Open Access Article
Open Access ArticleRotary manifold for automating a paper-based Salmonella immunoassay†
Cody S. Carrella,
Rachel M. Wydallisa,
Mridula Bonthac,
Katherine E. Boehlea,
J. Ross Beveridgec,
Brian J. Geiss b and
Charles S. Henry
b and
Charles S. Henry *a
*a
aDepartment of Chemistry, Colorado State University, USA. E-mail: chuck.henry@colostate.edu
bDepartment of Microbiology, Immunology & Pathology, Colorado State University, USA
cDepartment of Computer Science, Colorado State University, USA
First published on 17th September 2019
Abstract
Foodborne pathogens are responsible for hundreds of thousands of deaths around the world each year. Rapid screening of agricultural products for these pathogens is essential to reduce and/or prevent outbreaks and pinpoint contamination sources. Unfortunately, current detection methods are laborious, expensive, time-consuming and require a central laboratory. Therefore, a rapid, sensitive, and field-deployable pathogen-detection assay is needed. We previously developed a colorimetric sandwich immunoassay utilizing immuno-magnetic separation (IMS) and chlorophenol red-β-D-galactopyranoside for Salmonella detection on a paper-based analytical device (μPAD); however, the assay required many sample preparation steps prior to the μPAD as well as laboratory equipment, which decreased user-friendliness for future end-users. As a step towards overcoming these limitations in resource-limited settings, we demonstrate a reusable 3D-printed rotational manifold that couples with disposable μPAD layers for semi-automated reagent delivery, washing, and detection in 65 minutes. After IMS to clean the sample, the manifold performs pipette-free reagent delivery and washing steps in a sequential order with controlled volumes, followed by enzymatic amplification and colorimetric detection using automated image processing to quantify color change. Salmonella was used as the target pathogen in this project and was detected with the manifold in growth media and milk with detection limits of 4.4 × 102 and 6.4 × 102 CFU mL−1 respectively. The manifold increases user friendliness and simplifies immunoassays resulting in a practical product for in-field use and commercialization.
1. Introduction
Foodborne pathogens pose a major health risk around the world. Each year they are responsible for 600 million infections and over 400![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 deaths around the world (9.4 million and 1300 in the US).1,2 Screening for specific pathogens could prevent large and deadly outbreaks; however, traditional pathogen detection assays are inadequate for testing large volumes of samples in a timely manner.3 The most effective screening tests should be inexpensive, portable, rapid, and easy to use such that the assay can be performed in the field by an untrained end user.4
000 deaths around the world (9.4 million and 1300 in the US).1,2 Screening for specific pathogens could prevent large and deadly outbreaks; however, traditional pathogen detection assays are inadequate for testing large volumes of samples in a timely manner.3 The most effective screening tests should be inexpensive, portable, rapid, and easy to use such that the assay can be performed in the field by an untrained end user.4
Foodborne pathogen detection is typically accomplished using culture-based methods.5 Culture-based methods are well established, selective, sensitive, and benefit from decades of data to cross reference. Despite these advantages, culturing can take days to complete, requires trained laboratory personnel, transportation to a central laboratory for analysis, and is prone to false negatives due to viable but non-culturable pathogens.3,4,6 Therefore, culturing is an ill-suited tool to screen for foodborne pathogens, especially in resource-limited settings.
To decrease testing times, numerous rapid assays have been developed, including lateral flow immunoassays (LFAs), polymerase chain reaction (PCR), loop mediated isothermal amplification (LAMP), and enzyme linked immunosorbent assays (ELISAs). Other rapid assays have used nanomaterials and specific recognition elements like aptamers, phages, or antibodies for capture and detection.5,7 While faster than culture-based methods, PCR and ELISA are both expensive, require complex instrumentation, and are unsuitable for in-field testing.8 LAMP is a promising tool for resource limited settings, but it still requires heating to 65 °C, four sets of primers, and trained technicians for most detection techniques.9 Finally, LFAs have been used for decades to detect pathogens, antibodies, and small biomarkers.10,11 Although they are well established diagnostic tools, LFAs consistently suffer from poor detection limits without pre-enrichment steps such as culturing or PCR amplification.11,12 Additionally, complex food sample matrices are difficult to use in an LFA format without significant sample pre-treatment or cleaning.
Microfluidic paper-based analytical devices (μPADs) have become a common platform for point-of-care and field-based assays.13–17 μPADs are small, portable, inexpensive, easy to dispose of, frequently require no external instrumentation, and can effectively store reagents.14,16,18,19 Recently, μPADs have been used to detect multiple foodborne pathogens with the goal of creating a simple screening test.20–23 In these works, traditional antibody-based immunoassays, enzymatic detection, and electrochemical methods were adapted for use in μPADs. Although they are a promising technology, μPADs frequently suffer from inadequate sensitivity and high detection limits compared to traditional methods like PCR and ELISA.14 To improve detection limits and sensitivity in traditional immunoassays, washing steps and signal enhancement reagents are used.3,24,25 However, these steps require timed and sequential delivery of reagents and/or washing agents, which μPADs are not typically designed to do without significant manual intervention from the end user.
