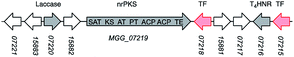Open Access Article
Open Access ArticleFunction of pathway specific regulators in the ACE1 and pyrichalasin H biosynthetic gene clusters†
Verena Hantke,
Chongqing Wang,
Elizabeth J. Skellam and
Russell J. Cox
and
Russell J. Cox *
*
Institute for Organic Chemistry and BMWZ, Leibniz Universität Hannover, Schneiderberg 38, 30167 Hannover, Germany. E-mail: russell.cox@oci.uni-hannover.de
First published on 4th November 2019
Abstract
Ectopic expression of BC1 which encodes a putative pathway specific transcription factor from the ACE1 biosynthetic gene cluster of the rice pathogen Pyricularia oryzae Guy11 did not lead to the production of ACE1-related compounds. However the known compound hinnulin A was formed. A putative partial gene cluster potentially involved in the biosynthesis of hinnulin A and DHN melanin was validated by RT-PCR and a possible biosynthetic pathway is proposed. Ectopic expression of pyiR which encodes a pathway specific transcription factor from the pyrichalasin H biosynthetic gene cluster in Magnaporthe grisea NI980 led to the apparent up-regulation of the pyi cluster and a 3-fold increase in pyrichalasin production under standard fermentation conditions, but did not lead to the formation of new compounds.
Introduction
The fungus Pyricularia oryzae (=Magnaporthe oryzae) is a major pathogen of rice (Oryza sativa) causing significant annual reduction in yields.1 The progress of rice blast disease caused by P. oryzae is well understood. Spores of the fungus are distributed by wind and germinate on rice leaf surfaces. A germ tube spreads across the cuticle, before forming an appressorium. The appressorium is a structure which mediates penetration of the fungus into the plant through the generation of turgor,2 the generation of specific hydrolytic enzymes,3 and the formation of 1,8-dihydroxynaphthalene (DHN) melanin.4 After penetration, the fungal mycelium multiplies through the plant leaves and stems, killing cells and causing visible lesions. Eventually the mycelium breaks out of the cells and sporulates on the plant surface, repeating the disease cycle.Rice resistance to P. oryzae infection is known, but resistant rice varieties only display this phenotype towards avirulent strains of P. oryzae. Work by the group of Lebrun has shown that avirulent strains of P. oryzae such as Guy11, contain the ACE1 (Avirulence Conferring Enzyme 1) biosynthetic gene cluster (BGC) which is responsible for the avirulence phenotype.5,6 It is hypothesised that the ACE1 BGC encodes the biosynthesis of a signal compound which is detected by resistant rice7 allowing the plant to mount an effective defence. The ACE1 BGC was shown to be expressed only under very specific conditions during appressorium penetration into plant cells.8 Under these conditions the ACE1 metabolite is produced in very low quantities for a very short period and it has therefore been impossible to isolate and identify the avirulence signal compound.9
Comparisons to related BGC suggest that the ACE1 metabolite is likely to be related to the cytochalasans10 and heterologous expression experiments of ACE1 and other genes from the BGC produce plausible cytochalasan intermediates or shunt metabolites.11 The BGC consists of genes encoding a typical highly programmed fungal polyketide synthase nonribosomal peptide synthetase12 hybrid (PKS-NRPS, ACE1) together with tailoring genes typical of cytochalasan biosynthesis (Fig. 1). The ACE1 BGC also encodes various transport proteins and the transcription factor (TF) BC2. A closely related BGC is that involved with the biosynthesis of pyrichalasin H 1 in the related organism Magnaporthe grisea NI980.13 The pyrichalasin (pyi) BGC also encodes a PKS-NRPS and includes a TF gene known as pyiR, but unlike the ACE1 BGC the pyi cluster is active under a variety of physiological conditions.
