's multiple range test. P-values of less than 0.05 were considered statistically significant.Results
Body weights gain and spleen index of mice
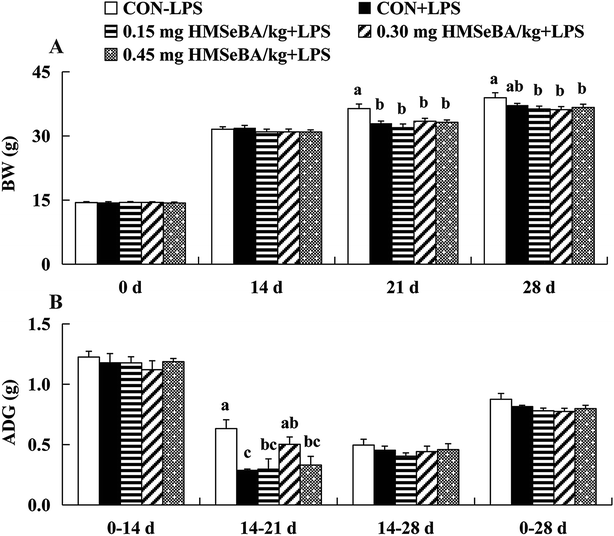
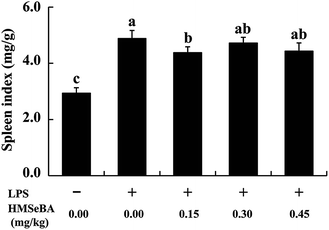
Mice fed on BD diet or BD supplied with three levels of HMSeBA had similar BW at 0 and 14 days, LPS injection depressed (P < 0.05) BW of mice at 21 days and HMSeBA supplementation did not improved the BW of mice after 1 week of LPS stimulation. It seems the effect of LPS on BW lasted 2 weeks, each LPS treatment group (including HMSeBA groups) exhibited lower (P < 0.05) BW than that of the CON-LOP group at 28th days, and HMSeBA supplementation did not improved BW of mice under LPS-induced stress (Fig. 1A). LPS (3 mg per kg BW) injection at 14 d shortly decreased (P < 0.05) ADG of mice during 14–21 d, while addition of HMSeBA alleviated the reduction of ADG affected by LPS at this stage, and 0.30 mg Se per kg HMSeBA exhibited (P < 0.05) good result (Fig. 1B). The mice of each group had similar ADG during 0–14 d, 14–28 d and 0–28 d. Compared with the CON-LPS group, the injection of LPS at 14 days significantly increased (P < 0.05) the spleen index of the mice at 28 d. Dietary HMSeBA supplementation alleviated the stimulation effect of LPS on spleen index and 0.15 mg Se per kg HMSeBA group exhibited lower (P < 0.05) spleen index than that of the CON+LPS group (Fig. 2).Plasma pro-inflammatory cytokine levels
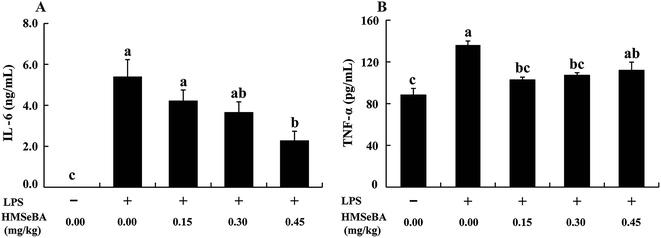
To determine whether LPS challenge effectively induces inflammatory response in mice and the protective effect of additional organic Se addition, we further investigated two pro-inflammatory cytokine levels in plasma (Fig. 3). Mice secreted a barely detectable amount of IL-6, whereas LPS stimulation significantly increased (P < 0.05) the concentration of IL-6 in plasma. Compared with the CON+LPS group, the addition of HMSeBA decreased plasma IL-6 levels in a dose dependent manner in stressed mice, and 0.45 mg Se per kg HMSeBA group exhibited lower (P < 0.05) plasma IL-6 (Fig. 3A). Similar results were found that LPS injection sharply increased (P < 0.05) plasma TNF-α, 0.15 and 0.30 mg Se per kg dietary HMSeBA supplementation effectively reduced (P < 0.05) plasma TNF-α induced by LPS (Fig. 3B). The results indicated additional organic Se in form of HMSeBA effective alleviates LPS-induced immunological response in mice.Expression of inflammation-related genes in spleen
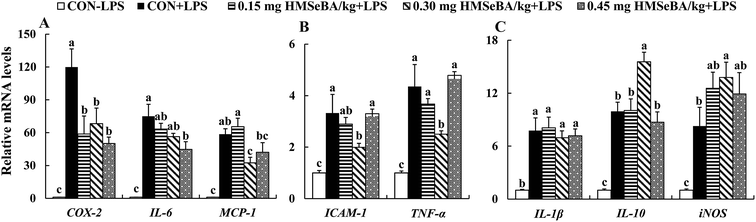