Several groups have successfully demonstrated sequential reagent delivery in μPADs. Fu et al. accomplished this in 2010 with a 2D μPAD containing multiple inlets,26 and Govindarajan et al. introduced folding, or origami, μPADs in 2012, where reagents are delivered in each folding step.27 Since then, many other devices have been proposed including key shaped devices, sliding μPADs, rotating devices, dissolving sugar bridges, and complex channel geometries.26,28–32 Although they accomplish the goal of sequential reagent delivery, many of these devices suffer from inconsistent results, as the end user is ultimately performing the folding or sliding, and others require multiple pipetting steps to continually add buffer.33 Additionally, the more complex channels necessary for sequential delivery in these systems can result in lost analyte to the cellulose fibers and decreased sensitivity.24 Instead of relying on paper alone for reagent delivery, several groups have developed paper–plastic hybrid devices for pathogen detection.34–36 In this work we propose a μPAD housed in a re-usable 3D-printed rotational manifold capable of reproducible sequential reagent delivery for the detection of Salmonella via an immuno-magnetic separation (IMS) sandwich immunoassay.
Among foodborne pathogens, Salmonella is the most harmful, causing more worldwide infections (>90 million) and deaths (∼155![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000) than any other foodborne pathogen.37 Therefore, Salmonella was chosen as the target in this study and the manifold was used to detect Salmonella in growth media and milk. Milk was used as a real-world sample as milk-borne infections are common in unpasteurized, raw milk.38 Specificity against E. coli and other bacteria naturally found in milk was also demonstrated. To simplify the assay, we employed colorimetric detection using an enzymatic reaction that cleaves chlorophenol red-β-D-galactopyranoside (CPRG) to chlorophenol red (CPR), which is indicated by a yellow to red color change. The color change can be qualitatively observed by the naked eye or quantitatively analyzed through image analysis. We used a flood fill algorithm and analysis system that lowered the limit of detection by an order of magnitude over previous color analysis techniques. Our target limit of detection (LOD) was ≤102 colony forming units (CFU) per mL, which has been reported as the infectious dose of Salmonella in food samples.39,40 This LOD was realized in milk and media samples and is also the lowest detection limit that has been realized for direct colorimetric detection of Salmonella on a paper-based sensor with no pre-enrichment.41,42 By combining a 3D-printed manifold with paper layers we have increased the capabilities of a μPAD without sacrificing user-friendliness. We believe that many of the pitfalls of traditional μPADs may be solved by integrating inexpensive plastic platforms or manifolds like the one described here.
000) than any other foodborne pathogen.37 Therefore, Salmonella was chosen as the target in this study and the manifold was used to detect Salmonella in growth media and milk. Milk was used as a real-world sample as milk-borne infections are common in unpasteurized, raw milk.38 Specificity against E. coli and other bacteria naturally found in milk was also demonstrated. To simplify the assay, we employed colorimetric detection using an enzymatic reaction that cleaves chlorophenol red-β-D-galactopyranoside (CPRG) to chlorophenol red (CPR), which is indicated by a yellow to red color change. The color change can be qualitatively observed by the naked eye or quantitatively analyzed through image analysis. We used a flood fill algorithm and analysis system that lowered the limit of detection by an order of magnitude over previous color analysis techniques. Our target limit of detection (LOD) was ≤102 colony forming units (CFU) per mL, which has been reported as the infectious dose of Salmonella in food samples.39,40 This LOD was realized in milk and media samples and is also the lowest detection limit that has been realized for direct colorimetric detection of Salmonella on a paper-based sensor with no pre-enrichment.41,42 By combining a 3D-printed manifold with paper layers we have increased the capabilities of a μPAD without sacrificing user-friendliness. We believe that many of the pitfalls of traditional μPADs may be solved by integrating inexpensive plastic platforms or manifolds like the one described here.