Previous work reported by Hertweck and coworkers has shown that transcription factors encoded in PKS-NRPS BGC are pathway-specific positive regulators of BGC expression and can make useful tools for the activation of previously silent BGC.14 For example, in the case of the aspyridone (apd) cluster the apdR TF was ectopically expressed in the native host A. nidulans under the control of the inducible alcohol dehydrogenase promoter PalcA. This ‘switched on’ the pathway and resulted in the production of aspyridones A 2a and B 2b (Fig. 1).15 Similarly, Tang and coworkers expressed ccsR which encodes a pathway-specific TF in the cytochalasin E 3 (ccs) BGC in Aspergillus clavatus. This resulted in an up-regulation of the cluster and a significant increase in the titre of 3.16
We reasoned that a strategy involving the ectopic expression of BC2 could be an effective way to activate the ACE1 BGC with the aim of isolating and structurally characterising the ACE1 signal compound from P. oryzae. In parallel, we also experimented with ectopic over-expression of pyiR with the aim of increasing the titre of pyrichalasin H 1 production in M. grisea NI980.
Results
P. oryzae Guy11 is sensitive to the antibiotics hygromycin B and glufosinate (Basta).17 In order to develop a reliable transformation method, genetic manipulation of P. oryzae Guy11 was established using the vector pTH-GS-hygR-egfp. The vector features a hygromycin B resistance cassette (hygR) as well as the egfp gene under the control of the strong inducible amyB promoter (PamyB) from Aspergillus oryzae.18,19Transformation of P. oryzae Guy11 was performed with vector pTH-GS-hygR-egfp using a PEG/CaCl2-mediated protoplast protocol resulting in 12 hygromycin B resistant transformants. Six transformants and the wild type were cultivated for 5 days in DPY medium before the samples were analyzed by fluorescence microscopy. Successful production of GFP was ascertained in all transformants, but not in the wild type control (Fig. 2). This indicates the suitability of the hygR/PamyB system for further manipulation of P. oryzae Guy11. BC2 was then amplified from P. oryzae Guy11 genomic DNA (gDNA) by PCR and placed downstream of PamyB and upstream of the terminator TamyB to create the vector pTYGS-hygR-BC2. The gene pyiR was amplified from M. grisea gDNA and inserted downstream of the strong constitutive PgpdA promoter in the vector pTYGS-bar which also contains a basta resistance gene (bar) under the control of the constitutive PtrpC promoter. This created the vector pTYGS-bar-pyiR. We previously showed that PgpdA is effective in M. grisea NI980.8
 | ||
| Fig. 2 Expression of egfp in P. oryzae Guy11. (A) Cartoon of expression system; (B) results of expression in P. oryzae Guy11. Left panels under white light, right panels under UV light. | ||
P. oryzae PEG/CaCl2 mediated protoplast transformation with pTYGS-hygR-BC2 resulted in the isolation of 14 hygromycin resistant transformants. In the case of M. grisea, transformation with pTYGS-bar-pyiR produced 40 basta-resistant transformants.
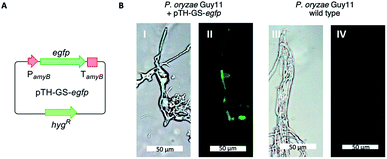
Eleven P. oryzae pTYGS-hygR-BC2 transformants were cultivated for 7 days in liquid DPY media (25 °C, 110 rpm) before cells and growth media were individually extracted and analysed by LCMS. In five transformants production of a new, dark brown compound 4 (MW = 348 g mol−1) was observed in the media extracts (Fig. 3). The production of 4 was not observed in extracts of P. oryzae Guy11 wild type (WT) strain. To test whether the PamyB BC2 construct is present in these transformants, a PCR experiment was designed to amplify the BC2 – TamyB junction. This showed the expected integration of BC2 into the genome of four genetically analysed transformants.