The mRNA expression of 8 inflammation-related genes (COX-2, ICAM-1, IL-1β, IL-6, IL-10, MCP-1, TNF-α and iNOS) in spleens were determined (Fig. 4). Compared with the CON-LPS group, LPS treatment up-regulated (P < 0.05) mRNA levels of 8 inflammation-related gene. LPS induced the up-regulation of inflammation-related genes in spleens can be modulated by additional dietary HMSeBA supplementation, which effectively reduced (P < 0.05) the effect of LPS on expression of COX-2, IL-6 and MCP-1 (Fig. 4A) in a dose dependent manner, decreased (P < 0.05) ICAM-1 and TNF-α (Fig. 4B) at 0.30 mg Se per kg dietary HMSeBA level, and further increased (P < 0.05) expression of IL-10 and iNOS (significant at level of 0.30 mg Se per kg) in spleen of the immunological stressed mice (Fig. 4C). In general, additional HMSeBA supplementation alleviates the LPS induced immunological stress and 0.30 mg Se per kg dietary HMSeBA exhibited the optimal protective effect in spleens of the mice.Expression of selenoprotein encoding genes in spleen
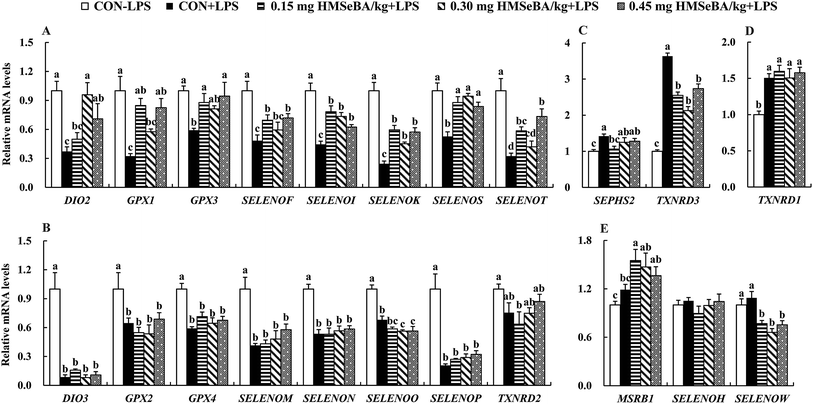
We further investigated expression of 24 selenoprotein encoding genes in spleens of the mice. As expected LPS challenge greatly affected expression of selenoprotein encoding genes in spleens, which globally decreased (P < 0.05) mRNA levels of 16 selenoprotein encoding genes (DIO2, DIO3, GPX1, GPX2, GPX3, GPX4, SELENOF, SELENOI, SELENOK, SELENOM, SELENON, SELENOO, SELENOP, SELENOS, SELENOT and TXNRD2) (Fig. 5A and B), further up-regulated (P < 0.05) 3 selenoprotein encoding genes (SEPHS2, TXNRD1 and TXNRD3) (Fig. 5C and D) and exhibited no effect on expression of MSRB1, SELENOH and SELENOW (Fig. 5E).Additional dietary HMSeBA supplementation exhibited impact on expression of selenoprotein encoding genes in spleens of mice under immunological stress. Firstly, dietary HMSeBA supplementation (0.15, 0.30 and 0.45 mg Se per kg) moderately recovered (P < 0.05) expression of 8 selenoprotein encoding genes (DIO2, GPX1, GPX3, SELENOF, SELENOI, SELENOK, SELENOS and SELENOT) (Fig. 5A), while showed limited effect (P > 0.05) on expression of DIO3, GPX2, GPX4, SELENOM, SELENON and SELENOP or even further decreased (P < 0.05) expression of SELENOO and TXNRD2 (Fig. 5B) in spleens of the stressed mice. Secondly, LPS up-regulated (P < 0.05) expression of selenoprotein encoding genes, while HMSeBA supplementation decreased or recovered (P < 0.05) their expression, those including SEPHS2 and TXNRD3 (Fig. 5C). Thirdly, LPS up-regulated (P < 0.05) expression of TXNRD3 and HMSeBA supplementation exhibited no effect (P > 0.05) on its expression (Fig. 5D). Finally, as described above, LPS exhibited no effect (P > 0.05) on expression of 3 selenoprotein encoding genes (MSRB1, SELENOH and SELENOW), and additional dietary HMSeBA further up-regulated (P < 0.05) MSRB1 and down-regulated (P < 0.05) SELENOW (Fig. 5E).
