Open Access Article
Open Access ArticleTrichoderma asperellum T42 induces local defense against Xanthomonas oryzae pv. oryzae under nitrate and ammonium nutrients in tobacco†
Bansh Narayan Singh ab,
Padmanabh Dwivedi*b,
Birinchi Kumar Sarmac and
Harikesh Bahadur Singhc
ab,
Padmanabh Dwivedi*b,
Birinchi Kumar Sarmac and
Harikesh Bahadur Singhc
aInstitute of Environment and Sustainable Development, Banaras Hindu University, Varanasi 221005, India
bDepartment of Plant Physiology, Institute of Agricultural Sciences, Banaras Hindu University, Varanasi 221005, India. E-mail: pdwivedi25@rediffmail.com
cDepartment of Mycology and Plant Pathology, Institute of Agricultural Sciences, Banaras Hindu University, Varanasi 221005, India
First published on 3rd December 2019
Abstract
Trichoderma has been explored and found to play a vital role in the defense mechanism of plants. However, its effects on host disease management in the presence of N nutrients remains elusive. The present study aimed to assess the latent effects of Trichoderma asperellum T42 on oxidative burst-mediated defense mechanisms against Xanthomonas oryzae pv. oryzae (Xoo) in tobacco plants fed 10 mM NO3− and 3 mM NH4+ nutrients. The nitrate-fed tobacco plants displayed an increased HR when Xoo infected, which was enhanced in the Trichoderma-treated plants. This mechanism was enhanced by the involvement of Trichoderma, which elicited NO production and enhanced the expression pattern of NO-modulating genes (NR, NOA and ARC). The real-time NO fluorescence intensity was alleviated in the NH4+-fed tobacco plants compared to that fed NO3− nutrient, suggesting the significant role of Trichoderma-elicited NO. The nitrite content and NR activity demonstration further confirmed that nitrate metabolism led to NO generation. The production of ROS (H2O2) in the plant leaves well-corroborated that the disease resistance was mediated through the oxidative burst mechanism. Nitrate application resulted in greater ROS production compared to NH4+ nutrient after Xoo infection at 12 h post-infection (hpi). Additionally, the mechanism of enhanced plant defense under NO3− and NH4+ nutrients mediated by Trichoderma involved NO, ROS production and induction of PR1a MEK3 and antioxidant enzyme transcription level. Moreover, the use of sodium nitroprusside (100 μM) with Xoo suspension in the leaves matched the disease resistance mediated via NO burst. Altogether, this study provides novel insights into the fundamental mechanism behind the role of Trichoderma in the activation of plant defense against non-host pathogens under N nutrients.
1. Introduction
Nitrogen (N) is an essential component for plant metabolism. It is the main component of chlorophyll, nucleic acid, and amino acid biosynthesis. Nitrogen plays a crucial role in plant development and disease management.1–6 The primary issue of N scarcity in soil is due to its slow mineralization, leaching, and de-nitrification process. Accordingly, a lack of N availability to plants results in several physiological and morphological responses.7 Thus, to minimize N deficiency, plants have a unique mechanism through which they overcome the problem of low N. Specifically, different transporters residing on the surface of the roots of plants can enhance the nitrogen uptake efficiency. These transporters have different affinities and subclasses for nitrate (NO3−) and ammonium (NH4+) uptake.5 N is applied in agricultural soil in the form of nitrate (NO3−), ammonium (NH4+) or combined. These forms of N activate several defense mechanisms in plant systems against pathogens.1,2 These defense mechanisms are associated with the involvement of PAMP or ETI (effector-triggered immunity), which reprograms different nuclear genes,8,9 and thus N is required for enhancing plant immunity.6 The PAMP defense response involving the rapid development of plant immunity in response to pathogen infections is referred to as the hypersensitive response (HR), which seems to appear after avirulent pathogen and plant interactions.10 A characteristic of this local HR is the rapid generation of reactive oxygen species (O2− and H2O2) and nitric oxide (NO). N is an essential component that facilitates the HR response against avirulent pathogens.1 However, N is not only involved in increasing the plant nutrient status, but its content affects disease defense. NO is a signaling molecule that was observed to be rapidly generated during plant pathogen-interactions and considered one of the important components in defense activators.11 The involvement of NO in the activation of defense signaling cascades has been well studied. The reactions of NO with cysteine-rich thiol groups and nitration of tyrosine-rich groups with different proteins are referred as S-nitrosylation, indicating the regulatory results mediated by NO.12,13 The modification of the SA marker protein (NPR1) and ROS generating complex (RBOH) in Arabidopsis during defense responses are considered good examples of the role of S-nitrosylation.14–16 In plants, RBOH production leads to localized ROS bursts to regulate plant development and immune responses under stress conditions. Induced plant immune responses are associated with the interaction of elicitors with the plant. Different response molecules and physiological changes occur during the interaction of elicitor molecules and plants.17,18 The interaction of Trichoderma as an elicitor with plants stimulates various defense molecules such as NO, reactive oxygen species, SA, and genes related to defense. Moreover, Trichoderma may induce multiple secondary metabolites, auxin production and phenolic compounds, which may enhance the defense mechanisms in plants.19–21Therefore, systems that can enhance the affinity for N uptake and promote disease resistance in plants are necessary. Several plant symbiotic microbes are well known as plant growth promoters, which facilitate nutrient availability to plants. However, the minimal use of these microbes in plant defense occurs in the presence of nitrate and ammonium or both supplements. Several species of Trichoderma have been demonstrated to play a key role in disease management against biotic stresses. They promote plant defense via various processes such as mycoparasitism, antibiosis, and activation of the basal defense mechanism. Colonization of Trichoderma within apoplastic cells led to an enhanced defense response in Arabidopsis6,22 and pea20 rapidly. Moreover, the colonization of Trichoderma with plant roots activated different defense-related enzyme activities and regulation of PR pathogenesis gene.6,12 Perhaps, the activation of these defense responses is due to the modulation of metabolites and defense genes involved in the resistance mechanism.20,23
If Trichoderma initiates an enhanced N uptake efficiency6 and NO accumulation,12 it may be implicated in plant defense via HR-mediated cell death. Trichoderma asperellum T42 as a biological control agent and Xanthomonas oryzae pv. oryzae (Xoo) were used in the present experiment. In the previous studies, the T42 strain was used as the most effective microbe in nitrogen utilization and root development in tobacco.4 Herein, we aimed to assess the effects of two forms of N (NO3− and NH4+) nutrients on the defense response in tobacco plants via HR-mediated cell death against the non-host bacterial pathogen Xoo induced by the symbiotic rhizospheric fungus Trichoderma asperellum T42. We demonstrated that Trichoderma asperellum T42 promotes the oxidative burst response in tobacco grown under nitrate and ammonium nutrient media via the involvement of both NO and ROS.
2. Experimental setup
2.1 The growth of Trichoderma asperellum T42 and Xanthomonas oryzae pv. oryzae and inoculation preparation
T. asperellum (T42 strain; Gene Bank Accession: JN128894) was routinely cultured on potato dextrose agar (PDA) plates at 28 ± 2 °C for 6–7 days until sporulation. Before treatment with the T42 strain, the spores of the fungus were washed with 0.80% NaCl solution and the spore suspension was diluted to 1 × 106 CFU mL−1. The Xanthomonas oryzae pv. oryzae strain was purchased from the Centre for Cellular & Molecular Biology (Hyderabad, India), and used as a non-host pathogen (incompatible) in tobacco plants. Before infiltration in the tobacco leaves, Xoo bacteria were grown for four days in peptone sugar agar (PSA) media. After incubation, the bacterial cell density was diluted to 1 × 107 CFU mL−1 to ensure each experiment contained an approximately accurate number of cells during infiltration.2.2 Plant material and experimental setup conditions
Six-week-old tobacco plants (Nicotiana tabacum cv. Xanthi) were used in the experiment. Before two-week-old tobacco plantlets were transferred to Perspex tubes, sterilized tobacco seeds were primed with a spore suspension of T42 strain according to the previously described method,4,24 and allowed to germinate without nitrogen supplements at 25 °C in the culture lab. After 2 weeks, the tobacco plants were transferred to Perspex tubes (capacity 1.8 L, 0.5 m high, and 15 cm inner diameter) containing a hydroponic solution for four weeks and provided continuous growth conditions (D/N, 16/8 h; temperature, 22 °C/20 °C; relative humidity, 70%; artificial light of photosynthetic photon flux density (PPDF), 350–400 μmol m−2 s−1). The hydroponic culture solution for nitrate nutrient (pH 6.3) consisted of 10 mM KNO3, 1 mM CaCl2, 1 mM MgSO4, 25 μM NaFe-EDTA, 0.5 mM K2HPO4, and 1 mM KH2PO4, while for ammonium nutrition (pH 6.0–6.2), 3 mM NH4Cl, 1 mM CaCl2, 1 mM MgSO4, 25 μM NaFe-EDTA, 0.5 mM K2HPO4, 1 mM KH2PO4 (ref. 4 and 25) and trace elements.26 The nutrient solutions were supplemented to the growing plants and routinely replaced every two days. Distilled water used in place of the nitrogen supplement was treated as the control.2.3 Infiltration of chemicals and bacterial strain in tobacco leaves
To study the modulation of endogenous NO in the HR response, 100 μM SNP (sodium nitroprusside) and 200 μM cPTIO (carboxy-2-phenyl-4,4,5,5-tetramethylimidazoline-1-oxyl-3-oxide; Alexis Biochemicals) were infiltrated with Xoo suspension. Stock solutions of SNP and cPTIO were prepared in distilled water (Alexis Biochemicals). The suspension culture containing Xoo inoculum was prepared in 10 mM MgCl2. There were four treatments: 10 mM MgCl2 (control), Xoo suspension, Xoo suspension + 100 μM SNP and Xoo suspension + 100 μM SNP + 200 μM cPTIO. Each treatment was infiltrated in a total of five groups of tobacco plants, T0: distilled water plants, T1: T42 strain inoculated plants, T2: NO3−-fed plants, T3: NO3−-fed + T42 strain inoculated plants, T4: NH4+-fed plants, and T5: NH4+-fed + T42 strain inoculated plants. Infiltration without Xoo cells and any chemical treatments was considered as the control (only 10 mM MgCl2). All the treatments were infiltrated on the abaxial side of the tobacco leaves.In addition, we studied the effect of SNP on the growth of Xoo under in vitro conditions. Different concentrations of SNP were freshly prepared and added to PSA media at 4 °C. Then, the spore suspension (0.05 OD600) of Xoo was spread on the PSA media and the growth of the Xoo colony was examined after 4–5 days (ESI Fig. 1†).