 | ||
| Fig. 3 LCMS traces of expression of BC2 in P. oryzae Guy11. (A) Extract of WT P. oryzae; (B–E) extracts of four representative strains of P. oryzae pTYGS-hygR-BC2. Diode array UV traces shown. | ||
1.3 mg of 4 were purified by preparative LCMS and the structure was elucidated by NMR. HRMS confirmed a molecular formula of C20H12NO6 ([M]H+ calculated 347.0542, found 347.0529). Couplings in the 1H–1H COSY spectrum revealed characteristic signals for aromatic regions. 4 contains 8 aromatic protons (H-1, H-7, H-8, H-12, H-13, H-18, H-19 and H-20, Fig. 4). Notably, the signal for H-1 is a singlet (δH 6.24 ppm); it demonstrated no coupling signal in the COSY spectrum. Based on the HMBC experiment, H-1 correlates to two carbonyl groups (C-6 at δC 193.5 ppm and C-3 at δC 182.6 ppm), one hydroxyl group (C-2 at δC 161.6 ppm) and one alkene (C-5 at δC 115.8 ppm) and is part of the first aromatic ring. Key HMBC signals for the adjacent aromatic ring originated from the hydroxyl group at C-10. Notably, only for 10-OH a signal (singlet at δH 12.86 ppm) was observed. Proton signals for the remaining three phenolic protons are missing (or too broad), presumably due to proton exchange in deuterated solvent. Starting from H-8, HMBC data revealed that the second and the third aromatic rings are connected by a C-9/C-11 carbon–carbon bond. Compound 4 was identified as the already known hinnulin A previously isolated from Nodulisporium hinnuleum.20
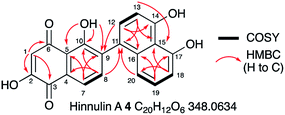 | ||
| Fig. 4 Summary of key 2D NMR correlations observed for 4. See ESI† for full data. | ||
Transcription of BC2 was determined by reverse transcriptase (RT) analysis of transformant VBI27-5. To this end, messenger RNA (mRNA) of VBI27-5 and WT P. oryzae Guy11 strains were extracted and converted into the corresponding complementary DNA (cDNA) before the samples were analyzed by PCR. RNA was used as negative control to ensure that no gDNA remained in the sample. As expected, expression of BC2 was confirmed for VBI27-5, but was absent in the WT strain.
In addition, expression of genes from the ACE1 BGC (ACE1, RAP1, ORFZ and ORF3) was tested. In accordance with the observed secondary metabolite profile, expression of all four genes was not observed. Apparently, expression of BC2 did not activate the biosynthetic pathway of the ACE1 metabolite under the tested conditions.
In the case of the M. grisea pTYGS-bar-pyiR transformants, 20 strains were tested by PCR of gDNA for the correct integration of the PgpdA-pyiR cassette. Five transformants were confirmed. One of these strains was grown in parallel to WT M. grisea NI980 in DPY liquid media for 7 days. After this time the liquid supernatant was extracted and the organic extract examined by LCMS. No differences in the chromatograms of the WT and pTYGS-bar-pyiR strains were observed (Fig. 5). However pyrichalasin H 1 was purified from each extract. The WT strain yielded 61.6 mg L−1 while the pTYGS-bar-pyiR strain yielded 190.4 mg L−1 (average over six flasks).
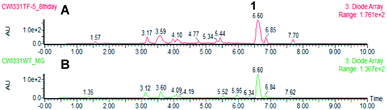 | ||
| Fig. 5 Results of expression of pyiR in M. grisea NI980. (A) Extract of M. grisea pTYGS-bar-pyiR; (B) extract of M. grisea NI980 WT. | ||
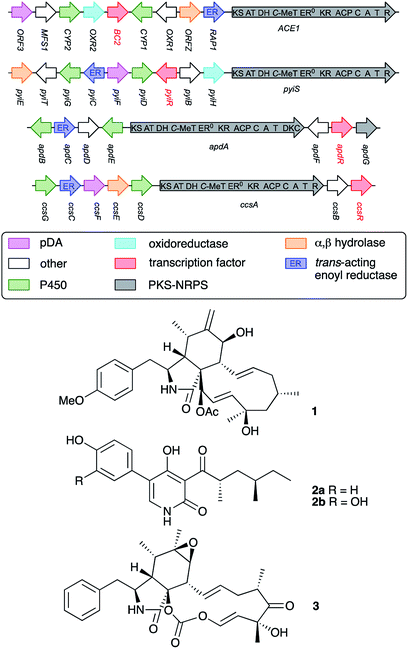
Remarkably, expression of BC2 in P. oryzae appeared to have influenced expression of another BGC, leading to the production of 4. Based on its structure, 4 seems to be related to fungal 1,8-dihydroxynaphthalene (DHN)-melanin. Therefore, the hinnulin BGC is likely to contain a gene encoding a PKS homologous to the alb1 gene (AFUA_2G17600) known to be involved in the synthesis of 1,3,6,8-tetrahydroxynaphthalene (T4HN) 5, the precursor of DHN in fungi. The Colletotrichum lagenarium PKS1 gene encodes a non-reducing-PKS (nrPKS) which is known to produce T4HN 5.21,22 A BLAST search was performed with the C. lagenarium PKS1 gene to identify a “melanin-like” BGC in P. oryzae Guy11. Two potential genes were identified: MGG_07219, (72% identity); and MGG_00428 (35% identify). MGG_07219 also shows 43% identity to the A. fumigatus Alb1 PKS which is also known to be involved in fungal melanin biosynthesis.23
Genes adjacent to MGG_07219 encode two transcription factors, a multicopper oxidase/laccase,25 a T4HN reductase23,26 and several hypothetical proteins of unknown function (Table 1). Significantly, laccase-like multicopper oxidases (MGG_07220) and T4HN reductases (MGG_07216) are known to be involved in melanin biosynthesis (vide infra).