Antioxidant measurements
We investigated the effect of HMSeBA supplementation on antioxidant measurements in plasma and spleens of mice under immunological stress (Table 2). Compared with the CON-LPS group, LPS challenge reduced (P < 0.05) MDA content in plasma, while exhibited no effect on activities of GSH-Px, SOD and T-AOC. Dietary supplementation with 0.15 and 0.45 mg Se per kg HMSeBA increased (P < 0.05) plasma GSH-Px in mice faced with LPS, while had no effect (P > 0.05) on activities of SOD, T-AOC and concentration of MDA in plasma. It seems that LPS induced oxidative stress in spleen of mice, which decreased (P < 0.05) activities of GSH-Px, SOD and T-AOC, and increased (P < 0.05) MDA in spleens compared with the CON-LPS group. Dietary supplementation with 0.15 and 0.30 mg Se per kg HMSeBA recovered (P < 0.05) activity of GSH-Px in spleens of mice challenged with LPS. As for the SOD, T-AOC and MDA, additional dietary HMSeBA supplementation exhibited limited impact (P > 0.05) on these parameters in spleens of mice under LPS-induced immune stress.| Parameters | Treatments | ||||
|---|---|---|---|---|---|
| CON-LPS | CON+LPS | 0.15 mg HMSeBA per kg + LPS | 0.30 mg HMSeBA per kg + LPS | 0.45 mg HMSeBA per kg + LPS | |
| a Value are means ± SE (n = 6), a,b,cmeans within each item differ (P < 0.05) without sharing a common superscript letter. | |||||
| Plasma | |||||
| GSH-Px (U mL−1) | 1013.3 ± 149.9b | 1045.6 ± 88.4b | 1193.1 ± 84.3a | 949.7 ± 122.8b | 1232.8 ± 79.3a |
| SOD (U mL−1) | 23.7 ± 3.8 | 24.1 ± 3.2 | 25.3 ± 1.7 | 23.2 ± 2.9 | 25.9 ± 1.8 |
| T-AOC (U mL−1) | 2.8 ± 0.7 | 2.2 ± 0.4 | 2.1 ± 0.4 | 2.1 ± 0.4 | 2.6 ± 1.1 |
| MDA (nmol mL−1) | 9.9 ± 2.1a | 6.8 ± 0.7b | 5.7 ± 0.5b | 6.4 ± 0.9b | 7.0 ± 1.4b |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||||
| Spleen | |||||
| GSH-Px (U per mg prot.) | 1621.8 ± 152.4a,b | 1422.0 ± 87.1c | 1576.5 ± 76.7b | 1765.9 ± 69.9a | 1556.3 ± 128.5b,c |
| SOD (U per mg prot.) | 7.7 ± 1.2a | 5.2 ± 1.1b | 5.3 ± 0.8b | 5.3 ± 0.7b | 4.8 ± 0.4b |
| T-AOC (U per mg prot.) | 4.1 ± 0.8a | 1.0 ± 0.2b,c | 0.7 ± 0.1c | 1.0 ± 0.2b,c | 1.5 ± 0.3b |
| MDA (nmol per mg prot.) | 1.6 ± 0.3b | 1.8 ± 0.2a | 1.8 ± 0.2a | 2.0 ± 0.2a | 1.8 ± 0.2a |
Discussion
Se is an essential dietary nutrient involved in immune regulation,30,31 and can alleviate toxic substance-caused inflammation of immune organs in animals.32 Our previous study indicated that Se supplementation suppresses LPS-induced inflammatory responses in RAW264.7 cells.11 LPS has been recognized as a strong inflammatory inducer.12,13 Here, we used LPS as an immune stress model in rodent and evaluated the protective effect of HMSeBA. LPS challenge decreases the ADG and average dietary feed intake (ADFI) of broiler and piglet.33,34 As expected, intraperitoneal injection of LPS in mice jeopardized the growth of mice with reduction of ADG, which was effectively prevented by dietary HMSeBA supplementation at 0.30 mg Se per kg (Fig. 1). Previous studies have shown that Se-enriched forage improves the growth rate in weaned beef calves and overdoes of Se enhances immune function.35,36 Our present study suggested that animal requires a greater Se supplementation to optimize the immune function in stress condition, compared with normal Se requirement. Immune stress induces edema, necrosis in immune organs, thus suppresses immune function of animals. Studies have shown that LPS or other immune challenges increase immune organs index, such as the spleen index in broiler.37 Lymphocyte proliferations may be responsible for the increase in immune organ index.38 LPS injection increased mouse spleen index, while dietary overdose of HMSeBA reduced the spleen index of mice (Fig. 2), indicating that HMSeBA supplementation attenuates LPS-induced inflammatory response in mice.IL-6 and TNF-α are important pro-inflammatory cytokines, used as a hallmark for evaluation of inflammatory responses.8,9,39 Consistent with previous studies in piglet,34 rat and immune cell models,11,40,41 LPS injection increased IL-6 and TNF-α levels in the plasma of mice, while dietary HMSeBA inhibited their production in plasma (Fig. 3), which agreed with that Se supplementation inhibits the expression of inflammatory factors IL-1β, IL-6 and TNF-α induced by staphylococci.42 Thus, HMSeBA protected Kunming mice from LPS-induced endotoxin shock.