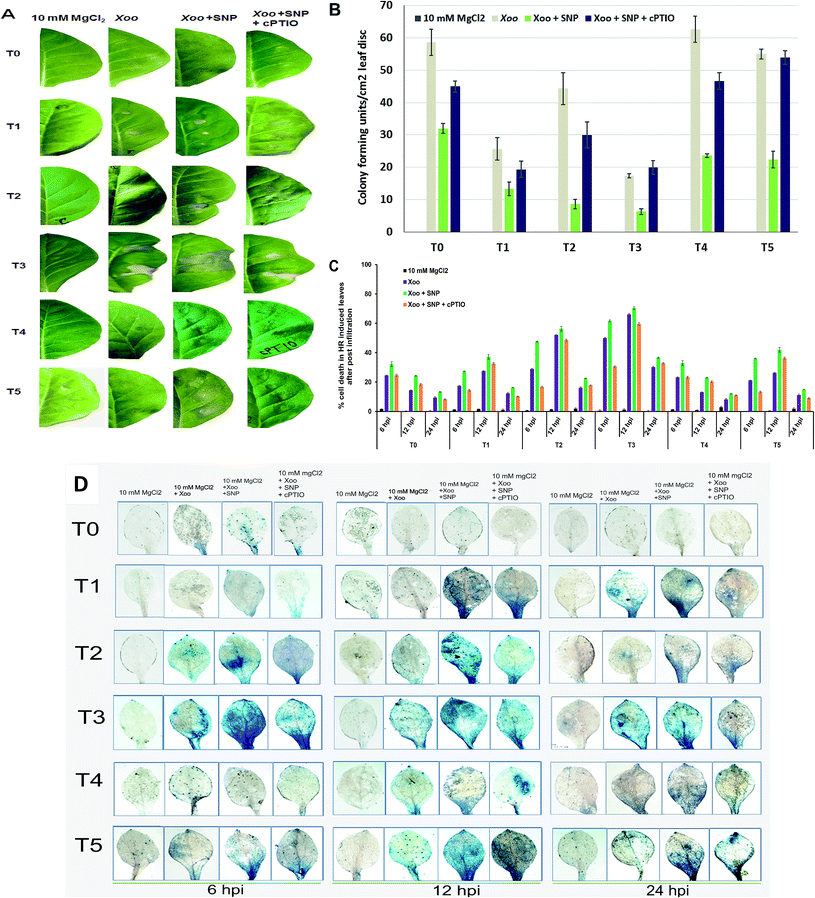
The lesion-induced leaves from each group of plants from each treatment were harvested. Twenty regions per leaf and 6 leaves per plant were selected from each experiment. The bacterial number in the leaves from each treatment during HR induction was counted according to the protocols mentioned in a previous study.27
2.4 Cell death assay
Cell death assay in the leaves of the different treatment groups of plants was estimated using Evan's blue solution with slight modification.28 The membrane damage or destabilized membrane is a hallmark of biotic stress. Evan's blue, an azo dye, is used to determine the cell viability of plant cells. Evan's blue quickly penetrates the ruptured membrane and stains cell. Accordingly, plant cells subjected to biotic stress exhibited a greater accumulation of Evan's blue dye compared to the control. In contrast, the healthy plant cells maintained their membrane integrity and did not allow blue protoplasmic staining, which could be easily quantified spectrophotometrically or visualized without the use of a camera.2.5 Assay for histochemical visualization and quantification of H2O2 content
The histochemical staining of H2O2 was performed as previously described29 with a few modifications. In the case of H2O2, HR lesions of leaves were dipped in 3,3′-diaminobenzidine (DAB; 1 mg ml−1, pH 3) and incubated for 6–8 h in the dark at 25 °C. Dechlorophylization was performed by transferring the HR samples to a bleaching solution of ethanol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) acetic acid
acetic acid![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) glycerol (3
glycerol (3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v) and boiling on a water bath for 10–15 min at 90 °C. The leaves were rinsed briefly in distilled water and mounted in lactic acid
1, v/v) and boiling on a water bath for 10–15 min at 90 °C. The leaves were rinsed briefly in distilled water and mounted in lactic acid![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) phenol
phenol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water (1
water (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v), then observed under a microscope. Estimation of the H2O2 content from the different treatment groups performed according to the previously described method.30
1, v/v), then observed under a microscope. Estimation of the H2O2 content from the different treatment groups performed according to the previously described method.30
2.6 Chlorophyll content determination
Leaf discs from HR-induced regions were collected, and the leaf tissues homogenized in 80% acetone. The total chlorophyll content was monitored spectrophotometrically at 645 and 663 nm in the inoculated and uninoculated leaves, respectively, and chlorophyll content was calculated as described in Arnon.312.7 Antioxidant enzyme assays
One gram of HR lesion sample was homogenized with a chilled mortar and pestle at 4 °C in 4 mL 50 mM phosphate buffer (pH 7.8) containing 1 mM EDTA and 2% PVP. The homogenate was centrifuged at 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 20 min at 4 °C and the supernatant was used for the subsequent enzyme activity assays. Total protein content was determined by the method described previously.32
000g for 20 min at 4 °C and the supernatant was used for the subsequent enzyme activity assays. Total protein content was determined by the method described previously.32
Superoxide dismutase (SOD; EC 1.15.1.1) activity was estimated by the method proposed previously.33 Samples without enzyme extract were taken as the controls. The absorbance was recorded at 560 nm and one unit of enzyme was taken as the quantity of enzyme that reduced the absorbance reading of the sample to 50% in comparison to the tubes lacking enzyme. Catalase activity (CAT: EC 1.11.1.6) was measured using the previously described method.34 Ascorbate peroxidase (APX: EC 1.11.1.11), which acts as an H2O2 scavenger, was measured using the previously described method.35
2.8 Assay for nitrite content
Nitrite level was measured according to the previously described method with some modifications.25 HR-induced leaf samples (1 g) were crushed in liquid N2 and 1200 μL (per sample) reaction mixture containing 500 μL sulphanilamide (1%), 500 μL N-(1)-(naphthyl) ethylenediamine-dihydrochloride (0.02%), and 200 μL zinc acetate (0.5 M) immediately added, and incubated at 25 °C for 3–4 h. Subsequently, the reaction mixture was centrifuged at 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 5 min. The absorbance of the chromophoric azo product was measured at 540 nm.
000g for 5 min. The absorbance of the chromophoric azo product was measured at 540 nm.