| Gene | Putative function | Homolog | Ident. (%) | Ref. |
|---|---|---|---|---|
| MGG_07221 | Hypothetical protein | TLD08818.1 | 94 | — |
| MGG_15883 | Hypothetical protein | N/A | — | — |
| MGG_07220 | Verticullium dahliae laccase | XP_009649861.1 | 59 | — |
| Cu dep. DHN polymerase | ABR1 | 40 | 16 | |
| Cu dep. DHN polymerase | ABR2 | 25 | 16 | |
| MGG_15882 | Hypothetical protein | QBZ57338.1 | 94 | — |
| MGG_07219 | C. lagenarium PKS1 | BAA18956.1 | 72 | 13 and 14 |
| A. fumigatus alb1 THN PKS | XP_756095.1 | 43 | 15 | |
| MGG_07218 | Transcriptional regulator | KLU88469.1 | 65 | — |
| MGG_15881 | Hypothetical protein | ELQ35579.1 | 98 | — |
| MGG_07217 | Hypothetical protein | QBZ57332.1 | 81 | — |
| MGG_07216 | T4HN reductase | 1JA9_A | 99 | 17 and 24 |
| MGG_07215 | Transcriptional regulator | AAF37291.1 | 99 | — |
| XP_009216426.1 | 72 | |||
The genes adjacent to the second nrPKS gene, MGG_00428, encode a short-chain dehydrogenase/reductase (SDR), an O-methyl transferase (O-MeT), a 1,2-dioxygenase and several other hypothetical proteins unlikely to be involved in the biosynthesis of DHN-melanin or 4. Based on these findings, the MGG_07219 BGC is the best bioinformatic hit and might be involved in the biosynthesis of 4. However, further experiments were conducted to experimentally validate this hypothesis.
Expression of both PKS genes, MGG_07219 and MGG_00428, was tested by RT-PCR. Expression of MGG_07219 was observed in the transformant VBI27-5 but not in the WT P. oryzae Guy11 control. Expression of MGG_00428 was not observed in either the transformant VBI27-5 or in the WT control. These findings support the hypothesis that MGG_07219 might be involved in the biosynthesis of 4. To gain more insight into the identified BGC containing MGG_07219, expression of the six adjacent genes was tested. These encode a T4HN reductase, two transcription factors (TF1 and TF2), two hypothetical proteins and a laccase-like multicopper oxidase. Expression of the T4HN reductase gene (MGG_07216) and the gene encoding TF1 (MGG_07215) was observed for the wild type and the transformant strain. However, expression of the other genes was not observed in either the wild type strain or in the transformant.
