Open Access Article
Open Access ArticleBulky cations greatly increase the turnover of a native hammerhead ribozyme†
Shu-ichi Nakano *a,
Hirofumi Yamashitaa,
Kazuya Tanabea and
Naoki Sugimotoab
*a,
Hirofumi Yamashitaa,
Kazuya Tanabea and
Naoki Sugimotoab
aDepartment of Nanobiochemistry, Faculty of Frontiers of Innovative Research in Science and Technology (FIRST), Konan University, 7-1-20, Minatojima-minamimachi, Chuo-ku, Kobe, 650-0047, Japan. E-mail: shuichi@konan-u.ac.jp
bFrontier Institute for Biomolecular Engineering Research (FIBER), Konan University, 7-1-20, Minatojima-minamimachi, Chuo-ku, Kobe, 650-0047, Japan
First published on 4th November 2019
Abstract
Methods to facilitate the catalytic turnover of ribozymes are required for advancing oligonucleotide-based technologies. This study examined tetraalkylammonium ions for their ability to increase the efficiency of catalytic turnover of a native hammerhead ribozyme. Kinetic analysis showed that large tetraalkylammonium ions significantly increased the turnover rate of the ribozyme and was much more effective than poly(ethylene glycol) (PEG) and urea. The magnitude of the rate increase depended on the concentrations of Mg2+ and tetrapentylammonium ions, and the rate was enhanced by more than 180-fold at the optimal concentrations of these salts. The results provide physical insights into interactions of ribozymes with large cationic molecules through electrostatic forces and steric hindrance.
Introduction
Self-cleaving RNA enzymes, called ribozymes, are found in the RNA satellites of plant viruses and in human genomes.1–8 The catalytic reaction of ribozymes proceeds in the presence of divalent metal ions, such as Mg2+, and even in the presence of Mg2+ complexes coordinated by amino acids or diphosphate molecules.9,10 Since ribozymes can selectively cleave RNA substrates, synthetic small constructs of trans-acting ribozymes are useful for developing oligonucleotide-based technologies, for example, for the in vivo inhibition of gene functions and for the construction of biosensor probes.11–13 To maximize applications of ribozymes for biotechnology, it is important to develop methods to enhance their catalytic performance. The strength and specificity of substrate binding depend on the number of base pairs formed between the ribozyme and its substrate, and thus ribozyme constructs containing a long substrate recognition sequence are useful for the recognition of target RNA. However, the turnover of ribozymes with long sequences is intrinsically difficult because of high thermodynamic stability and slow dissociation of base pairs formed between the ribozyme and cleaved products.14,15 In addition, formation of the ribozyme–substrate complex is inhibited when the RNA sequences form interfering intramolecularly-folded structures, which is crucial for long sequences. It is supposed that the turnover of ribozymes can be enhanced under conditions that decrease the stability of RNA base pairing without significant disrupting the catalytically active conformation.The hammerhead ribozyme constitutes a class of small ribozymes that exhibit high RNA substrate cleavage activity in the presence of metal ions, such as Mg2+. The structure and catalytic mechanism of this ribozyme have well been studied, and metal ions are known to play both structural and catalytic roles: structural ions stabilize base pairing and the catalytically active conformation by electrostatic interactions, and catalytic ions that bind to specific sites participate in chemical reactions and electrostatic stabilization of a transition state.16–22 The catalytic activity of hammerhead ribozymes also depends on organic cations that affect RNA–RNA interactions and catalytic mechanism. Polyamines, such as spermidine, can increase the catalytic rate by effective electrostatic screening of RNA charges.23 Conversely, polyamines also act as an inhibitor by inducing RNA misfolding.24 Inhibitory RNA-polycation interactions are also reported for coacervates made with polycations, such as poly(diallyldimethylammonium chloride) and oligoarginine.25 Furthermore, cationic additives that stabilize RNA–RNA interactions are expected to disfavor the dissociation of cleaved products, making the rate of multiple-turnover cleavage slow down. There have been reports of several-to several tens-fold enhancement in the turnover of hammerhead ribozymes using RNA-binding proteins,26 oligonucleotide facilitators,27,28 locked nucleic acid nucleotides,29 poly(ethylene glycol) (PEG),30,31 and aqueous droplet-based microfluidics.32 In this report, we show further improvements of the turnover rate of a long hammerhead ribozyme construct by the use of tetraalkylammonium ions, with increases of greater than a hundred-fold at optimal salt concentrations.