Spleen is the important immune organs in animal. We measured expressions of 8 inflammation-related genes in the spleen, and found all these genes were up-regulated in response to LPS stimulation (Fig. 4). Similar results are reported in macrophages.11,43 In this study, extra dietary Se supplementation modestly suppressed expressions of 5 inflammation-related genes (COX-2, IL-6, MCP-1, ICAM-1 and TNF-α) in the spleens of stressed mice (Fig. 4A and B). As a hallmark for inflammatory responses, IL-6 modulates host defense and induces the acute phase response.44 High levels of TNF-α cause organ damage and induce oxidative stress.45 COX-2 mediates the production of various prostaglandins,46 which regulate inflammation response. ICAM-1, belonged to the immunoglobulin superfamily, is up-regulated in response to a variety of inflammatory mediators.47 The down-regulation of these inflammation-related genes suggests the protective effect of overdoes of Se.11 It has been reported the addition of Se ameliorates Staphylococcus aureus-induced inflammation in bovine mammary epithelial cells by inhibiting the expression of pro-inflammatory factors (IL-1β, IL-6, IL-8 and TNF-α).48 Se also exhibited impact on expression of anti-inflammatory genes. As an anti-inflammatory gene, IL-10 inhibits the release of immune mediator by monocytes/macrophages and the production of pro-inflammatory cytokines.49 Therefore, the up-regulation of IL-10 (Fig. 4C) may contribute to enhance the anti-inflammatory capacity of the host.50 In general, 0.30 mg Se per kg HMSeBA exhibited optimal protective effect based on above described results.
Se regulates immune function in the form of selenoproteins,51 which involved in the activation, proliferation, and differentiation of cells that drive immune responses.15 We further investigated mRNA abundance of 24 selenoprotein encoding genes in spleens. Consist with previous finding,11,52 LPS treatment down-regulated 16 selenoprotein encoding genes (Fig. 5) in parallel with significant increases in expressions of 8 inflammation-related genes (Fig. 4). Supplementation of HMSeBA moderately recovered expressions of 8 selenoprotein encoding genes (DIO2, GPX1, GPX3, SELENOF, SELENOI, SELENOK, SELENOS and SELENOT) in the spleens of mice against LPS stimulation (Fig. 5A). Among the affected genes, GPX1 is the most expressed selenoprotein genes in specific immune cells or organs,30 which involves in immune homeostasis and inflammatory responses.30,53 Mice with GPX1 knockout display senescent-like features and are susceptible to H2O2-mediated cell death.54 GPX1 is sensitive to Se levels that the increase in supplementation of Se enhances its activity in mouse T cells and human lymphocytes.55–57 Enhanced GPX3 expression has the potential to protect organs from oxidative stress through removing hydrogen peroxide and other oxygen free radicals.58 Our results showed that LPS injection suppressed the antioxidant capacity in plasma and spleens of mice while HMSeBA reinforces the GSH-Px activity (Table 2). The up-regulation of GPX1 and GPX3 was consistent with the recovered GSH-Px activity in spleen and plasma (Table 2), while the deficiency in GPX activity affects the immune response to infection.59 SELK, SEPS1, and SELT are endoplasmic reticulum (ER) transmembrane proteins that involves in inflammatory response. SELK regulates ER homeostasis and SELENOK gene knockout impairs immune responses.60,61 SEPS1 influences cytokine production in macrophage cells and SELENOS RNAi increased IL-6 and TNF-α production in mice.62,63 SELT and SEPW1 belong to the same protein family and SELT deficiency elevates SEPW1 expression in murine fibroblast cells.64 Therefore, the recovered expression of those 8 selenoprotein encoding genes may indicate reinforced immunity by extra dietary Se (Fig. 5A). The roles of DIO2, SELENOF and SELENOI in immune regulation are still unknown. Up-regulation of DIO2, a selenoprotein encoding gene responsible for thyroid hormone regulation,65 indicates enhanced metabolism, which may help alleviate inflammatory response. LPS affected expression of selenoprotein encoding genes in the spleens, indicating dietary Se alleviates immunological stress mainly via up-regulation of a subset of selenoprotein encoding genes.11 LPS decreased expressions of 8 selenoprotein encoding genes (DIO3, GPX2, GPX4, SELENOM, SELENON, SELENOP, SELENOO and TXNRD2) in the spleens, while dietary HMSeBA supplementation exhibited limited impact on their expression (Fig. 5B).