Discussion
Our results show that upon expression of BC2 in P. oryzae Guy11, transcription of key genes from the ACE1 BGC including ACE1, RAP1, ORF3 and ORFZ was not observed. This corresponds with the absence of production of PKS-NRPS-related metabolites which are the known products of the ACE1 BGC.7 Apparently, this TF was not able to activate the ACE1 BGC under the tested conditions. One reason for this might be that BC2 is insufficient alone to initiate transcription. Another explanation might be that BC2 was not able to activate the ACE1 BGC if its expression is inhibited by the formation of heterochromatin in the region of the ACE1 BGC.27Notably, overexpression of BC2 in P. oryzae Guy11 does lead to the expression of the nrPKS encoded by MGG_07219 and to the production of hinnulin A 4 which was first identified in the fungus Nodulisporium hinnuleum. 4 belongs to the class of 1,2′-binaphthyl natural products,15 and would appear to be structurally related to fungal DHN-melanin. Most fungal melanins are derived from T4HN 5.28 The pathway involves reduction of T4HN 5 to scytalone 6 by T4HN reductase (T4HNR) and then dehydration of scytalone 6 by scytalone dehydratase (SDH) to form 1,3,8-trihydroxynaphthalene (T3HN) 7. This is then reduced again to form vermelone 8,29 at which point SDH acts again to form 1,8-DHN 9. 1,8-DHN 9 is then polymerised by various laccases to form melanin (Scheme 1). Oxidation of T3HN 7 is also known to lead to 2-hydroxy juglone 10 (ref. 30) although the molecular basis for this step in fungi is not known.
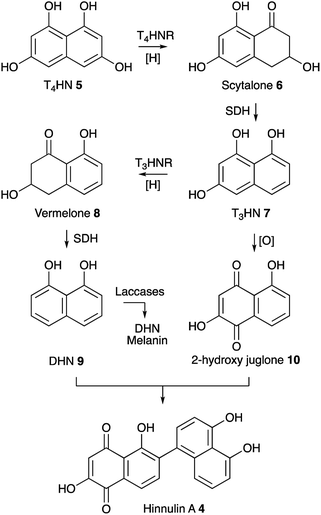 | ||
| Scheme 1 Known pathway to DHN melanin in fungi and proposed biosynthesis of 4. Abbreviations: T4HNR, T4HN reductase; SDH, scytalone dehydratase; T3HNR, trihydroxynaphthalene reductase. | ||
The MGG_07219 BGC also encodes a T4HNR and a laccase related to ABR1 and ABR2 which are proteins known to be involved in melanin formation in Aspergillus fumigatus. However, genes encoding SDH, T3HNR and other laccases are not present close to MGG_07219. Some fungi such as Talaromyces marneffei do contain a complete melanin BGC,31 but in other organisms such as Cladosporium fulvum the melanin biosynthetic genes appear not to be fully clustered.32 In the case of P. oryzae Guy11 suitable candidates for SDH-, T3HNR- and laccase-encoding genes are present elsewhere on the genome (see ESI† for details). Under the conditions tested MGG_07219 (nrPKS) and MGG_07220 (T4HNR) are expressed, likely giving scytalone 6 which is a known precursor of 9 and 10 and presumably 4. However further work will be required to find the missing pathway steps.
Interestingly it is known that the ACE1 signal molecule is produced in the appressoria of P. oryzae, which are also heavily melanized.2 The linkage between the ACE1 BGC and a putative melanin BGC in A. oryzae is therefore probably not coincidental. However further detailed experiments will be required to unpick these relationships. Our experiments also show that the pyiR encoded TF acts as expected as a pathway-specific activator in M. grisea NI980, increasing the titre of 1 by 3-fold. The differences between the actions of BC2 and pyiR emphasise the fact that the operation of regulatory genes in closely related BGC (i.e. the pyi and ACE1 clusters) are surprisingly different and remain cryptic to bioinformatic analysis. Again, significant future work will be required to untangle the differing control pathways responsible for the activation of these pathways.