2.9 Assay of nitrate reductase activity
NR activity in the HR lesion samples was determined through the previously described method.36 The intensity of the pink color was measured with a spectrophotometer (SpectraMax Me2, Molecular Devices, USA) at 540 nm.2.10 Spectrofluorometry-based detection of NO production
For fluorometric NO determination, the fluorophore 4,5-diaminofluorescein-FM diacetate (DAF-FM DA) (Alexis Biochemicals, Gruenberg, Germany) was used. DAF-FM DA (10 μM) probing dye (stock prepared in 100 mM HEPES-KOH, pH 7.5) was preloaded in the leaves before infiltration. The leaves were kept in the dark at 25 °C for 30 min. Then, the different treatment plant groups were re-infiltrated in the same region where DAF-FM DA was preloaded. MgCl2 (10 mM) was used as a control. Then, 100 mg leaves was excised from the HR-induced leaves with the help of a cork borer (size 0.79 cm2) and immediately crushed in liq. N2. Then, 700 μL of pre-chilled 100 mM HEPES-KOH (pH 7.5) buffer was added and centrifuged (16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g, 15 min, at 4 °C). The reaction product (supernatant) was used for the measurement of NO production at 488 nm excitation and 500–515 nm emission wavelengths (2 nm bandwidth) with spectrofluorimetry (Spectra-Max 2, Molecular Devices, USA). Relative fluorescence was expressed as arbitrary units (AU) with some modifications.25
000g, 15 min, at 4 °C). The reaction product (supernatant) was used for the measurement of NO production at 488 nm excitation and 500–515 nm emission wavelengths (2 nm bandwidth) with spectrofluorimetry (Spectra-Max 2, Molecular Devices, USA). Relative fluorescence was expressed as arbitrary units (AU) with some modifications.25
2.11 In vivo measurement of NO emission
The HR-induced leaves were incubated in 10 μM DAF-FM DA probe (4-amino-5-methylamino-2′,7′-difluorofluorescein-diacetate) in 100 mM HEPES-KOH buffer (pH 7.5) and placed in cavity glass slides and incubated for 5 min in the dark at 25 °C.37 The procedure for DAF-FM DA loading and infiltration was similar to that described above. Then, the leaf samples were rinsed three times for 5 min with fresh 10 mM HEPES-KOH (pH 7.5) buffer and transferred onto a slide and visualized (Nikon Eclipse 90i fluorescence microscope, Nikon Instruments Inc. America, excitation at 488 nm, emission at 500–515 nm).2.12 RNA extraction, cDNA preparation and qRT-PCR
Total RNA was harvested from the HR-induced leaf samples from each group of plants after post-infiltration at 6, 12, and 24 h according to the method described previously.20 cDNA preparation and qPCR were performed according to the method by Singh et al.4 The synthesized cDNA used as a template for the analysis of targeted genes is listed in ESI Table 1.† qRT-PCR was carried out in an iQ5 Real-Time PCR Detection System (Bio-Rad Laboratories, Munchen, Germany) using SYBR Green chemistry (Affymetrix SYBR® Green Supermix Kit). The transcript level of mRNA was normalized and determined with the level of housekeeping gene β-tubulin for tobacco. The data obtained by real-time PCR of the different treatment groups was normalized with the 2−ΔΔCT value method.38 Three biological replicates were used for each group of plants for each treatment.2.13 Statistical analysis
The data obtained from the different experiments is shown as mean ± standard error (SE) and subjected to analysis of variance (ANOVA). The treatment mean values were compared by Duncan's multiple range test at the p < 0.05 significance level and analyzed by SPSS ver. 16 (SPSS Inc., Chicago, IL). ClustVis, an online web tool, was used for heat map development.3. Result and discussion
Crop production is challenging for farmers in the 21st century.39 The continuous increment of microbes in the ecosystem increases pressure on agricultural production. Nitrogen availability in different forms directly affects the growth and development of plants.7 Since N is an essential component of macronutrients, it plays a major role in the defense mechanisms of plants.1 Ammonium ions are readily available to plants in comparison to nitrate ions, but an excess amount cause toxic effects to plant.1,6 However, plants prefer NO3− as the N source for growth and disease resistance.1,5 In addition, nitrate availability in the soil can help charge balance.40 However, due to leaching, plants are unable to uptake sufficient amounts of nitrate. Several investigations have highlighted the role of nitrate in different physiological aspects, for example, root differentiation in tobacco4 and local defense response against Pseudomonas syringae,6 but there has been much less work done concerning HR-mediated disease resistance. Currently, the continuous increase in disease incidence and nutrient deficiency make plants more prone to pathogen invasion, especially since the R-gene-mediated HR-linked defense is often ephemeral.41 Therefore, alternative strategies including microbial recruitment by plant roots may help solve this problem. Accordingly, it is necessary to characterize this type of pathological approach, which may compromise or increase HR-mediated disease resistance. In the present investigation, we present the effects of NO3− and NH4+ nutrition on the defense response with or without Trichoderma asperellum T42 in tobacco plants.3.1 Nitrogen nutrients and Trichoderma recruitment affect defense response via HR-mediated cell death
Plant resistance is a complex process and exploring the signal mechanisms is the primary goal to understand host responses against invading pathogens. Oxidative burst in the plasma membrane is a primary and rapid event occurring in the defense mechanism of plants.28 Evidence has supported that the growth of incompatible or compatible pathogens is restricted by early host cell death during the HR reaction.42 The HR is frequently triggered by the rapid generation of reactive oxygen species (O2−, H2O2, OH−, etc.), particularly superoxide, which is a very unstable free radical and rapidly mutated into H2O2 and molecular oxygen. In this study, we demonstrate the HR in the infiltrated region after challenged with Xoo (Fig. 1). According to the comparison in Fig. 1A (T0, T2 and T4 plants) 12 hpi, the NO3− (T2)-fed plants showed a higher level of HR compared to NH4+ (T4). In contrast, no HR was observed at the sites of infiltration in the plants grown without nutrient (T0). Also, the induction of cell death was found to be more significant in the NO3− grown plants at 12 hpi in comparison to the control (T0) and NH4+-fed (T4) plants (compare Fig. 1C and D, respectively). We further found that maximum cell death increased in the NO3− grown plants (Fig. 1C). These results suggest that N is essential for the defense response, and the NO3− nutrient is better than NH4+ for the activation of the local defense response against non-host pathogens. Further, we studied the effect of Trichoderma asperellum T42 in the activation of the local defense response. We found that the HR was greater in the plants grown in Trichoderma plus NO3− (T3) than Trichoderma plus NH4+ (T5). While, in the case of the T42 grown plants (T1), the HR was greater than the control plants (T0) (Fig. 1A). Thus, these results further support that plants require N for growth and disease development and Trichoderma asperellum T42 inoculation can help in increasing the N utilization efficiency.4 Similarly, the inoculation of some groups with Trichoderma promoted their nutrient absorption capacity, which elevated the local defense mechanism.22 It was evidenced that Trichoderma asperelloides inoculation elevated the local defense mechanisms in plants against both biotic and abiotic stresses.43Furthermore, the CFU count and cell death observations suggest that the NH4+-grown plants displayed compromised disease resistance both in the plants inoculated with and without Trichoderma (Fig. 1B and C, respectively). After evaluating the effects of N nutrients and Trichoderma in the local defense response against Xoo, we further assessed the impact of the NO donor (SNP) in HR induction. Co-infiltration of SNP with Xoo increased the intensities of the HR compared to without NO donor in all the treated plants (Fig. 1). The application of cPTIO confirmed the importance of the NO donor in the induction of HR. These findings indicate that Trichoderma asperellum T42 inoculation sustained the HR against bacterial pathogen Xoo interaction, and an especially greater response was found in NO3− nutrient. However, there are scarce reports available on demonstrating the role of Trichoderma asperellum T42 in the activation of the local defense mechanism in plants grown under different forms of N nutrients. Hence, in this experiment, we studied the comparative impact of Trichoderma on enhancing the local defense response against Xoo stress under NO3− and NH4+ nutrients.