2. Materials and methods
2.1. Rotational manifold
The 3D-printed manifold was printed using a FormLabs Form2 printer with their Clear V2 resin. All 3D-printed parts were designed in OnShape, a cloud-based CAD software. The paper channels, waste-pads, and sample layers are made of Fusion 5 paper, a conjugate-release membrane from GE healthcare (Waukesha, WI). The reagent layer and sample layers were made from Fusion 5 pieces, 3M transparency sheets, and 10 mil Fellowes® lamination sheets. The two layers are sealed using a TruLam laminator at 360 °F. The surfaces of the reagent and sample layers are coated in Never-Wet®, a hydrophobic spray-on product from Rust-Oleum®. All designs for the two layers and paper pieces were created in CorelDraw (Fig. S1†) and were cut with a 30 W CO2 laser cutter (Epilog Model 10000).2.2. IMS assay reagents
The reagents used in the Salmonella immunoassay include: Dynabeads™ M-280 Tosylactivated (Invitrogen™, purchased from Thermo Fisher Scientific, Milwaukee, WI); anti-Salmonella typhimurium 0–4 monoclonal mouse antibody (Abcam, Cambridge, MA ab8274); Salmonella antibody–biotin conjugate (Invitrogen™, purchased from Thermo Fisher Scientific); streptavidin–β-galactosidase conjugate (Invitrogen™, purchased from Thermo Fisher Scientific), and chlorophenol red-β-D-galactopyranoside (purchased from Sigma Aldrich, St. Louis, MS). The bacterial strains used in this work were Salmonella enterica serovar typhimurium and Escherichia coli DH5α. The cells were grown in a Difco (Salmonella) and LB (E. coli) media for 12 h to a final concentration ∼108 CFU mL−1. After each growth the cell concentration was quantified by serial dilution and plating. The cells were spiked into and diluted in growth media or milk at the desired concentrations before running the assay with the manifold. The milk used was DairyGold ultra-high pasteurized whole milk. The buffer used in the manifold was a 1× solution of phosphate buffered saline (PBS) at pH 7.4 diluted in Milli-Q water. 2.5–4.5 μm pink fluorescent magnetic beads from SpheroTech (Product #FP-4058-2) were used to perform the bead retention studies with a Dino-Lite EDGE fluorescent digital microscope at an excitation wavelength of 575 nm.2.3. Assay steps
One mL of liquid sample is incubated with 5 μL of 5 mg mL−1 magnetic bead–Salmonella antibody conjugate for 15 min at room temperature in a microcentrifuge tube. After a washing step using an external DynaMag-2 (Thermo Fisher Scientific) magnet for IMS, the magnetic bead–antibody-Salmonella conjugate is reconstituted in 60 μL of PBS buffer. A 15 μL aliquot of the concentrated solution was placed on the sample layer and the remainder of the assay (enzyme labeling and detection) is performed in the rotational manifold.2.4. Device operation
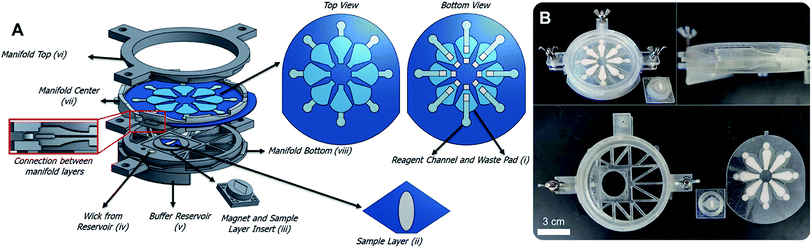
The manifold consists of four 3D printed pieces: the manifold top, center, and bottom, and the sample layer insert (Fig. 1). The top, center, and bottom pieces are all held together by bolts and springs. The springs allow the center piece to rotate while the top and bottom pieces are held in place. The center piece contains a slot for the reagent card. Within the reagent card are eight reagent channels and waste pads. The reagent card is made of five layers: a transparency sheet with a hydrophobic coating, the paper reagent channels, a 3 mil lamination sheet, the paper waste pads, and a 10 mil lamination sheet. The waste pads and sample layers are stacked on top of each other but are separated by the 3 mil lamination sheet. Four reagent channels store reagents and four are empty channels used for washing. Two reagent and two washing channels are needed to run the assay in the current format, so eight total channels allow two tests to be run with one reagent card. In the four channels needed, the first channel contains 0.4 μg of biotin-labeled Salmonella antibody that is deposited and dried prior to the assay; the second is empty for washing; the third contains 0.2 μg of dried streptavidin β-galactosidase; and the fourth is empty. 5 μL aliquots of 40 μg mL−1 and 80 μg mL−1 streptavidin β-galactosidase and biotin-ab are added to the reagent channels to obtain 0.2 μg and 0.4 μg of dried reagent respectively. These reagents are all added and dried before the assay is run so the end-user does not need to perform the reagent addition steps.When the manifold is assembled, the reagent layer rotates with the center piece and the sample layer insert remains stationary. As the center piece rotates, the top and bottom pieces move vertically while keeping pressure on the center piece. After a 45° rotation the center piece clicks into a slot and the top and bottom pieces collapse down with it. A video in the ESI† shows the device rotation, clicking mechanism, and flow through the layers.
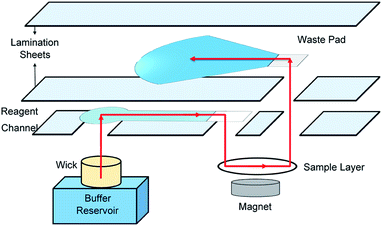
With each rotation, an opening in the reagent layer is aligned with a wick in the bottom piece of the manifold and the next reagent layer and waste pad are aligned with the sample layer. The wick sits in a buffer reservoir and, buffer flows through the reagent channel, through sample layer, and into a waste pad. The flow through the system either delivers a reagent to the sample layer or simply washes excess reagent away from the sample layer. Magnetic beads, and anything conjugated to them, will not wash through the sample layer because of a magnet that's fitted underneath the sample layer in the sample layer insert. The end-user knows to rotate the device after the waste pad is saturated (∼2 min).