Experimental
Organisms
P. oryzae Guy11 and M. grisea NI980 were obtained from Professor Marc-Henri Lebrun.Analytical and preparative methods
Analytical LCMS data was obtained using a Waters LCMS system comprising of a Waters 2767 autosampler, Waters 2545 pump, a Phenomenex Kinetex column (2.6 μm, C18, 100 Å, 4.6 × 100 mm) equipped with a Phenomenex Security Guard precolumn (Luna, C5, 300 Å) and a flow rate of 1 mL min−1. Detection was carried out by a diode array detector (Waters 2998) in the range 210 to 600 nm and an ELSD detector (Waters 2424) together with a mass spectrometer, Waters SQD-2 mass detector, operating simultaneously in ES+ and ES− modes between 150 and 1000 m/z. A solvent gradient was run over 15 min starting at 10% acetonitrile/90% HPLC grade water (0.05% formic acid) and ramping to 90% acetonitrile.Preparative LCMS was used to purify compounds from a raw extract or from a reaction mixture. This consisted of the same system described above with a Phenomenex Kinetex Axia column (5 μm, C18, 100 Å, 21.2 × 250 mm) and Phenomenex Luna, C5, 300 Å precolumn. A solvent gradient was run over 15 min starting at 10% acetonitrile/90% HPLC grade water (0.05% formic acid) and ramping to 90% acetonitrile. The flow was set to 20 mL min−1 and the post-column flow was split (100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) and the minority flow was made up with HPLC grade MeOH + 0.045% formic acid to 1 mL min−1 for simultaneous analysis by diode array detector (Waters 2998) in the range 210 to 600 nm and an ELSD detector (Waters 2424) together with a mass spectrometry, Waters SQD-2 mass detector, operating simultaneously in ES+ and ES− modes between 150 and 1000 m/z. Detected peaks were collected into glass test tubes. Combined tubes were evaporated (vacuum centrifuge) and residues analysed directly.
1) and the minority flow was made up with HPLC grade MeOH + 0.045% formic acid to 1 mL min−1 for simultaneous analysis by diode array detector (Waters 2998) in the range 210 to 600 nm and an ELSD detector (Waters 2424) together with a mass spectrometry, Waters SQD-2 mass detector, operating simultaneously in ES+ and ES− modes between 150 and 1000 m/z. Detected peaks were collected into glass test tubes. Combined tubes were evaporated (vacuum centrifuge) and residues analysed directly.
NMR measurements were acquired on Bruker DRX 400, Bruker Avance 500 or Bruker Avance 600 MHz spectrometers (Institute for Organic Chemistry, Leibniz Universität Hannover). Chemical shifts are shown in parts per million (ppm) in comparison to the TMS (Tetramethylsilane) standard. Coupling constants J are quoted in Hz.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
DFG is thanked for the provision of NMR and LCMS equipment (INST 187/621-1, INST 187/686-1). VH was funded by DFG (CO 1328/2-1) and CW was funded by the China Scholarship Council (2016 08310143). Dr Colin Lazarus, University of Bristol, is thanked for the gifts of pTHGS-eGFP and pTYGS-ade. The publication of this article was funded by the Open Access Fund of the Leibniz Universität Hannover.Notes and references
- R. Dean, J. Kan, Z. Pretorius, K. Hammond-Kosack, A. Pietro, P. Spanu, J. Rudd, M. Dickman, R. Kahmann, J. Ellis and G. Foster, Mol. Plant Pathol., 2012, 13, 414–430 CrossRef PubMed
.
- R. Howard and B. Valent, Annu. Rev. Microbiol., 1996, 50, 491–512 CrossRef CAS
.
- P. Skamnioti and S. J. Gurr, Plant Cell, 2007, 19, 2674–2689 CrossRef CAS
.
- R. Wilson and N. Talbot, Nat. Rev. Microbiol., 2009, 7, 185–195 CrossRef CAS PubMed
.
- H. Bohnert, I. Fudal, W. Dioh, D. Tharreau, J. Notteghem and M.-H. Lebrun, Plant Cell, 2004, 16, 2499–2513 CrossRef
.
- J. Collemare, M. Pianfetti, A.-E. Houlle, D. Morin, L. Camborde, M.-J. Gagey, C. Barbisan, I. Fudal, M.-H. Lebrun and H. U. Bohnert, New Phytol., 2008, 179, 196–208 CrossRef CAS
.
- R. Berruyer, H. Adreit, J. Milazzo, S. Gaillard, A. Berger, W. Dioh, M.-H. Lebrun and D. Tharreau, Theor. Appl. Genet., 2003, 107, 1139–1147 CrossRef CAS PubMed
.
- I. Fudal, J. Collemare, H. U. Bohnert, D. Melayah and M.-H. Lebrun, Eukaryotic Cell, 2007, 6, 546–554 CrossRef CAS
.
- J. Collemare, R. O'Connell and M. Lebrun, New Phytol., 2019, 232, 590–596 CrossRef
.
- E. J. Skellam, Nat. Prod. Rep., 2017, 34, 1252–1263 RSC
.