We also observed cell death efficiency through histochemical staining using the Evan's blue method (Fig. 1D). The cell death efficiency was observed to be higher in the NO3−-grown plants compared to NH4+ at 12 hpi. Meanwhile, T42 inoculation in the plants further enhanced the cell death efficiency and the maximum efficiency was observed in NO3− nutrient after challenged with Xoo pathogen. A reduction in cell death was observed in the case of the plants grown without any treatments (Fig. 1D, upper panel). This efficiency was further increased through SNP application, suggesting that NO3− nutrient has a greater effect in the local defense response (HR induction), where even inoculation with Trichoderma promoted HR induction. Previous reports have studied the induced local defense response with Trichoderma virens inoculation in tomato leaves after exposure to Rhizoctonia solani.12,44 Here, the Trichoderma asperellum T42-treated tobacco plants activated local defense mechanisms, which influenced Xoo invasion. Our results suggest that the suppression of local disease symptoms in the Trichoderma asperellum T42 plus NO3− plants may be correlated with the activation of the plant defense response through other signaling networks, as demonstrated by Wany et al. (2018) in Arabidopsis plants protected by Trichoderma strains against Pseudomonas syringae.6
3.2 H2O2 production and RBOH transcript accumulation helps HR induction in Xoo challenged plants
ROS generation is a continuous process, but in the presence of elicitors/or incompatible pathogens, it drastically increases. ROS generation is a result of constant metabolism processes and may involve NADPH oxidase/RBOH activities.45 Furthermore, ROS are by-products generated from different part of plant organs such as chloroplast, mitochondria, and peroxisome; however, ROS production participates in HR-mediated cell death.1 Evidence has highlighted that Trichoderma spp. induces ROS against several biotic stresses46 and together with NO, plays an important role in HR-mediated cell death.47 H2O2 is the most critical ROS molecule, which acts as a signal transducer in HR induction.48 Herein, we investigated whether H2O2 production participating in cell death is linked to disease resistance, which is regulated by plant homologs membrane-associated RBOHs in plants against pathogens. We found that the induction of H2O2 production was significantly greater after Xoo challenged, and particularly greater in the NO3−-fed plant leaves (T2) compared to the NH4+-fed plants (T4) (Fig. 2A), similarly to H2O2 production in Arabidopsis.42 However, it was low without any treatment (T0), even after Xoo challenged. H2O2 production was found to be maximum in the plant leaves grown under Trichoderma plus NO3− treatment (3.72 μmol g−1 FW) at 6 hpi among the treatments (Fig. 2A). DAB staining also followed a similar pattern of observations (Fig. 2B; H2O2 staining images at 6 and 24 hpi not shown). A similar view was also reported in maize, where H2O2 production was generated in the early stage of infection of a fungal hypha of Colletotrichum graminicola.20,49 Moreover, H2O2 production was also observed in Arabidopsis after colonization with T. virens43 and in pea by Trichoderma asperellum.20 The higher accumulation of H2O2 production in the Trichoderma-inoculated plants grown in N nutrients provides evidence that Trichoderma can promote H2O2 production under N nutrients during Xoo infection. A previous study demonstrated that the T42 strain increased the nitrogen use efficiency in tobacco plants.4 However, H2O2 production was reduced after co-infiltration of 100 μM SNP with Xoo, and the lowest production was observed in the control (T0) plant leaves (0.079 μmol g−1 FW). Interestingly, cPTIO reversed the effect of SNP in all the treatment groups.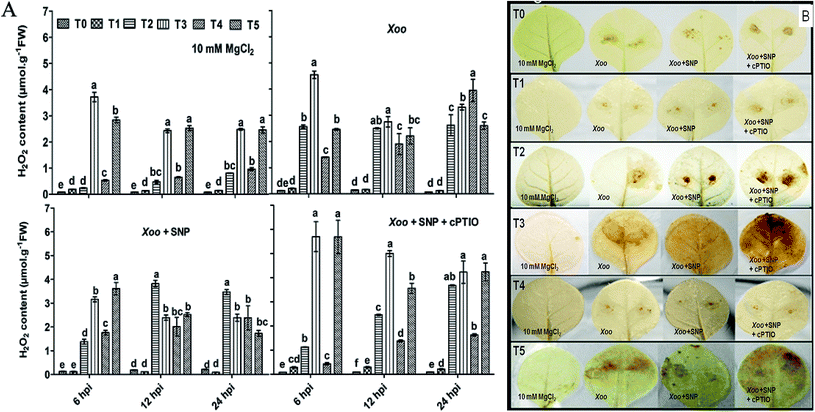 | ||
| Fig. 2 Detection of ROS production in HR-induced tobacco leaves. Reactive oxygen species (H2O2) generation in HR-induced tobacco leaves modulated by Xoo, as analyzed spectrophotometrically at 6 to 24 hpi (Fig. 2A). Histochemical staining of H2O2 production with DAB dye in tobacco leaves at 12 hpi (Fig. 2B). H2O2 accumulation in tobacco leaves grown in different treatments: T0 = dist. H2O, T1 = T42 treatment, T2 = NO3− nutrition, T3 = T42 + NO3− nutrition, T4 = NH4+ nutrition, and T5 = T42 + NH4+ nutrition at 6, 12 and 24 hpi. Suspension cultures of Xoo infiltrated with 100 μM SNP and 200 μM cPTIO. Suspension culture of Xoo was prepared in 10 mM MgCl2 as a control. Error bars represent SEs from the mean of three measurements. Different superscript letters indicate significant differences from other treatments (p ≤ 0.05; Duncan's multiple range test) (Fig. 2A). Brown color represents H2O2 deposition. The color intensity represents higher ROS (H2O2) generation during plant–pathogen interactions (Fig. 2B). | ||
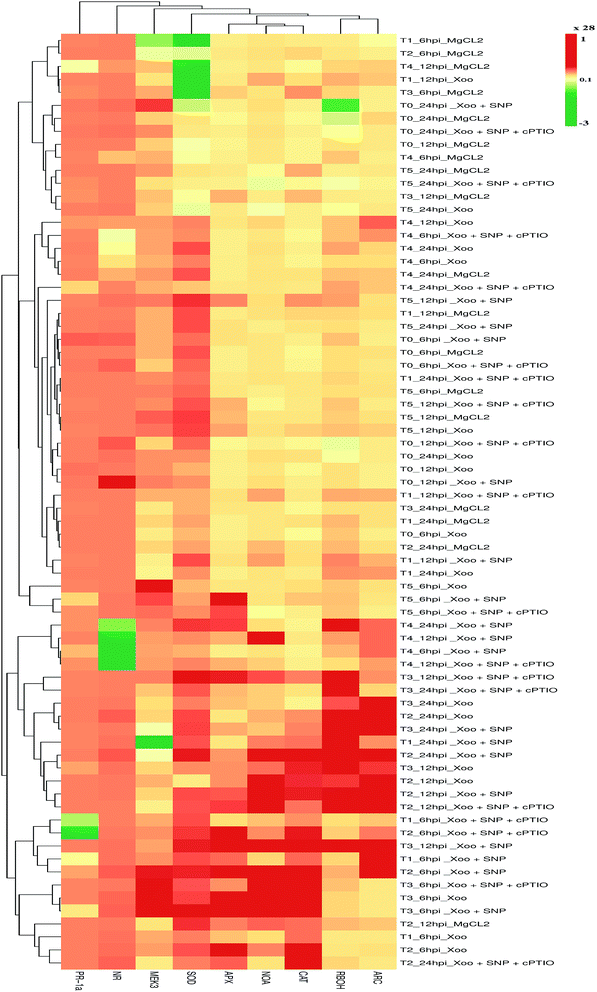
RBOHs are one of the primary sources for ROS generation upon pathogen elicitation, which participate in defense responses mediated through cell death in tobacco50,51 and pea leaves.52 Here, we also found that the higher accumulation of NtRBOH transcripts was activated at a late stage of infection, i.e., 24 hpi in the T1, T2, and T4 treatment groups, except for T3 and T5, where Trichoderma association promoted earlier transcript accumulation at 12 hpi (comparison in Fig. 9). A reduction in NtRBOH transcript expression was observed in the NH4+ (T4) fed plants (0.8 fold) compared to that in NO3− (T2) nutrient (5.3 folds) after Xoo challenged, indicating the first cue that the NO3− form of N nutrient promotes the RBOH level during incompatible Xoo pathogen interaction. These results are consistent with the H2O2 production and staining results (compare Fig. 2 with Fig. 9), where ROS production was demonstrated to be higher in the NO3−-fed leaves as compared to NH4+. The use of SNP further elevated the NtRBOH transcripts level, suggesting that RBOH is involved in plant cell death.
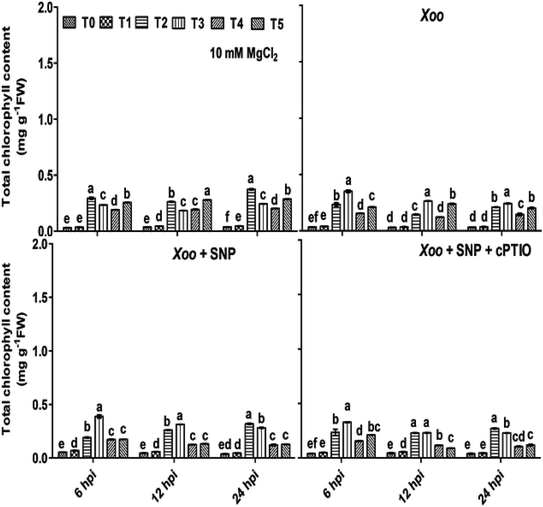
3.3 Trichoderma and N nutrients overcome chlorophyll degradation
Besides, the activation of RBOH-dependent ROS generation leads to continuous oxidative reaction in plants systems, causing membrane instability, and thus chlorophyll degradation is the first visual sign of oxidative damage. It was evidenced that nitrogen metabolism plays important in role in preventing chlorophyll degradation in potato leaves infected with Phytophthora infestans.53Considering the putative biological roles of N fertilizers and Trichoderma in disease management, a study was conducted to determine if N fertilizers and Trichoderma have any visual effects in tobacco leaves infected by bacterial Xoo pathogen using seven-week-old plant leaves. We found that the chlorophyll content was significantly degraded/reduced after Xoo infiltrated the tobacco leaves compared to in the absence of pathogen infection at 6–24 hpi (Fig. 3). It was observed that at 6 hpi, the chlorophyll content in the infiltrated tobacco leaves was reduced, and maximum reduction was observed in the control plants (T0) (0.03 mg g−1 FW) where Xoo was challenged, while the lowest reduction was observed in the Trichoderma plus NO3− fed plants (T3) (0.35 mg g−1 FW). No significant difference was found in the NO3− and NH4+ fed plant leaves during 6–24 hpi, indicating that Trichoderma prevented chlorophyll degradation only in the plants grown in NO3− nutrient. Considering that NO donors play diverse roles in biological systems and help in chlorophyll development, 100 μM SNP was infiltrated with Xoo, and it was observed that the loss of chlorophyll degradation was reverted with time (Fig. 3).
3.4 Induction of antioxidant enzymes activities and gene expression-promoted defense in Trichoderma-treated tobacco plants grown under N nutrients
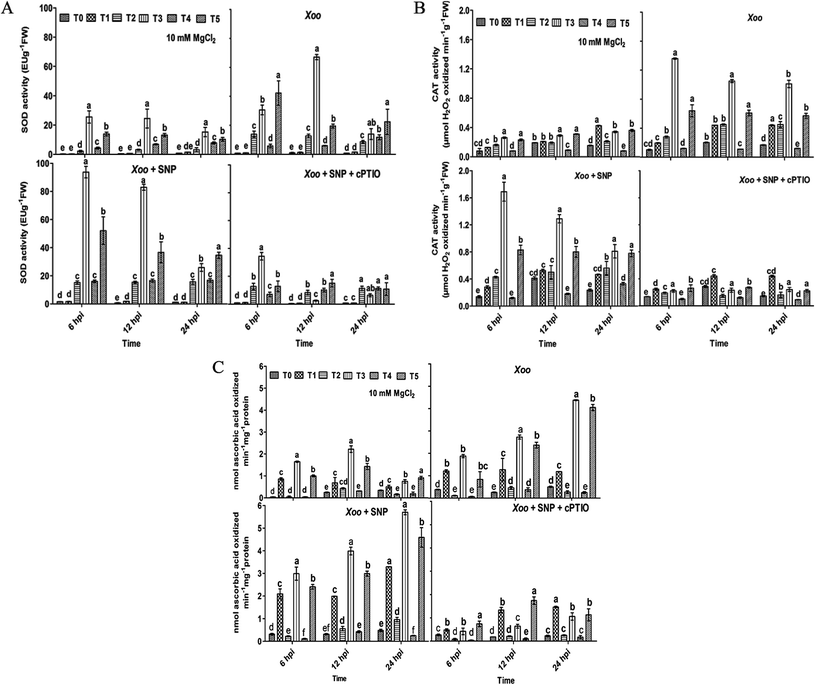
Overall, these studies demonstrate that the Trichoderma asperellum T42 strain is efficient in promoting plant immunity against Xoo. The T42-induced local defense response may be essential in Xoo resistance in tobacco. The T42 strain treatment of tobacco plants stimulated ROS molecules (H2O2), which may activate other defense-related pathways, leading to an increase antioxidant enzymatic activity. ROS are naturally occurring signal molecules involved in plant defense.11 They are rapidly generated at the site of infection caused by biotrophic or non-biotrophic pathogens in plants.42 Previously, it was reported that several antioxidant enzymatic activities increased during Ralstonia solani-induced disease in sunflower.54 These antioxidant properties were further elevated when sunflower seeds were treated with Trichoderma. Moreover, SOD, CAT, and APX elevation during oxidative stress is actively involved in plant cell wall strengthening and preventing cellular damage.44,55 Several pieces of evidence have been demonstrated that plant growth-promoting microbes induce the generation of antioxidant enzymes such as SOD and POx during plant host–pathogen interactions.56,57The present experiment revealed the key role of the Trichoderma asperellum T42 strain in triggering antioxidant enzymatic pathways, including SOD, CAT and APx. This is similar to previous reports, where the antioxidant potential increased in mustard and tomato after Trichoderma inoculation.58,59 Herein, we observed that compared to the untreated plants, NO3−-fed (T2) tobacco showed greater SOD activity (13.9 EU g−1 FW) than the NH4+-fed plants (T4; 11.8 EU g−1 FW) at 6–24 hpi after Xoo challenged (Fig. 4A). Meanwhile, Trichoderma inoculation significantly increased SOD activity, similar to tomato plants,60 and this effect was comparatively greater in the NO3−-fed (T3) plants (67.1 EU g−1 FW) than the NH4+-fed (T5) plants (42.3 EU g−1 FW) 6–24 hpi. No significant difference in the untreated (T0) and Trichoderma (T1) treatment groups were noticed during the infection period (Fig. 4A). Moreover, enhanced SOD activity was observed in the presence of SNP in the NO3− and Trichoderma plus NO3− fed plants compared to the other treatments, indicating that NO may be an essential factor for the induction of a local defense response against Xoo in tobacco.