Consistent with the up-regulation of TXNRD1 and TXNRD3 in murine macrophage upon LPS stimulation,11,66 SEPHS2, TXNRD1 and TXNRD3 were up-regulated by LPS in the spleens of mice (Fig. 5C and D). SEPHS2 is responsible for selenoprotein biosynthesis,67 and the up-regulation of this gene may indicate a demand of selenoprotein synthesis in spleen upon LPS stimulation. Thioredoxin reductase (TXNRD) enzymes play key roles in regulating redox tone or restoring oxidative damage.68,69 TXNRD1 is the most abundant selenoprotein in mouse macrophages,66 indicating its importance in immune regulation. Dietary HMSeBA supplementation resulted in the up-regulation of MSRB1 and the down-regulation of SELENOW, while LPS treatment did not affect expression of these genes. MSRB1 plays roles in protecting cells against oxidative damage,70,71 and the increases in its expression may help to resist inflammatory injury in immune organs.
Conclusion
LPS injection induced inflammatory responses in Kunming mice accompanied with the up-regulation expression of inflammation-related genes and down-regulation expression of selenoprotein encoding genes in spleens. Dietary supplementation with the new type of organic Se source (HMSeBA) exhibited immune protective effect on mice, which mainly through regulation expression of a subset selenoprotein encoding genes in spleens. Our results provide a new target for further exploration roles of these genes involving in immune response in animals.Author contributions
Hua Zhao and Jiayong Tang designed the research; Jiayong Tang and Longqiong Wang conducted the experiments; Gang Jia, Guangmang Liu, Xiaoling Chen, Gang Tian, Jingyi Cai, Haiying Shang and Hua Zhao collected sample and analyzed the data; Hua Zhao and Jiayong Tang wrote the paper; and Hua Zhao had primary responsibility for the final content. All authors read and approved the final manuscript.Abbreviations
| HMSeBA | Hydroxy-analogue of selenomethionine |
| LPS | Lipopolysaccharide |
| Se | Selenium |
| ADG | Average daily gain |
| BD | Basal diet |
| Sec | Selenocysteine |
| qPCR | Real-time quantitative PCR |
| ER | Endoplasmic reticulum |
| IVC | Individually ventilated cages |
| BW | Body weight |
| GSH-Px | Glutathione peroxidase |
| SOD | Superoxide dismutase |
| T-AOC | Total antioxidant capability |
| MDA | Malondialdehyde |
Conflicts of interest
The authors have declared that no conflict of interests exists.Acknowledgements
This work was supported partly by the Natural Science Foundation of China (No. 31772643 and 31272468), the Foundation of Educational Department of Sichuan (No. 17ZA0318), the Special Research Funding for Discipline Construction in Sichuan Agricultural University (No. 03570126), and the Sichuan Longda Animal Husbandry Science and Technology Co., Ltd No. 2015SCLD001.References
- T. J. Guzik, R. Korbut and T. Adamek-Guzik, J. Physiol. Pharmacol., 2003, 54, 469–487 CAS.
- S. F. Zheng, H. Y. Song, H. Gao, C. P. Liu, Z. W. Zhang and J. Fu, Biol. Trace Elem. Res., 2016, 173, 177–184 CrossRef CAS PubMed.
- X. Jin, T. T. Jia, R. H. Liu and S. W. Xu, J. Hazard. Mater., 2018, 357, 355–362 CrossRef CAS PubMed.
- T. Glaros, M. Larsen and L. Li, Front. Biosci., 2009, 14, 3988–3993 CrossRef CAS PubMed.
- M. van Deuren, A. S. Dofferhoff and J. W. van der Meer, J. Pathol., 1992, 168, 349–356 CrossRef CAS PubMed.
- Q. Xie and C. Nathan, J. Leukocyte Biol., 1994, 56, 576–582 CrossRef CAS PubMed.
- C. Whitfield and M. S. Trent, Annu. Rev. Biochem., 2014, 83, 99–128 CrossRef CAS PubMed.