2.5. Colorimetric detection
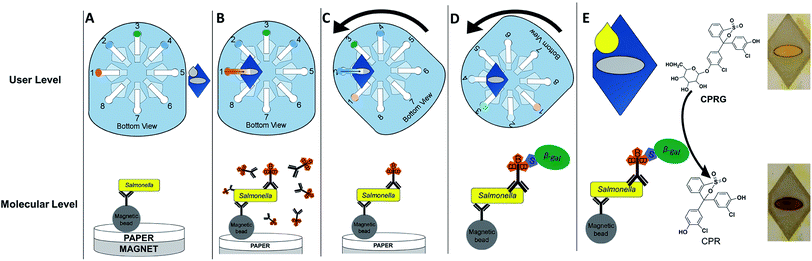
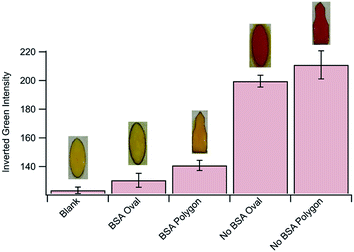
The final step of the assay is to add 25 μL of a colorimetric substrate (CPRG) to the sample layer with a pipette. In the presence of β-galactosidase, and therefore Salmonella, CPRG (Yellow) will be turned over to CPR (Red). The [CPR] can be monitored via image capture and analysis. The image of the colored sample layer is taken inside a light box. The light box consists of two shells. The inner shell houses the sample layer and is made of clear acrylic. The acrylic was sanded until cloudy to diffuse light and prevent glare on the sample. The outer shell covers the first and is made of black acrylic so no ambient light can reach the sample layer. The outer shell contains 35 white and warm white LEDs built into the walls and powered by a 9 V battery. The inner and outer shells contain an opening for a smartphone to capture an image (Fig. S2†).In this work we used a Samsung Galaxy S7 smartphone to capture static images of the sample layer at different time points. In the past, our group has frequently used manual processing with NIH ImageJ to quantify color changes.42–45 To decrease the manual labor associated with ImageJ we used a flood-fill algorithm available in the OpenCV computer vision library.46 Flood-fill identifies the color pixels in our sample region by employing a recursive region growing process. The regions grow out from a seed known to be in the sample. Flood-fill adds pixels relatively similar in color until no more similar pixels are adjacent to the region. In our case this means it stops at the edge of the oval paper that bounds our sample. The average RGB color values in the resulting set of pixels is then analyzed for each sample.47 When compared to manual analysis using NIH ImageJ, the automated algorithm returned nearly identical results (Fig. S3†).
We define the signal in each trial as the difference in color between the blank and the sample. To quantify this difference the vector length in 3D RGB space between our blank RGB coordinates and the sample RGB coordinates (ΔRGB) was calculated using eqn (1) as previously described.48,49
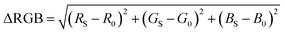 | (1) |
3. Results and discussion
3.1. Assay steps and design
In a previous publication we presented an immuno-magnetic separation sandwich immunoassay for Salmonella detection.42 The chemistry of the immunoassay remains the same in this work and is discussed in depth in our previous publication. Briefly, Salmonella is isolated from a sample using Salmonella-antibody labeled magnetic beads. After a short incubation with the sample the beads are removed from solution with an external magnet, washed, and reconstituted in a small volume of buffer to concentrate the sample. A secondary Salmonella-antibody labeled with biotin is then introduced to the Salmonella–magnetic bead complex solution. After an incubation and washing step, streptavidin-labeled β-galactosidase is added to the Salmonella–magnetic bead–biotin conjugate solution. Finally, after another incubation and washing step a small volume of the Salmonella–magnetic bead–biotin–β-galactosidase conjugate solution reacts with the substrate CPRG. After a set reaction time, a color change from yellow to red indicates the presence of β-galactosidase, and therefore Salmonella. Previously, all steps of the assay were performed in solution in a microcentrifuge tube until the final step, where a small aliquot of magnetic bead–Salmonella–enzyme conjugate was added to a paper spot with dried substrate. The assay worked but required thirteen pipetting steps. To improve ease-of-use, we designed a rotational manifold to minimize pipetting steps by washing and delivering reagents to the test zone (Fig. 2). Here, the initial step of sample incubation with antibody-labeled magnetic beads remains in-solution to ensure a sufficient number of bacteria are available for conjugation to the magnetic beads. After the bacteria is isolated from the sample, a 15 μL aliquot of the magnetic bead–bacteria complex is added to a small paper sample-layer and the remaining steps are completed in the device.3.2. Device design and operation
Schematic drawings and a photograph of the manifold are shown in Fig. 1. An important feature of the device is that the buffer volume delivered in each step can be controlled by changing the size of the waste pad. Fusion 5 paper holds 0.422 ± 0.006 μL mm−2 (Fig. S5†), so by changing the surface area of the waste pad the volume of buffer used in each step can be customized. Volume control of each reagent delivery and washing step is critical when using the device for different assays that may need more thorough washing. Flow through each layer of the device is illustrated in Fig. 3. The sample layer sits on the 3D-printed sample layer insert, which fits into the bottom manifold piece with lego-style fittings. A magnet screws into the bottom of the insert and is positioned directly underneath the sample layer. The magnet ensures that the magnetic beads are not washed away during reagent delivery and washing steps (Fig. S6†). Using magnetic beads and magnets to create a test zone allowed us to use membranes other than nitrocellulose, which is expensive and has a short shelf life.50 Additionally, the sample in the manifold is stationary on the sample layer and therefore will not be lost to the membrane during flow through the device as is the case in traditional LFAs. Sample loss to paper channels is a problem in paper-based devices,24,51 and a stationary sample can improve detection limits.After the final washing step, the insert and sample layer are removed from the device. 25 μL of 2.5 mM CPRG is pipetted onto the sample layer containing the conjugated system and the substrate reacts with any enzyme present for 40 min. A picture is then taken of the sample layer inside the light box. The image is analyzed as described in the materials and methods to determine the Salmonella concentration. Using the rotational manifold, the number of pipetting steps needed to complete the IMS sandwich immunoassay was decreased from thirteen to four, which significantly reduces labor and complexity for the end user. Each sample addition step requires ∼1 second of user input (rotating the device) compared to traditional pipetting and IMS that can take multiple minutes per step. Additionally, the waste generated during each step of a traditional in-solution IMS assay exceeds 1 mL, while our assay uses roughly 0.1 mL per step, reducing cost and storage needs.