Then, we further checked the role of T42 strain-elicited transcript accumulation of the SOD gene in the T2 and T4 treatment groups. The expression level of SOD level increased 0.2- and 0.15-fold in the T2 and T4 treated plants, respectively and no significant difference was recorded in the T0 and T1 groups (Fig. 9). Trichoderma inoculation elevated the transcript accumulation of the SOD gene similarly to SOD enzyme activity and maximum folding occurred in Trichoderma plus NO3− nutrient (T3; 0.23 folds) followed by T42 plus NH4+ (T5; 0.21 folds) 6–24 hpi (compare Fig. 4A with Fig. 9). Thus, these findings suggest that Trichoderma inoculation enhanced the SOD activity in the presence of NO3− compared to without any treatment (control). Co-infiltration of 100 μM SNP with Xoo followed the same pattern as SOD activity. Interestingly, after SNP use, the SOD expression level was greater in the T2 plants compared to the T4 plants (Fig. 4A), while the Trichoderma-treated plants showed a greater SOD expression level compared to the T0 plants (Fig. 4A). The effects of SNP were further abolished using 200 μM cPTIO.
During the local defense response assay, Trichoderma and N nutrients treatments also affected the CAT activity in the plant host and Xoo interactions. Herein, CAT activity was significantly increased in T2 compared to that in the T4-treated plants 6–24 hpi (Fig. 4B). A contrasting report was earlier demonstrated where the maximum CAT activity was observed in NH4+-fed plants in comparison to NO3−.61 The CAT activity was enhanced more in the Trichoderma treatment group (T1) compared to the untreated plants (T0). Consistent with our results, Zhang et al. (2016) found that the overexpression of Tachi (a chitinase gene form Trichoderma asperellum) in Glycine max abolished disease infection caused by Sclerotinia sclerotiorum by enhancing CAT activity.62 A similar result was also reported in tomato plants, where Trichoderma inoculation enhanced CAT activity under water-deficit conditions.60 In the present study, the effect of Trichoderma treatment was evaluated in N nutrient conditions. The CAT activity was found to be greater in the Trichoderma plus NO3− fed (T3) plants compared to the Trichoderma plus NH4+ fed (T5) after Xoo challenged. Transcriptomic profiling of the CAT gene followed a similar pattern as CAT activity, and the expression level increased in tobacco after challenged with Xoo (compare Fig. 4B with Fig. 9). The transcript level of the CAT gene in the NO3−-fed plants was greater than that in the NH4+-fed plants compared to the untreated plants (0.25 fold). Moreover, Trichoderma inoculation elevated the transcript accumulation of the CAT gene and it was found to be significantly higher in the T3 group (8.9 folds) among the treatment groups 6–24 hpi. However, the CAT expression level increased 3.7-fold in the Trichoderma inoculated plant grown under N-deprived conditions (T1; in the presence of Trichoderma) compared to T0 treatment. The higher accumulation of CAT transcripts and activity in the Trichoderma-grown NO3− fed plants in comparison to the other treatments in the presence of NO donor (SNP) again provides evidence that Trichoderma can enhance the local defense response under nitrogen (compare Fig. 4B with Fig. 9).
Consequently, in case of APx activity, Trichoderma inoculation enhanced the APx activity in comparison to the untreated plants, similarly to that in an earlier report in tomato plants under water-deficit conditions.60 This effect was observed to be remarkably higher in the T3 plants (0.35 nmol) followed by T5 (0.13 nmol) compared to the untreated plants (T0) after Xoo challenged (Fig. 4C). No significant difference in the T2 and T4 treated plants were noticed after Xoo infection, suggesting that Trichoderma can enhance APx activity in the presence of NO3− nutrient. Further, the transcript level of APx gene after Xoo challenged was observed. The maximum APx transcript level after Xoo challenged in the plant leaves was observed to be 0.38, 0.87, 4.5, 4.6, 1.2 and 3.8 fold in the T0, T1, T2, T3, T4, and T5 treatment groups, respectively, 6–24 hpi (Fig. 9). These findings indicate that the maximum APx expression level increased in the Trichoderma plus NO3− fed plants (T3). The application of SNP significantly affected APx activity and transcript accumulation in all the treatment groups, which was especially higher in the T3 treatment group compared to the control (T0) (compare Fig. 4C with Fig. 9).
Consistent with the above results, Trichoderma treatment increased the SOD, CAT, and APx activity in tobacco plants after Xoo challenged. As observed earlier, Trichoderma inoculation resulted in greater antioxidant activity in cucumber roots63 and in tomato60 under different biotic and abiotic stresses. The increased activity of SOD, CAT and APx may result from the additional protection of cells through the involvement of NO in the plant cell defense mechanism.56,64 During colonization, the overproduction of antioxidant enzymes in nitrate nutrient occurs as a result of the use of SNP, which was demonstrated previously in Cassia tora roots65 and leaves of tall fescue66 under different stresses as a mechanism to protect host plants.
3.5 NO-mediated defense is augmented by NO3− nutrition and Trichoderma asperellum T42 against Xoo
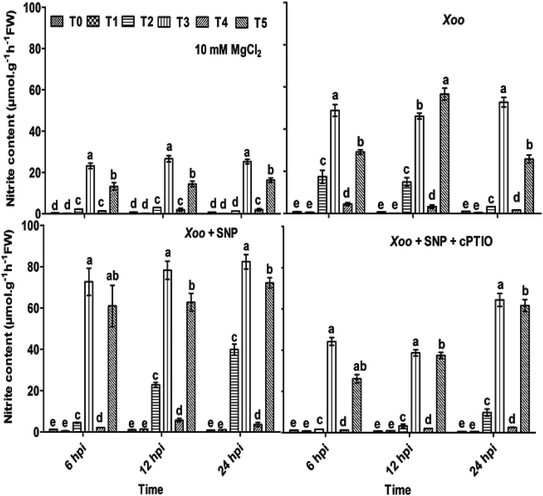
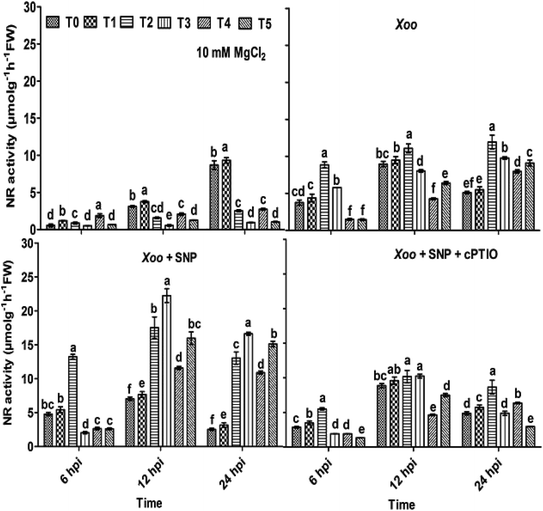
Here, we further checked the mechanism behind the elevated local defense response mediated through Trichoderma. It has been demonstrated that the key features of Trichoderma is the induction of reactive oxygen species and NO-dependent defense. The induction of ROS plays a key role in defense responses.67 Recently, short-term NO production was induced by elicitation upon Trichoderma,6,68 most probably mediated by an increase in the expression of nitrate transporters. A mutation studied with the nia 1,2 tobacco mutant revealed that NO production is mediated through nitrate reductase in plants. Several arguments suggested that nitrate reductase is a key enzyme for NO generation in plants through NR-dependent pathways.69 However, the mechanism of NO generation in plants is still a debate. The main source of NO in plants is nitrate reductase or nitric oxide associated genes, which was clearly identified in plants.68,70,71 In contrast, nitric oxide synthase (NOS) is a key source for NO generation in animals. Nitrate is reduced to nitrite by nitrate reductase, and thus an increased level of nitrite in the system is sign that NO conversion occurred.72 The study by Modolo et al. (2005) demonstrated the production of NO from nitrite in Arabidopsis as a result of nitrate reductase activity, which showed disease resistance against Pseudomonas syringae.73 Also, NO production was detected in other plants upon subjected to nitrite treatment.74In our experiments, to check whether the nitrite content and nitrate reductase activity can help in NO production and defense, the plants were treated with Trichoderma. We studied the nitrite content and nitrate reductase activity in all the treated plants. It was found that in the absence of Xoo infiltration, the nitrite content was 0.58, 0.53, 19.95, 21.42, 1.32 and 16.61 μmol g−1 h−1 FW in the T0, T1, T2, T3, T4, and T5 plants, respectively (Fig. 5). These findings indicate that the nitrite content was higher in the NO3−-fed plants (T2) in comparison to the NH4+-fed plants (T4). However, Trichoderma inoculation increased the nitrite content, especially in the plants grown in NO3− nutrient. No significant difference in the untreated plants (T0) and Trichoderma-treated plants (T1, alone) was observed. Meanwhile, a similar pattern as the nitrite content was also recorded with NR activity in all the treated plant leaves except in the T2 and T3 treatment groups, where no significance difference was observed (Fig. 6). To study the effect of Xoo infection, nitrite content and NR activity were further analyzed in the Trichoderma and N nutrient-treated tobacco plants (Fig. 5 and 6, respectively). The nitrite content significantly increased more in the NO3− grown plants (T2) than NH4+ (T4) 6–24 hpi. No significant difference in the T0 and T1 groups was noticed. However, in the presence of Trichoderma, the nitrite content increased in the NO3− fed (T3) and NH4+ fed (T5) nutrient groups during Xoo infection (Fig. 5), suggesting that Trichoderma has potential to assimilate nitrate.6 Meanwhile, the NR activity followed the same pattern as nitrite content. The maximum NR activity was noticed in Trichoderma plus NO3− treated plants (T3) (Fig. 6). No significant difference was observed in the T0 and T1 plants, providing evidence of the role Trichoderma in nitrate acquisition. Co-infiltration of SNP with Xoo significantly increased both nitrite content and NR activity in the tobacco leaves grown in all the treatment groups 6–24 hpi, indicating that NO may play a role in the mechanism for promoting nitrate acquisition from roots, but this mechanism is still unclear.