- J. F. Miao, Y. M. Fa, B. B. Gu, W. Zhu and S. X. Zou, Cytokines, 2012, 59, 35–40 CrossRef CAS PubMed.
- D. P. Li, N. S. Zhang, Y. G. Cao, W. Zhang, G. L. Su, Y. Sun, Z. C. Liu, F. Y. Li, D. J. Liang, B. Liu, M. Y. Guo, Y. H. Fu, X. C. Zhang and Z. T. Yang, Eur. J. Pharmacol., 2013, 705, 79–85 CrossRef CAS PubMed.
- J. Y. Kim, S. J. Park, K. J. Yun, Y. W. Cho, H. J. Park and K. T. Lee, Eur. J. Pharmacol., 2008, 584, 175–184 CrossRef CAS PubMed.
- L. Q. Wang, J. Z. Jing, H. Yan, J. Y. Tang, G. Jia, G. M. Liu, X. L. Chen, G. Tian, J. Y. Cai, H. Y. Shang and H. Zhao, Biol. Trace Elem. Res., 2018, 186, 505–513 CrossRef CAS PubMed.
- A. Rakhshandeh and C. F. de Lange, Animals, 2012, 6, 305–310 CAS.
- D. Escribano, P. H. Campos, A. M. Gutiérrez, N. Le Floc'h, J. J. Cerón and E. Merlot, Vet. J., 2014, 200, 393–397 CrossRef CAS PubMed.
- P. R. Hoffmann and M. J. Berry, Mol. Nutr. Food Res., 2008, 52, 1273–1280 CrossRef CAS PubMed.
- Z. Huang, A. H. Rose and P. R. Hoffmann, Antioxid. Redox Signaling, 2012, 16, 705–743 CrossRef CAS PubMed.
- M. L. Dylewski, A. M. Mastro and M. F. Picciano, Neonatology, 2002, 82, 122–127 CrossRef CAS PubMed.
- W. Zhang, R. X. Zhang, T. C. Wang, H. C. Jiang, M. Y. Guo, E. S. Zhou, Y. Sun, Z. T. Yang, S. W. Xu, Y. G. Cao and N. S. Zhang, Inflammation, 2014, 37, 478–485 CrossRef CAS PubMed.
- G. V. Kryukov, S. Castellano, S. V. Novoselov, A. V. Lobanov, O. Zehtab, R. Guigó and V. N. Gladyshev, Science, 2003, 300, 1439–1443 CrossRef CAS PubMed.
- A. V. Lobanov, D. L. Hatfield and V. N. Gladyshev, Biochim. Biophys. Acta, 2009, 1790, 1424–1428 CrossRef CAS PubMed.
- L. H. Sun, N. Y. Zhang, M. K. Zhu, L. Zhao, J. C. Zhou and D. S. Qi, J. Nutr., 2016, 146, 655–661 CrossRef CAS PubMed.
- D. C. Mahan, T. R. Cline and B. Richert, J. Anim. Sci., 1999, 77, 2172–2179 CrossRef CAS PubMed.
- R. L. Payne and L. L. Southern, Poult. Sci., 2005, 84, 898–902 CrossRef CAS PubMed.
- M. Jlali, M. Briens, F. Rouffineau, P. A. Geraert and Y. Mercier, J. Anim. Sci., 2014, 92, 182–188 CrossRef CAS PubMed.
- M. Jlali, M. Briens, F. Rouffineau, F. Mercerand, P. A. Geraert and Y. Mercier, J. Anim. Sci., 2013, 91, 1745–1752 CrossRef CAS PubMed.
- M. Briens, Y. Mercier, F. Rouffineau, V. Vacchina and P. A. Geraert, Br. J. Nutr., 2013, 110, 617–624 CrossRef CAS PubMed.
- H. Zhao, K. Li, J. Y. Tang, J. C. Zhou, K. N. Wang, X. J. Xia and X. G. Lei, J. Nutr., 2015, 145, 1394–1401 CrossRef CAS PubMed.
- Y. Liu, H. Zhao, Q. S. Zhang, J. Y. Tang, K. Li, X. J. Xia, K. N. Wang, K. Li and X. G. Lei, J. Nutr., 2012, 142, 1410–1416 CrossRef CAS PubMed.
- X. F. Huang, J. Y. Tang, J. Y. Xu, G. Jia, G. M. Liu, X. L. Chen, J. Y. Cai, H. Y. Shang and H. Zhao, RSC Adv., 2016, 6, 84990–84998 RSC.