3.3. Assay optimization and parameters
3.4. Colorimetric Salmonella detection
A Salmonella detection assay was used to test the functionality of the manifold. Here, Salmonella cultured in Difco nutrient broth was diluted to concentrations spanning from 102 to 107 CFU mL−1. The results are shown in Fig. 5 (n = 3). The data was fit to a 4-parameter logistic curve (Equation ESI†) with a χ2 = 13.02 (α = 0.05, χcritical = 15.51).55 The logistic fit was chosen because the antibody binding kinetics of the sandwich immunoassay are the limiting steps. The LOD was calculated by finding 3 × SD + mean of the blank and plugging it into the fit equation. An LOD of 4.40 × 102 CFU mL−1 was realized using the manifold. In our previous entirely in-solution assay, we found a LOD on the same order of magnitude, which demonstrates the capabilities of our device to perform a multi-step immunoassay on a μPAD.42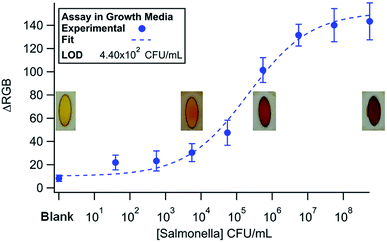 | ||
| Fig. 5 Dose response curve (n = 3) for Salmonella in media detected using the rotational manifold. The curve was fit to a 4-parameter logistic model and a LOD of 2.9 × 103 was achieved. | ||
3.5. Specificity and real-world samples
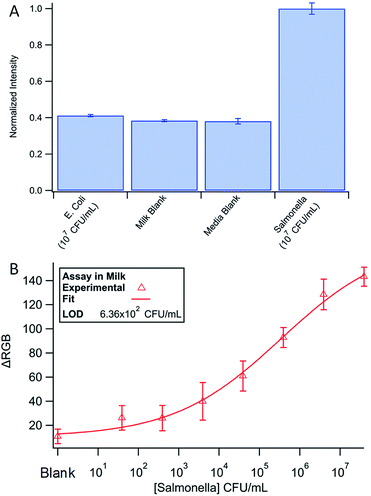
To confirm the specificity of our Salmonella assay, 107 CFU mL−1 DH5α E. coli in media was tested with the device using Salmonella specific antibodies (Fig. 6A). The low signal for E. coli samples demonstrates the specificity of the assay. A low signal also indicates efficient washing throughout the system as E. coli produces β-galactosidase naturally, and any excess enzyme would increase the signal.Finally, the assay was run using milk spiked with Salmonella (Fig. 6B). In milk, we found a detection limit of 6.36 × 102 CFU mL−1. The data was once again fit to a 4-parameter logistic curve (χ2 = 4.13, α = 0.05, χcritical = 14.07) and the LOD found using the fit equations and 3 × SD + the mean of the blank. Although the detection limit is slightly higher, the assay performed nearly as well in milk versus growth media. The small difference in LOD could be attributed to decreased antibody binding efficiency in the first step of the assay due to non-specific adsorption of other biomolecules in milk onto the magnetic beads. The international microbiological criteria for dairy products states that Salmonella should have a concentration of <1 cell per mL.56 The only way to meet the required LOD with our device, and any other inexpensive point-of-care system, is through some form of pre-enrichment like culturing. Although we believe that the rotary manifold is an innovative way to perform IMS as a rapid screening tool, further technologies will need to be developed to meet the strict requirements of the food safety industry.