To confirm NO production by tobacco after infiltration with Xoo, spectrofluorimetry-based detection of NO intensity and microscopy experiments with DAF-FM dye were performed.12 Previously, NO was identified as a regulator of physiological response in animals, but now, it is a crucial component for plant immune response.1,47 The interaction of NO with H2O2 leads to the activation of HR-mediated cell death during non-host–pathogen interactions.42,75,76 NO played a crucial role in the activation of the defense mechanism during the interaction between plants and the bacterial pathogen Pseudomonas syringae,77,78 but the contribution of NO against necrotrophic and biotrophic fungi has also been demonstrated.79–82
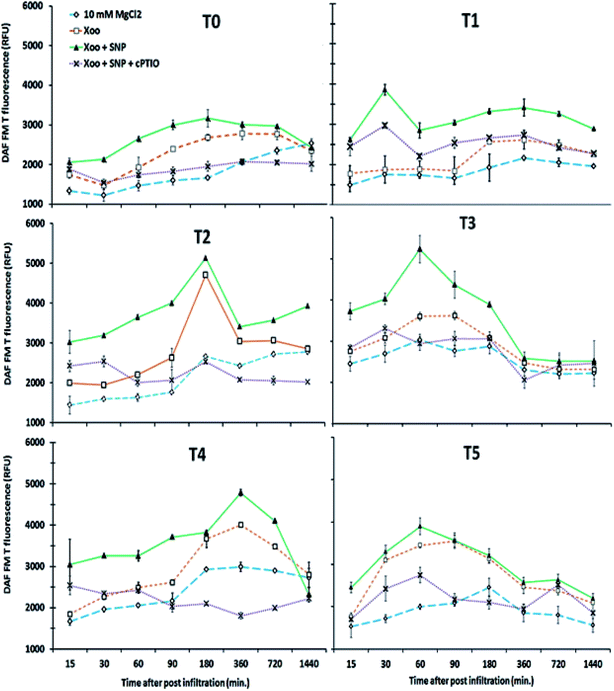
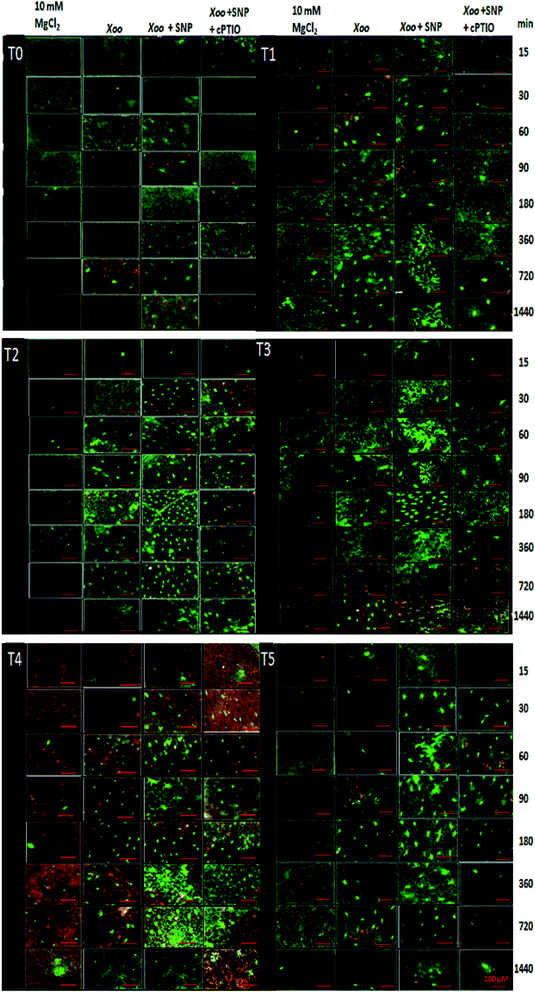
Our findings demonstrate that NO production was induced in the tobacco leaves after infiltration with Xoo. The real-time spectrofluorimetry-based detection of NO intensity showed NO production continuously increased in the tobacco leaves (Fig. 7). The maximum NO fluorescence intensity was observed at 90 min in the T2-treated plants after Xoo challenged, while in the T4-treated plants, at 180 min. Further, Trichoderma inoculation elevated the NO production in the plants (Fig. 7). Significant differences in NO intensity were detected in the T3 and T5 plants after they were infected with Xoo, suggesting that Trichoderma inoculation enhanced the NO production. However, in the absence of Trichoderma inoculation, the relative NO fluorescence intensity was lower in the T0-treated plants (Fig. 7). Moreover, the results of the DAF-FM T intensities in the HR leaves closely match the fluorescence signal observed in the fluorescence microscopy study with all the treatments except the infiltration of 10 mM MgCl2 in the T0 tobacco leaves (control) (Fig. 8). The microscopic analysis showed that in the NO3− and Trichoderma plus NO3− grown plants, stronger fluorescence signals of NO were detected compared to the control (T0) and T1 plants (Fig. 8). In all the treated plants, NO accumulation was detected in the tobacco leaves at a different time intervals after Xoo challenged. The intensities of NO production in the tobacco leaves were enhanced by co-infiltration of SNP together with Xoo in all the treatment groups. However, the effect of SNP was confirmed using an NO scavenger. The reduction in the fluorimetry-based NO signal and microscopy DAF-FM T intensity was associated with the cPTIO application (Fig. 8).
Consistent with this, we demonstrated that NO generation showed comparatively higher disease resistance in the NO3− nutrient group as compared to NH4+ due to the possibility of utilizing nitrate as a substrate.25 In our experimental approach, NO generation in the presence of Trichoderma asperellum T42 was activated, which may be an essential component of signaling of the defense response induced in the presence of nitrate. The combination with other signaling molecules, including H2O2 formation during pathogen interaction, may mutually influence their activities. NO accumulation was enhanced in the presence of nitrate nutrient in the Trichoderma-inoculated tobacco plants. Thus, the question arises, how is NO3− nutrition involved in the cell death mediated response? One of the likely pathways was explained by Romero-Puertas et al.52 Several well-known pathways have been identified for the production of NO such as L-arginine-, polyamine- or NAD(P)H-linked reduction of NO3− by cytosolic NR.74 Our assessment of NO production from the nia30 mutant plant demonstrated that the NO3− nutrition was the major source of NO production,4 similarly to that in the tobacco plant.1 Considering this, we observed that significant NO was generated in the NO3−-fed tobacco leaves after elicitation with Xoo, whereas a lower intensity was observed in the NH4+-fed tobacco plants. Previously, Shoresh and Harman et al.23 demonstrated that Trichoderma inoculation induced NO generation. Our observation showed that NO generation activated by Trichoderma asperellum T42 inoculation may influence the Xoo infection greater in nitrate nutrient; however, the tobacco plants were compromised to infection against Xoo in ammonium nutrient. Consistent with the presence of Xoo-induced NO emission in tobacco leaves, NO donor (SNP) had a more significant effect on Xoo-induced cell death. The NO3−-fed plant leaves showed greater induced HR compared to the NH4+-fed tobacco leaves. Notably, the inoculation of tobacco plants by T. asperellum resulted in the ability to counter HR in the NO3−-fed leaves in infiltrated with Xoo pathogen. This result suggests that priming with T. asperellum equipped the plants to better respond to pathogen infection,23,83 especially more in NO3− nutrition.
3.6 T. asperellum T42 recruitment favors activation of NO-modulating, PR1a and MEK3 genes in N nutrition, suppressing incompatible pathogen Xoo
The nitrite content and NR activity levels in the tobacco leaves increased in the presence of Xoo infection in the different treatment groups, which are indicative of the fact that NO production depends on nitrite content and NR activity. A previous study reported that NR expression was highly induced by N nutrient in plants.5 In the present study, the expression level of NR and NOA was found to increase in comparison to unchallenged Xoo in all the treatments (Fig. 9). The maximum expression level of NR and NOA increased at 12 hpi in all the tobacco-treated plants after Xoo challenged except T0 and T2, where the maximum NR expression level was observed at 24 hpi. NR expression was induced at an early stage, i.e. at 6 hpi in the T1-treated plants. Nitrate nutrient had a significant effect and induced greater NR and NOA expression in the NO3− grown plants (T2) compared to the NH4+ grown plants (T4). Down-regulation of NR expression in the NH4+ (T4) tobacco leaves was noticed. Interestingly, Trichoderma inoculation in the tobacco plants significantly up-regulated the NR and NOA expression in the plants grown in NO3− nutrient (T3 plants) compared to the NH4+-fed tobacco leaves (T5 plants). Co-infiltration of SNP with Xoo further induced a higher expression level by several folds in the NO-modulating genes in all the treatments except the T5 treatment, where SNP did not affect the expression of the NO-modulating gene. These results were further confirmed by the use of an NO scavenger with Xoo and SNP. Meanwhile, the transcript accumulation of ARC increased in all the treatment groups after Xoo infiltration except T0 (Fig. 9), where ARC was down-regulated. The maximum ARC transcript accumulation occurred in the Trichoderma plus NO3− grown plants (T3) (3.3-fold) followed by the NH4+ grown plants (T4) (2-fold) 6–24 hpi. The application of SNP with Xoo negatively affected the ARC expression level. Thus, these findings suggest that Trichoderma inoculation may enhance ARC accumulation, particularly in the presence of NO3− nutrient.To further characterize the N nutritional effects and T. asperellum T42 recruitment on defense, we focused on the MAP kinase gene response in the defense mechanism against Xoo in tobacco leaves. It was also remarkable that in our assessments of the defense gene, the expression of NO3− effects seemed to be confirmed by MEK3-regulated oxidative burst. According to Asai et al. (2008), the expression of MEK2-SIPK is crucial for the induction of RBOH in N. benthamiana, which is elicited by INF1, causing oxidative burst.79 In our experiment, the transcript accumulation of MEK3 was correlated with the regulation of RBOH expression in response to Xoo infection in the tobacco leaves (Fig. 9). We found that the transcript accumulation of MEK3 increased more in T0 (0.4-fold) compared to T1 (0.14 folds) 6–24 hpi. No significant difference in MEK3 expression level in the T2 and T4 grown plants was observed after Xoo challenged (Fig. 9). On the other hand, upon T42 recruitment, MEK3 expression was found earlier (6 hpi) in T42 plus NO3− compared to T42 plus NH4+ treatment indicating that Trichoderma priming involves activation of MEK3/RBOH and, promotes cell death during infection. However, the transcript accumulation of MEK3 increased several folds in the T5-grown plants (7.1-fold) than the T3-grown plants (3.8-fold) at a later stage. Co-infiltration of SNP with Xoo in tobacco leaves up-regulated the MEK3 expression in all the treated plants except in the T5 plants, where a negative effect of SNP on MEK3 was observed. The lack of complete information about the correlation of NO and MEK3 in NO3− or NH4+ feeding modifying HR in the presence of T. asperellum T42 requires further characterization to determine how Trichoderma recruitment changes HR against infection of non-host–pathogens through NO3− or NH4+.