- J. Y. Xu, L. Q. Wang, J. Y. Tang, G. Jia, G. M. Liu, X. L. Chen, J. Y. Cai, H. Y. Shang and H. Zhao, PLoS One, 2017, 12, e0182079 CrossRef PubMed.
- B. A. Carlson, M. H. Yoo, R. K. Shrimali, R. Irons, V. N. Gladyshev, D. L. Hatfield and J. M. Park, Proc. Nutr. Soc., 2010, 69, 300–310 CrossRef CAS PubMed.
- R. K. Shrimali, R. D. Irons, B. A. Carlson, Y. Sano, V. N. Gladyshev, J. M. Park and D. L. Hatfield, J. Biol. Chem., 2008, 283, 20181–20185 CrossRef CAS PubMed.
- X. Y. Jiao, K. Yang, Y. An, X. J. Teng and X. H. Teng, Environ. Sci. Pollut. Res. Int., 2017, 24, 7555–7564 CrossRef CAS PubMed.
- L. Liu, D. K. Qin, X. F. Wang, Y. Feng, X. J. Yang and J. H. Yao, Food Agric. Immunol., 2015, 26, 194–203 CrossRef CAS.
- D. M. Webel, B. N. Finck, D. H. Baker and R. W. Johnson, J. Anim. Sci., 1997, 75, 1514–1520 CrossRef CAS PubMed.
- J. A. Hall, G. Bobe, W. R. Vorachek, K. Kasper, M. G. Traber, W. D. Mosher, G. J. Pirelli and M. Gamroth, Biol. Trace Elem. Res., 2014, 161, 272–287 CrossRef CAS PubMed.
- H. P. Leite, P. C. Nogueira, S. B. Iglesias, S. V. de Oliveira and R. O. Sarni, Nutrition, 2015, 31, 485–490 CrossRef CAS PubMed.
- X. J. Yang, W. L. Li, Y. Feng and J. H. Yao, Poult. Sci., 2011, 90, 2740–2746 CrossRef CAS PubMed.
- W. B. Gross, Avian Dis., 1990, 34, 759–761 CrossRef CAS PubMed.
- M. Geivelis, D. W. Turner, E. D. Pederson and B. L. Lamberts, J. Periodontol., 1993, 64, 980–983 CrossRef CAS PubMed.
- M. M. Jepson, J. M. Pell, P. C. Bates and D. J. Millward, Biochem. J., 1986, 235, 329–336 CrossRef CAS PubMed.
- A. Rhule, S. Navarro, J. R. Smith and D. M. Shepherd, J. Ethnopharmacol., 2006, 106, 121–128 CrossRef PubMed.
- C. L. Bi, H. Wang, Y. J. Wang, J. Sun, J. S. Dong, X. Meng and J. J. Li, Eur. J. Pharmacol., 2016, 780, 159–165 CrossRef CAS PubMed.
- K. V. Ramana, A. A. Fadl, R. Tammali, A. B. Reddy, A. K. Chopra and S. K. Srivastava, J. Biol. Chem., 2006, 281, 33019–33029 CrossRef CAS PubMed.
- R. J. Simpson, A. Hammacher, D. K. Smith, J. M. Matthews and L. D. Ward, Protein Sci., 1997, 6, 929–955 CrossRef CAS PubMed.
- C. X. Wang and A. Shuaib, Prog. Neurobiol., 2002, 67, 161–172 CrossRef CAS PubMed.
- M. Amaravani, N. Prasad and V. Ramakrishna, SpringerPlus, 2012, 1, 58 CrossRef CAS PubMed.
- A. K. Hubbard and R. Rothlein, Free Radical Biol. Med., 2000, 28, 1379–1386 CrossRef CAS.
- H. Wang, C. Bi, Y. Wang, J. Sun, X. Meng and J. Li, BMC Vet. Res., 2018, 14, 197 CrossRef PubMed.
- T. V. Nguyen, L. Yuan, M. S. Azevedo, K. I. Jeong, A. M. Gonzalez and L. J. Saif, Vet. Immunol. Immunopathol., 2007, 117, 236–248 CrossRef CAS PubMed.
- M. Kubo and Y. Motomura, Front. Immunol., 2012, 3, 275 Search PubMed.
- T. C. Stadtman, Ann. N. Y. Acad. Sci., 2000, 899, 399–402 CrossRef CAS PubMed.
- L. H. Sun, D. A. Pi, L. Zhao, X. Y. Wang, L. Y. Zhu, D. S. Qi and Y. L. Liu, Biol. Trace Elem. Res., 2017, 177, 90–96 CrossRef CAS PubMed.