4. Conclusions and future directions
The field of microfluidic paper-based devices has grown tremendously over the last 10 years in academic settings. There are many reasons for the disconnect between published works and commercial products, but the most commonly cited argument is poor sensitivity and specificity in μPADs.14,19 Attempts to improve these criteria are often successful but result in assays that are far too complicated for their intended use at the point-of-care with untrained end-users. We believe that coupling μPADs with other inexpensive materials could enhance sensitivity and specificity without sacrificing usability. In this work we designed a 3D-printed rotational manifold to perform multiple reagent delivery and washing steps in a sequential order. The washings steps increase sensitivity over traditional LFAs and the manifold ensures that the additional steps do not infringe on usability. The manifold was used to detect Salmonella, however with a simple change of reagents could be used to detect a large host of other pathogens, proteins, and/or biomarkers. In the future we will continue working with this device to detect additional biomolecules in a multiplexed format. We also recognize that although we simplified testing and reduced manual input, the four pipetting steps still required are not ideal. Some liquid transfer will always be necessary in an IMS assay, however, future work will be dedicated to decreasing user intervention further. For example, work has begun to implement a motorized rotational mechanism and further automate image capture and analysis with a web-based interface. With devices like the one described here, we anticipate an increase in new μPADs on the market.Conflicts of interest
There are no conflicts to declare.Acknowledgements
Funding for this project was provided by the United States Department of Agriculture, Grant Number #16-74000589 through the National Wildlife Research Center. The authors would also like to thank the members of the Henry and Geiss laboratories and Dr Alan Franklin for their guidance throughout the project.References
- E. Scallan, R. M. Hoekstra, F. J. Angulo, R. V. Tauxe, M. A. Widdowson, S. L. Roy, J. L. Jones and P. M. Griffin, Foodborne Illness Acquired in the United States-Major Pathogens, Emerging Infect. Dis., 2011, 17(1), 7–15 CrossRef.
- Group, F. D. B. E. R., WHO Estimates of the Global Burden of Foodborne Diseases, Organization, W. H., 2015 Search PubMed.
- J. W. F. Law, N. S. Ab Mutalib, K. G. Chan and L. H. Lee, Rapid methods for the detection of foodborne bacterial pathogens: principles, applications, advantages and limitations, Front. Microbiol., 2015, 5, 19 CrossRef PubMed.
- V. Velusamy, K. Arshak, O. Korostynska, K. Oliwa and C. Adley, An overview of foodborne pathogen detection: In the perspective of biosensors, Biotechnol. Adv., 2010, 28(2), 232–254 CrossRef CAS PubMed.
- V. M. Bohaychuk, G. E. Gensler, R. K. King, J. T. Wu and L. M. McMullen, Evaluation of detection methods for screening meat and poultry products for the presence of foodborne pathogens, J. Food Prot., 2005, 68(12), 2637–2647 CrossRef.
- P. K. Mandal, A. K. Biswas, K. Choi and U. K. Pal, Methods for Rapid Detection of Foodborne Pathogens: An Overview, Am. J. Food Technol., 2011, 6(2), 15 Search PubMed.
- J. Chen, S. M. Andler, J. M. Goddard, S. R. Nugen and V. M. Rotello, Integrating recognition elements with nanomaterials for bacteria sensing, Chem. Soc. Rev., 2017, 46(5), 1272–1283 RSC.
- X. Zhao, C. W. Lin, J. Wang and D. H. Oh, Advances in Rapid Detection Methods for Foodborne Pathogens, J. Microbiol. Biotechnol., 2014, 24(3), 297–312 CrossRef CAS.
- Y. Mori and T. Notomi, Loop-mediated isothermal amplification (LAMP): a rapid, accurate, and cost-effective diagnostic method for infectious diseases, J. Infect. Chemother., 2009, 15(2), 62–69 CrossRef CAS.
- Y. Zhao, H. R. Wang, P. P. Zhang, C. Y. Sun, X. C. Wang, X. R. Wang, R. F. Yang, C. B. Wang and L. Zhou, Rapid multiplex detection of 10 foodborne pathogens with an up-converting phosphor technology-based 10-channel lateral flow assay, Sci. Rep., 2016, 6, 21342 CrossRef CAS.
- G. A. Posthuma-Trumpie, J. Korf and A. van Amerongen, Lateral flow (immuno) assay: its strengths, weaknesses, opportunities and threats. A literature survey, Anal. Bioanal. Chem., 2009, 393(2), 569–582 CrossRef CAS PubMed.
- J. Hu, S. Q. Wang, L. Wang, F. Li, B. Pingguan-Murphy, T. J. Lu and F. Xu, Advances in paper-based point-of-care diagnostics, Biosens. Bioelectron., 2014, 54, 585–597 CrossRef CAS.
- A. W. Martinez, S. T. Phillips, M. J. Butte and G. M. Whitesides, Patterned paper as a platform for inexpensive, low-volume, portable bioassays, Angew. Chem., Int. Ed., 2007, 46(8), 1318–1320 CrossRef CAS.
- K. Yamada, H. Shibata, K. Suzuki and D. Citterio, Toward practical application of paper-based microfluidics for medical diagnostics: state-of-the-art and challenges, Lab Chip, 2017, 17(7), 1206–1249 RSC.
- A. W. Martinez, S. T. Phillips, G. M. Whitesides and E. Carrilho, Diagnostics for the Developing World: Microfluidic Paper-Based Analytical Devices, Anal. Chem., 2010, 82(1), 3–10 CrossRef CAS PubMed.
- Y. Y. Yang, E. Noviana, M. P. Nguyen, B. J. Geiss, D. S. Dandy and C. S. Henry, Paper-Based Microfluidic Devices: Emerging Themes and Applications, Anal. Chem., 2017, 89(1), 71–91 CrossRef CAS PubMed.
- Y. Y. Xia, J. Si and Z. Y. Li, Fabrication techniques for microfluidic paper-based analytical devices and their applications for biological testing: A review, Biosens. Bioelectron., 2016, 77, 774–789 CrossRef CAS.