NO is known to trigger the salicylic acid marker gene,47 and it has the ability to induce oxidative burst to augment cell death during HR.80 Thus, the induction of PR1a gene activation in response to Xoo infection in the NO3−-fed tobacco leaves is most likely linked to induced NO production (Fig. 9), which was further investigated. The expression level of PR1a increased 0.3-fold under nutrient-deprived (control; T0) conditions at 6 hpi (Fig. 9). The PR1a expression was high in the NO3−-fed plants compared to NH4+ nutrient, suggesting that the plants fed in NO3− nutrient showed more PR-mediated disease resistance compared to NH4+ nutrient against incompatible Xoo, similarly to the findings of Gupta et al.,1 where the form of N nutrition affected cell death linked to resistance in response to elicitation of HR against Pseudomonas syringe in tobacco leaves. However, switching of PR1a expression by T. asperellum T42 inoculum in the N nutrient-fed tobacco plants depends on the mode of infection mechanism. Its expression level was down-regulated in the T1, T3, T4, and T5 grown plants (Fig. 9). The induction of PR1a expression in the presence of NO donor (SNP) linked with disease resistance mediated through NO and PR1a was demonstrated in tobacco leaves, similarly to another observation.25
4. Conclusion
In conclusion, in the present experiment, we found that Trichoderma asperellum T42, saprophytic filamentous fungi, could significantly promote the plant defense response against Xanthomonas oryzae pv. oryzae by modulating reactive oxygen species and nitric oxide molecules in tobacco plants. Trichoderma-induced disease resistance to Xanthomonas oryzae pv. oryzae was demonstrated to induce NO via the NR and NOA pathways, and accumulation of H2O2 by an increase in antioxidant enzyme activity. Moreover, the application of nitrate as a nitrogen source was promoted by Trichoderma inoculation, which helped the plants maintain NO and H2O2 generation during the hypersensitive response against Xanthomonas infection. Our study provides new insight into the mechanistic approach of Trichoderma, which reprograms disease resistance against non-host–pathogen Xanthomonas oryzae via NO and H2O2 production in response to nitrogen nutrition. Further, these findings will open new gateways for researchers to study how and which mechanisms are associated with Trichoderma for increasing N acquisition. It may be possible that certain novel signaling molecules are volatile compounds; however, additional experiments are essential to determine this.Abbreviations
| DAF-FM | Diacetate 4,5-diaminofluorescein-FM |
| HR | Hypersensitive response |
| MAPK | Mitogen-activated protein kinase |
| NR | Nitrate reductase |
| NO | Nitric oxide |
| ONOO− | Peroxynitrite ion |
| PCD | Programmed cell death |
| PR-1 | Pathogenesis-related protein |
| ROS | Reactive oxygen species |
| SA | Salicylic acid |
| SOD | Superoxide dismutase |
| SNP | Sodium nitroprusside |
| cPTIO | μM cPTIO (carboxy-2-phenyl-4,4,5,5-tetramethylimidazoline-1-oxyl-3-oxide) |
Author contributions
Contributor BNS performed laboratory experiments, data collection, and statistical analysis. PD designed the research, revised the manuscript for publication and supervised the work. Contributed reagents/materials/analysis tools/equipment facilities: PD, BKS, HBS.Conflicts of interest
The authors declare that research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.Acknowledgements
We also thank SERB-DST, Government of India, New Delhi for providing financial grant [SR/SO/PS-23/10(G)] in the form of a research project to the corresponding author. Authors also acknowledged to Dr Mahendra Vikram Singh Rajawat, ICAR-National Bureau of Agriculturally Important Microorganisms, Kushmaur, Mau for his assistance in graph preparation.References
- K. J. Gupta, Y. Brotman, S. Segu, T. Zeier, J. Zeier, S. T. Persijn, S. M. Cristescu, F. J. Harren, H. Bauwe and A. R. Fernie, J. Exp. Bot., 2012, 64, 553–568 CrossRef PubMed.
- L. A. Mur, C. Simpson, A. Kumari, A. K. Gupta and K. J. Gupta, Ann. Bot., 2017, 119, 703–709 CAS.
- S. Ruffel, A. Poitout, G. Krouk, G. M. Coruzzi and B. Lacombe, J. Integr. Plant Biol., 2016, 58, 226–229 CrossRef CAS.
- B. N. Singh, P. Dwivedi, B. K. Sarma, G. S. Singh and H. B. Singh, Front. Plant Sci., 2018, 9, 163 CrossRef PubMed.
- H. Sun, J. Li, W. Song, J. Tao, S. Huang, S. Chen, M. Hou, G. Xu and Y. Zhang, J. Exp. Bot., 2015, 66, 2449–2459 CrossRef CAS.
- A. Wany, P. K. Pathak and K. J. Gupta, bioRxiv, 2018, 502492 Search PubMed.
- B. Landrein, P. Formosa-Jordan, A. Malivert, C. Schuster, C. W. Melnyk, W. Yang, C. Turnbull, E. M. Meyerowitz, J. C. Locke and H. Jönsson, Proc. Natl. Acad. Sci. U. S. A., 2018, 115, 1382–1387 CrossRef CAS PubMed.
- M. Alves, S. Dadalto, A. Gonçalves, G. de Souza, V. Barros and L. Fietto, Proteomes, 2014, 2, 85–106 CrossRef CAS.
- J. D. Jones and J. L. Dangl, Nature, 2006, 444, 323 CrossRef CAS.
- M. Delledonne, Y. Xia, R. A. Dixon and C. Lamb, Nature, 1998, 394, 585 CrossRef CAS.
- L. A. Mur, E. Prats, S. Pierre, M. A. Hall and K. H. Hebelstrup, Front. Plant Sci., 2013, 4, 215 Search PubMed.
- J. Nawrocka, A. Gromek and U. Małolepsza, Front. Plant Sci., 2019, 10, 421 CrossRef PubMed.
- D. Spadaro, B. W. Yun, S. H. Spoel, C. Chu, Y. Q. Wang and G. J. Loake, Physiol. Plant., 2010, 138, 360–371 CrossRef CAS PubMed.
- S.-C. Chen, J.-J. Ren, H.-J. Zhao, X.-L. Wang, T.-H. Wang, S.-D. Jin, Z.-H. Wang, C.-y. Li, A.-R. Liu and X.-M. Lin, Phytopathology, 2019, 109, 972–982 CrossRef PubMed.
- A. Sivakumaran, A. Akinyemi, J. Mandon, S. M. Cristescu, M. A. Hall, F. J. Harren and L. A. Mur, Front. Plant Sci., 2016, 7, 709 Search PubMed.
- G. E. Ti-da, S. Shi-wei, C. H. I. Ming-han, H. Dan-feng and K. Iwasaki, Agric. Sci. China, 2008, 7, 1308–1317 CrossRef.
- A. C. Kushalappa, K. N. Yogendra and S. Karre, Crit. Rev. Plant Sci., 2016, 35, 38–55 CrossRef CAS.
- M. Oliveira, C. Varanda and M. Félix, Phytochem. Lett., 2016, 15, 152–158 CrossRef CAS.
- G. E. Harman, A. H. Herrera-Estrella, B. A. Horwitz and M. Lorito, Microbiology, 2012, 158, 1 CrossRef CAS PubMed.
- J. S. Patel, B. K. Sarma, H. B. Singh, R. S. Upadhyay, R. N. Kharwar and M. Ahmed, Front. Plant Sci., 2016, 6, 1206 CrossRef.
- B. N. Singh, A. Singh, G. S. Singh and P. Dwivedi, J. Pure Appl. Microbiol., 2015, 9, 1069–1074 CAS.
- Y. Brotman, J. Lisec, M. Méret, I. Chet, L. Willmitzer and A. Viterbo, Microbiology, 2012, 158, 139–146 CrossRef CAS PubMed.
- M. Shoresh and G. E. Harman, BMC Plant Biol., 2010, 10, 136 CrossRef.
- S. K. Yadav, A. Dave, A. Sarkar, H. B. Singh and B. K. Sarma, Int. J. Agric. Environ. Biotechnol., 2013, 6, 255 Search PubMed.