- T. H. Zhuang, H. B. Xu, S. Hao, F. Ren, X. X. Chen, C. L. Pan and K. H. Huang, Res. Vet. Sci., 2015, 98, 59–65 CrossRef CAS PubMed.
- J. B. de Haan, C. Bladier, M. Lotfi-Miri, J. Taylor, P. Hutchinson, P. J. Crack, P. Hertzog and I. Kola, Free Radical Biol. Med., 2004, 36, 53–64 CrossRef CAS PubMed.
- R. A. Sunde, A. M. Raines, K. M. Barnes and J. K. Evenson, Biosci. Rep., 2009, 29, 329–338 CrossRef CAS PubMed.
- F. W. Hoffmann, A. C. Hashimoto, L. A. Shafer, S. Dow, M. J. Berry and P. R. Hoffmann, J. Nutr., 2010, 140, 1155–1161 CrossRef CAS PubMed.
- C. S. Broome, F. Mcardle, J. A. Kyle, F. Andrews, N. M. Lowe, C. A. Hart, J. R. Arthur and M. J. Jackson, Am. J. Clin. Nutr., 2004, 80, 154–162 CrossRef CAS PubMed.
- Y. G. Li, D. F. Ji, S. Zhong, L. G. Shi, G. Y. Hu and S. Chen, Alcohol Alcohol., 2010, 45, 320–331 CrossRef CAS PubMed.
- P. A. Sheridan, N. Zhong, B. A. Carlson, C. M. Perella, D. L. Hatfield and M. A. Beck, J. Nutr., 2007, 137, 1466–1471 CrossRef CAS PubMed.
- V. A. Shchedrina, R. A. Everley, Y. Zhang, S. P. Gygi, D. L. Hatfield and V. N. Gladyshev, J. Biol. Chem., 2011, 286, 42937–42948 CrossRef CAS PubMed.
- S. Verma, F. W. Hoffmann, M. Kumar, Z. Huang, K. Roe, E. Nguyen-Wu, A. S. Hashimoto and P. R. Hoffmann, J. Immunol., 2011, 186, 2127–2137 CrossRef CAS PubMed.
- J. E. Curran, J. B. Jowett, K. S. Elliott, Y. Gao, K. Gluschenko, J. Wang, D. M. Abel Azim, G. Cai, M. C. Mahaney, A. G. Comuzzie, T. D. Dyer, K. R. Walder, P. Zimmet, J. W. MacCluer, G. R. Collier, A. H. Kissebah and J. Blangero, Nat. Genet., 2005, 37, 1234–1241 CrossRef CAS PubMed.
- L. He, B. Wang, Y. Yao, M. S. Su, H. X. Ma and N. Jia, Mol. Med. Rep., 2014, 9, 1869–1876 CrossRef CAS.
- A. Sengupta, B. A. Carlson, V. M. Labunskyy, V. N. Gladyshev and D. L. Hatfield, Biochem. Cell Biol., 2009, 87, 953–961 CrossRef CAS.
- M. J. Schneider, S. N. Fiering, S. E. Pallud, A. F. Parlow, D. L. St Germain and V. A. Galton, Mol. Endocrinol., 2001, 15, 2137–2148 CrossRef CAS PubMed.
- B. A. Carlson, M. H. Yoo, Y. Sano, A. Sengupta, J. Y. Kim, R. Irons, V. N. Gladyshev, D. L. Hatfield and J. M. Park, BMC Immunol., 2009, 10, 57 CrossRef PubMed.
- T. S. Kim, M. H. Yu, Y. W. Chung, J. Kim, E. J. Choi, K. Ahn and I. Y. Kim, Mol. Cells, 1999, 9, 422–428 CAS.
- A. A. Turanov, S. Kehr, S. M. Marino, M. H. Yoo, B. A. Carlson, D. L. Hatfield and V. N. Gladyshev, Biochem. J., 2010, 430, 285–293 CrossRef CAS PubMed.
- H. A. Hansson, B. Rozell, S. Stemme, Y. Engström, L. Thelander and A. Holmgren, Exp. Cell Res., 1986, 163, 363–369 CrossRef CAS PubMed.
- J. Y. Tang, L. Cao, Q. Li, J. Y. Xu, G. Jia, G. M. Liu, X. L. Chen, H. Y. Shang, J. Y. Cai and H. Zhao, Biol. Trace Elem. Res., 2016, 173, 71–78 CrossRef CAS PubMed.
- J. Y. Tang, A. H. He, G. Jia, G. M. Liu, X. L. Chen, J. Y. Cai, H. Y. Shang, J. Q. Liao and H. Zhao, Biol. Trace Elem. Res., 2018, 181, 44–53 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9ra07260h |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2019 |