- A. M. Lopez-Marzo and A. Merkoci, Paper-based sensors and assays: a success of the engineering design and the convergence of knowledge areas, Lab Chip, 2016, 16(17), 3150–3176 RSC.
- C. Carrell, A. Kava, M. Nguyen, R. Menger, Z. Munshi, Z. Call, M. Nussbaum and C. Henry, Beyond the lateral flow assay: A review of paper-based microfluidics, Microelectron. Eng., 2019, 206, 45–54 CrossRef CAS.
- K. Kant, M.-A. Shahbazi, V. P. Dave, T. A. Ngo, V. A. Chidambara, L. Q. Than, D. D. Bang and A. Wolff, Microfluidic devices for sample preparation and rapid detection of foodborne pathogens, Biotechnol. Adv., 2018, 36, 1003–1024 CrossRef CAS PubMed.
- J. C. Jokerst, J. A. Adkins, B. Bisha, M. M. Mentele, L. D. Goodridge and C. S. Henry, Development of a Paper-Based Analytical Device for Colorimetric Detection of Select Foodborne Pathogens, Anal. Chem., 2012, 84(6), 2900–2907 CrossRef CAS PubMed.
- J. Bhardwaj, S. Devarakonda, S. Kumar and J. Jang, Development of a paper-based electrochemical immunosensor using an antibody-single walled carbon nanotubes bio-conjugate modified electrode for label-free detection of foodborne pathogens, Sens. Actuators, B, 2017, 253, 115–123 CrossRef CAS.
- J. Park, J. H. Shin and J. K. Park, Pressed Paper-Based Dipstick for Detection of Foodborne Pathogens with Multistep Reactions, Anal. Chem., 2016, 88(7), 3781–3788 CrossRef CAS PubMed.
- M. P. Nguyen, N. A. Meredith, S. P. Kelly and C. S. Henry, Design considerations for reducing sample loss in microfluidic paper-based analytical devices, Anal. Chim. Acta, 2018, 1017, 20–25 CrossRef CAS PubMed.
- E. Fu, P. Kauffman, B. Lutz and P. Yager, Chemical signal amplification in two-dimensional paper networks, Sens. Actuators, B, 2010, 149(1), 325–328 CrossRef CAS PubMed.
- E. Fu, B. Lutz, P. Kauffman and P. Yager, Controlled reagent transport in disposable 2D paper networks, Lab Chip, 2010, 10(7), 918–920 RSC.
- A. V. Govindarajan, S. Ramachandran, G. D. Vigil, P. Yager and K. F. Bohringer, A low cost point-of-care viscous sample preparation device for molecular diagnosis in the developing world; an example of microfluidic origami, Lab Chip, 2012, 12(1), 174–181 RSC.
- R. B. Channon, M. P. Nguyen, A. G. Scorzelli, E. M. Henry, J. Volckens, D. S. Dandy and C. S. Henry, Rapid flow in multilayer microfluidic paper-based analytical devices, Lab Chip, 2018, 18(5), 793–802 RSC.
- A. Apilux, Y. Ukita, M. Chikae, O. Chailapakul and Y. Takamura, Development of automated paper-based devices for sequential multistep sandwich enzyme-linked immunosorbent assays using inkjet printing, Lab Chip, 2013, 13(1), 126–135 RSC.
- H. Liu, X. Li and R. M. Crooks, Paper-Based SlipPAD for High-Throughput Chemical Sensing, Anal. Chem., 2013, 85(9), 4263–4267 CrossRef CAS PubMed.
- B. Lutz, T. Liang, E. Fu, S. Ramachandran, P. Kauffman and P. Yager, Dissolvable fluidic time delays for programming multi-step assays in instrument-free paper diagnostics, Lab Chip, 2013, 13(14), 2840–2847 RSC.
- X. E. Sun, B. W. Li, C. Y. Tian, F. B. Yu, N. Zhou, Y. H. Zhan and L. X. Chen, Rotational paper-based electrochemiluminescence immunodevices for sensitive and multiplexed detection of cancer biomarkers, Anal. Chim. Acta, 2018, 1007, 33–39 CrossRef CAS PubMed.
- E. Fu and C. Downs, Progress in the development and integration of fluid flow control tools in paper microfluidics, Lab Chip, 2017, 17(4), 614–628 RSC.
- L. Lafleur, D. Stevens, K. McKenzie, S. Ramachandran, P. Spicar-Mihalic, M. Singhal, A. Arjyal, J. Osborn, P. Kauffman, P. Yager and B. Lutz, Progress toward multiplexed sample-to-result detection in low resource settings using microfluidic immunoassay cards, Lab Chip, 2012, 12(6), 1119–1127 RSC.
- R. H. Tang, H. Yang, J. R. Choi, Y. Gong, J. Hu, T. Wen, X. J. Li, B. Xu, Q. B. Mei and F. Xu, Paper-based device with on-chip reagent storage for rapid extraction of DNA from biological samples, Microchim. Acta, 2017, 184(7), 2141–2150 CrossRef CAS.