- Z. Song, W. Bakeer, J. Marshall, A. Yakasai, R. Khalid, J. Collemare, E. J. Skellam, D. Tharreau, M.-H. Lebrun, C. M. Lazarus, A. M. Bailey, T. J. Simpson and R. J. Cox, Chem. Sci., 2015, 6, 4837–4845 RSC
.
- X.-L. Yang, S. Friedrich, S. Yin, O. Piech, K. Williams, T. J. Simpson and R. J. Cox, Chem. Sci., 2019, 10, 8478–8489 RSC
.
- C. Wang, V. Hantke, R. J. Cox and E. J. Skellam, Org. Lett., 2019, 21, 4163–4167 CrossRef CAS
.
- S. Bergmann, J. Schümann, K. Scherlach, C. Lange, A. A. Brakhage and C. Hertweck, Nat. Chem. Biol., 2007, 3, 213–217 CrossRef CAS
.
- Z. Wasil, K. Pahirulzaman, C. P. Butts, T. J. Simpson, C. M. Lazarus and R. J. Cox, Chem. Sci., 2013, 4, 3845–3856 RSC
.
- K. Qiao, Y.-H. Chooi and Y. Tang, Metab. Eng., 2011, 13, 723–732 CrossRef CAS
.
- W. I. M. M. Bakeer, PhD thesis, University of Bristol, School of Chemistry, 2011
.
- G. Zhang, V. Gurtu and S. R. Kain, Biochem. Biophys. Res. Commun., 1996, 227, 707–711 CrossRef CAS
.
- K. Tsuchiya, S. Tada, K. Gomi, K. Kitamoto, C. Kumagai and G. Tamura, Biosci., Biotechnol., Biochem., 1992, 56, 1849–1853 CrossRef CAS PubMed
.
- G. Schlingman, S. Luckman, L. A. McDonald and F. E. Koehn, Rec. Nat. Prod., 2011, 5, 252–260 Search PubMed
.
- Y. Takano, Y. Kubo, K. Shimizu, K. Mise, T. Okuno and I. Furusawa, Mol. Gen. Genet., 1995, 249, 162–167 CrossRef CAS
.
- A. Vagstad, E. Hill, J. Labonte and C. Townsend, Chem. Biol., 2012, 19, 1525–1534 CrossRef CAS
.
- A. Watanabe, I. Fujii, H. Tsai, Y. C. Chang, K. J. Kwon-Chung and Y. Ebizuka, FEMS Microbiol. Lett., 2000, 192, 39–44 CrossRef CAS
.
- J. Thompson, S. Fahnestock, L. Farrall, D.-I. Liao, B. Valent and D. Jordan, J. Biol. Chem., 2000, 275, 34867–34872 CrossRef CAS
.
- S. Upadhyay, G. Torres and X. Lin, Eukaryotic Cell, 2013, 12, 1641–1652 CrossRef CAS
.
- D. I. Liao, J. E. Thompson, S. Fahnestock, B. Valent and D. B. Jordan, Biochemistry, 2001, 40, 8696–8704 CrossRef CAS
.
- J. Collemare and M. Seidl, FEMS Microbiol. Rev., 2019, 1–17 Search PubMed
.
- D. Conradt, M. Schätzle, S. Husain and M. Müller, ChemCatChem, 2015, 7, 3116–3120 CrossRef CAS
.
- T. J. Simpson and M. Weerasooriya, J. Chem. Soc., Perkin Trans. 1, 2000, 2771–2775 RSC
.
- M. H. Wheeler and R. D. Stipanovic, Arch. Microbiol., 1985, 142, 234–241 CrossRef CAS
.
- P. Woo, E. Tam, K. Chong, J. Cai, E. Tung, A. Ngan, S. Lau and K. Yuen, FEBS J., 2010, 277, 3750–3758 CrossRef CAS
.
- S. A. Griffiths, R. J. Cox, E. J. R. Overdijk, C. H. Mesarich, B. Saccomanno, C. M. Lazarus, P. J. G. M. de Wit and J. Collemare, PLoS One, 2018, 13, e0209600 CrossRef
.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9ra07028a |
| This journal is © The Royal Society of Chemistry 2019 |