- E. Planchet, K. Jagadis Gupta, M. Sonoda and W. M. Kaiser, Plant J., 2005, 41, 732–743 CrossRef CAS PubMed.
- C. Johnson, P. Stout, T. C. Broyer and A. B. Carlton, Plant Soil, 1957, 8, 337–353 CrossRef CAS.
- B. Singh and P. Dwivedi, International Journal of Plant Reproductive Biology, 2016, 8, 88–93 Search PubMed.
- A. Levine, R. Tenhaken, R. Dixon and C. Lamb, Cell, 1994, 79, 583–593 CrossRef CAS.
- H. Thordal-Christensen, Z. Zhang, Y. Wei and D. B. Collinge, Plant J., 1997, 11, 1187–1194 CrossRef CAS.
- V. Velikova, I. Yordanov and A. Edreva, Plant Sci., 2000, 151, 59–66 CrossRef CAS.
- D. I. Arnon, Plant Physiol., 1949, 24, 1 CrossRef CAS.
- M. M. Bradford, Anal. Biochem., 1976, 72, 248–254 CrossRef CAS.
- I. Fridovich, External Resources Pubmed/Medline (NLM) CrossRef (DOI) Chemical Abstracts Service (CAS) ISI Web of Science, 1981 Search PubMed.
- Y. Teranishi, A. Tanaka, M. Osumi and S. Fukui, Agric. Biol. Chem., 1974, 38, 1213–1220 CrossRef CAS.
- Y. Nakano and K. Asada, Plant Cell Physiol., 1981, 22, 867–880 CAS.
- H. Srivastava, Phytochemistry, 1980, 19, 725–733 CrossRef CAS.
- F. J. Corpas, J. B. Barroso, A. Carreras, R. Valderrama, J. M. Palma, A. M. León, L. M. Sandalio and L. A. Del Río, Planta, 2006, 224, 246–254 CrossRef CAS.
- T. D. Schmittgen and K. J. Livak, Nat. Protoc., 2008, 3, 1101 CrossRef CAS PubMed.
- D. Tilman, K. G. Cassman, P. A. Matson, R. Naylor and S. Polasky, Nature, 2002, 418, 671 CrossRef CAS PubMed.
- S. Boudsocq, A. Niboyet, J. C. Lata, X. Raynaud, N. Loeuille, J. Mathieu, M. Blouin, L. Abbadie and S. Barot, Am. Nat., 2012, 180, 60–69 CrossRef CAS PubMed.
- D. D. Stuthman, Euphytica, 2002, 124, 253–258 CrossRef CAS.
- M. Schlicht and E. Kombrink, Front. Plant Sci., 2013, 4, 351 Search PubMed.
- H. A. Contreras-Cornejo, L. Macías-Rodríguez, E. Beltrán-Peña, A. Herrera-Estrella and J. López-Bucio, Plant Signaling Behav., 2011, 6, 1554–1563 CrossRef CAS PubMed.
- U. Małolepsza, J. Nawrocka and M. Szczech, Biocontrol Sci. Technol., 2017, 27, 180–199 CrossRef.
- M. A. Torres, J. D. Jones and J. L. Dangl, Plant Physiol., 2006, 141, 373–378 CrossRef CAS PubMed.
- L. Asmawati, A. Widiastuti and C. Sumardiyono, in Proceeding of the 1st International Conference on Tropical Agriculture, ed. A. Isnansetyo and T. Nuringtyas, Springer, Cham, 2017, pp. 139–146 Search PubMed.
- J. Durner and D. F. Klessig, Curr. Opin. Plant Biol., 1999, 2, 369–374 CrossRef CAS PubMed.
- C. Pieterse, S. Van Wees, E. Hoffland, J. A. Van Pelt and L. C. Van Loon, Plant Cell, 1996, 8, 1225–1237 CAS.
- F. Vargas, Z. González, R. Sánchez, L. Jiménez and A. Rodríguez, BioResources, 2012, 7, 4161–4170 Search PubMed.
- Y. Kadota, K. Shirasu and C. Zipfel, Plant Cell Physiol., 2015, 56, 1472–1480 CrossRef CAS PubMed.
- H. Yoshioka, N. Numata, K. Nakajima, S. Katou, K. Kawakita, O. Rowland, J. D. Jones and N. Doke, Plant Cell, 2003, 15, 706–718 CrossRef CAS PubMed.
- M. C. Romero-Puertas, M. Perazzolli, E. D. Zago and M. Delledonne, Cell. Microbiol., 2004, 6, 795–803 CrossRef CAS.
- A. Lazalt, M. V. Beligni and L. Lamattina, Eur. J. Plant Pathol., 1997, 103, 643–651 CrossRef.
- B. N. Singh, A. Singh, S. P. Singh and H. B. Singh, Eur. J. Plant Pathol., 2011, 131, 121–134 CrossRef CAS.
- M. D. Domínguez-Valdivia, P. M. Aparicio-Tejo, C. Lamsfus, C. Cruz, M. A. Martins-Loução and J. F. Moran, Physiol. Plant., 2008, 132, 359–369 CrossRef PubMed.
- A. Singh, B. K. Sarma, R. S. Upadhyay and H. B. Singh, Microbiol. Res., 2013, 168, 33–40 CrossRef CAS.
- P. Singhai, B. Sarma and J. Srivastava, Biol. Control, 2011, 57, 150–157 CrossRef.
- P. Bernat, J. Nykiel-Szymańska, E. Gajewska, S. Różalska, P. Stolarek, J. Dackowa and M. Słaba, J. Plant Physiol., 2018, 229, 158–163 CrossRef CAS PubMed.
- A. Zehra, M. Meena, M. K. Dubey, M. Aamir and R. Upadhyay, Bot. Stud., 2017, 58, 44 CrossRef.
- F. Mastouri, T. Björkman and G. E. Harman, Mol. Plant-Microbe Interact., 2012, 25, 1264–1271 CrossRef CAS.
- O. Polesskaya, E. Kashirina and N. Alekhina, Russ. J. Plant Physiol., 2004, 51, 615–620 CrossRef CAS.
- F. Zhang, X. Ruan, X. Wang, Z. Liu, L. Hu and C. Li, Appl. Biochem. Biotechnol., 2016, 180, 1542–1558 CrossRef CAS.
- Y. Yu, Z. Yang, K. Guo, Z. Li, H. Zhou, Y. Wei, J. Li, X. Zhang, P. Harvey and H. Yang, Curr. Microbiol., 2015, 70, 618–622 CrossRef CAS.
- M. Shoresh and G. E. Harman, Plant Physiol., 2008, 147, 2147–2163 CrossRef CAS PubMed.
- Y.-S. Wang and Z.-M. Yang, Plant Cell Physiol., 2005, 46, 1915–1923 CrossRef CAS.
- Y. Xu, X. Sun, J. Jin and H. Zhou, J. Plant Physiol., 2010, 167, 512–518 CrossRef CAS PubMed.
- K. J. Gupta, L. A. Mur and Y. Brotman, Mol. Plant-Microbe Interact., 2014, 27, 307–314 CrossRef CAS PubMed.
- K. J. Gupta, A. R. Fernie, W. M. Kaiser and J. T. van Dongen, Trends Plant Sci., 2011, 16, 160–168 CrossRef CAS PubMed.
- L. Lamattina, C. García-Mata, M. Graziano and G. Pagnussat, Annu. Rev. Plant Biol., 2003, 54, 109–136 CrossRef CAS PubMed.
- L. A. del Rio, J. Exp. Bot., 2015, 66, 2827–2837 CrossRef CAS.
- J. Santolini, F. André, S. Jeandroz and D. Wendehenne, Nitric Oxide, 2017, 63, 30–38 CrossRef CAS.
- L. V. Modolo, O. Augusto, I. M. Almeida, C. A. Pinto-Maglio, H. C. Oliveira, K. Seligman and I. Salgado, Plant Sci., 2006, 171, 34–40 CrossRef CAS.
- L. V. Modolo, O. Augusto, I. M. Almeida, J. R. Magalhaes and I. Salgado, FEBS Lett., 2005, 579, 3814–3820 CrossRef CAS PubMed.
- P. Rockel, F. Strube, A. Rockel, J. Wildt and W. M. Kaiser, J. Exp. Bot., 2002, 53, 103–110 CrossRef CAS PubMed.
- D. Bellin, S. Asai, M. Delledonne and H. Yoshioka, Mol. Plant-Microbe Interact., 2013, 26, 271–277 CrossRef CAS PubMed.
- H. Yoshioka, K. Mase, M. Yoshioka, M. Kobayashi and S. Asai, Nitric Oxide, 2011, 25, 216–221 CrossRef CAS PubMed.
- H. C. Oliveira, E. E. Saviani, J. F. Oliveira and I. Salgado, Tropical Plant Pathology, 2010, 35, 104–107 CrossRef.
- E. Zago, S. Morsa, J. F. Dat, P. Alard, A. Ferrarini, D. Inzé, M. Delledonne and F. Van Breusegem, Plant Physiol., 2006, 141, 404–411 CrossRef CAS PubMed.
- S. Asai, K. Ohta and H. Yoshioka, Plant Cell, 2008, 20, 1390–1406 CrossRef CAS PubMed.
- L. A. Mur, T. L. Carver and E. Prats, J. Exp. Bot., 2005, 57, 489–505 CrossRef.
- L. Perchepied, C. Balagué, C. Riou, C. Claudel-Renard, N. Rivière, B. Grezes-Besset and D. Roby, Mol. Plant-Microbe Interact., 2010, 23, 846–860 CrossRef CAS.
- J. Piterkova, M. Petřivalský, L. Luhova, B. Mieslerova, M. Sedlářová and A. Lebeda, Mol. Plant Pathol., 2009, 10, 501–513 CrossRef CAS PubMed.
- A. Nagaraju, J. Sudisha, S. M. Murthy and S.-i. Ito, Australas. Plant Pathol., 2012, 41, 609–620 CrossRef.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9ra06802c |
| This journal is © The Royal Society of Chemistry 2019 |