- U. M. Jalal, G. J. Jin and J. S. Shim, Paper-Plastic Hybrid Microfluidic Device for Smartphone-Based Colorimetric Analysis of Urine, Anal. Chem., 2017, 89(24), 13160–13166 CrossRef CAS.
- S. E. Majowicz, J. Musto, E. Scallan, F. J. Angulo, M. Kirk, S. J. O'Brien, T. F. Jones, A. Fazil, R. M. Hoekstra and Int. Collaboration Enteric Dis., B., The Global Burden of Nontyphoidal Salmonella Gastroenteritis, Clin. Infect. Dis., 2010, 50(6), 882–889 CrossRef PubMed.
- S. P. Oliver, K. J. Boor, S. C. Murphy and S. E. Murinda, Food Safety Hazards Associated with Consumption of Raw Milk, Foodborne Pathog. Dis., 2009, 6(7), 793–806 CrossRef.
- M. J. Blaser and L. S. Newman, A Review of Human Salmonellosis. 1. Infective Dose, Rev. Infect. Dis., 1982, 4(6), 1096–1106 CrossRef CAS PubMed.
- M. H. Kothary and U. S. Babu, Infective dose of foodborne pathogens in volunteers: A review, J. Food Saf., 2001, 21(1), 49–73 CrossRef.
- C. Z. Li, K. Vandenberg, S. Prabhulkar, X. N. Zhu, L. Schneper, K. Methee, C. J. Rosser and E. Almeide, Paper based point-of-care testing disc for multiplex whole cell bacteria analysis, Biosens. Bioelectron., 2011, 26(11), 4342–4348 CrossRef CAS PubMed.
- M. Srisa-Art, K. E. Boehle, B. J. Geiss and C. S. Henry, Highly Sensitive Detection of Salmonella typhimurium Using a Colorimetric Paper-Based Analytical Device Coupled with Immunomagnetic Separation, Anal. Chem., 2018, 90(1), 1035–1043 CrossRef CAS PubMed.
- K. E. Boehle, J. Gilliand, C. R. Wheeldon, A. Holder, J. A. Adkins, B. J. Geiss, E. P. Ryan and C. S. Henry, Utilizing Paper-Based Devices for Antimicrobial-Resistant Bacteria Detection, Angew. Chem., Int. Ed., 2017, 56(24), 6886–6890 CrossRef CAS PubMed.
- K. E. Boehle, C. S. Carrell, J. Caraway and C. S. Henry, Paper-Based Enzyme Competition Assay for Detecting Falsified beta-Lactam Antibiotics, ACS Sens., 2018, 3(7), 1299–1307 CrossRef CAS PubMed.
- J. A. Adkins, K. Boehle, C. Friend, B. Chamberlain, B. Bisha and C. S. Henry, Colorimetric and Electrochemical Bacteria Detection Using Printed Paper- and Transparency-Based Analytic Devices, Anal. Chem., 2017, 89(6), 3613–3621 CrossRef CAS.
- G. Bradski, The OpenCV library, Dr. Dobb's Journal, 2000, 25(11), 120–125 Search PubMed.
- K. E. Boehle, E. Doan, S. Henry, J. R. Beveridge, S. L. Pallickara and C. S. Henry, Single board computing system for automated colorimetric analysis on low-cost analytical devices, Anal. Methods, 2018, 10(44), 5282–5290 RSC.
- R. C. Murdock, L. Shen, D. K. Griffin, N. Kelley-Loughnane, I. Papautsky and J. A. Hagen, Optimization of a Paper-Based ELISA for a Human Performance Biomarker, Anal. Chem., 2013, 85(23), 11634–11642 CrossRef CAS PubMed.
- L. Shen, J. A. Hagen and I. Papautsky, Point-of-care colorimetric detection with a smartphone, Lab Chip, 2012, 12(21), 4240–4243 RSC.
- M. Sajid, A. N. Kawde and M. Daud, Designs, formats and applications of lateral flow assay: A literature review, J. Saudi Chem. Soc., 2015, 19(6), 689–705 CrossRef.
- X. Li, D. R. Ballerini and W. Shen, A perspective on paper-based microfluidics: Current status and future trends, Biomicrofluidics, 2012, 6(1), 011301 CrossRef PubMed.
- K. Jones, FUSION 5: A New Platform for Lateral Flow Immunoassay Tests, Lateral Flow Immunoassay, 2009, pp. 115–129 Search PubMed.
- J. A. Adkins, E. Noviana and C. S. Henry, Development of a Quasi-Steady Flow Electrochemical Paper-Based Analytical Device, Anal. Chem., 2016, 88(21), 10639–10647 CrossRef CAS.
- D. M. Zhang and Q. J. Liu, Biosensors and bioelectronics on smartphone for portable biochemical detection, Biosens. Bioelectron., 2016, 75, 273–284 CrossRef CAS.
- T. K. Christopoulos and E. P. Diamandis, Theory of immunoassays, Immunoassay, 1996, 25–50 CAS.
- Scientific Criteria to Ensure Safe Food, National Academies Press, Washington, DC, 2003, https://www.ncbi.nlm.nih.gov/books/NBK221563/ Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9ra07106g |
| This journal is © The Royal Society of Chemistry 2019 |