Experimental
Materials
The HH10 ribozyme was prepared by run-off transcription from a DNA template duplex (the 60-mer ribozyme sequence linked to a 17-mer T7 RNA polymerase promoter sequence) using T7 RNA polymerase (GE Healthcare; Chicago, USA). The DNA oligonucleotides and the RNA substrate labeled with 6-fluorescein at the 5′-end were purchased from Hokkaido System Science (Hokkaido, Japan). All reagents used to prepare buffer solutions were purchased from Wako Chemicals (Osaka, Japan), except for 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) and disodium salt of ethylenediamine-N,N,N′,N′-tetraacetic acid (Na2EDTA) which were from Dojindo (Kumamoto, Japan).Kinetic analysis of substrate cleavage
The rate of substrate cleavage was measured using 5.0 nM ribozyme and 100 nM substrate for multiple-turnover or 100 nM ribozyme and 2.0 nM substrate for single-turnover reactions. The reaction was performed in a volume of 20 μL of a buffered solution consisting of 50 mM HEPES, 50 mM NaCl, and 0.10 mM Na2EDTA, adjusted to pH 7.0 at 37 °C.The solution was annealed at 70 °C and subsequently incubated at 37 °C for 10 min, followed by the addition of MgCl2 (the final concentration of 10 mM) to initiate substrate cleavage, unless otherwise stated. Reactions were quenched by mixing with 90% formamide solution containing 100 mM Na2EDTA, and then loaded on 20% polyacrylamide gel containing 7.0 M urea. After electrophoresis, fluorescence emission of fluorescently labeled RNA fragments was visualized and the amounts of uncleaved and cleaved fragments ([U] and [C], respectively) were quantified, using a fluorescent scanner at 473 nm excitation and 520 nm emission (FLA-5100, Fujifilm; Tokyo, Japan).
The cleavage rate and turnover frequency of ribozyme-catalyzed substrate cleavage (molecules of cleaved substrate per molecules of ribozyme, per hour) were analyzed by plotting the fraction of RNA cleaved at the correct site to the total amount of RNA substrate, [C]/([U] + [C]), against time. The single-turnover rate was determined by fitting the slow phase data to a single exponential function, and the rate of multiple-turnover kinetics and turnover frequency were determined by fitting to a linear function.
Results and discussion
Catalytic activity of the long hammerhead ribozyme construct
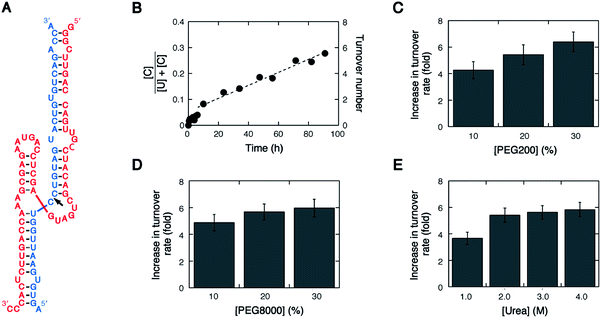
Native hammerhead ribozymes are composed of a conserved nucleotide core flanked by long stretches of base pairs, and the stem regions consisting of base-paired and unpaired nucleotides participate in tertiary interactions essential for fast cleavage under physiological conditions.33–36 We investigated the trans-acting ribozyme derived from an intronic ribozyme in the human genome, HH10.7 This ribozyme forms long base pairing with its RNA substrate, and the substrate is cleaved at the scissile bond between two cytosines located at the junction of three stems in the presence of Mg2+ (Fig. 1A).The substrate cleavage under the single-turnover condition conducted with 10 mM MgCl2 at 37 °C was fast, yielding a cleaved fraction of ≈0.4 within 10 seconds and reaching greater than 0.8 after a few minutes (Fig. S1, ESI†). The kinetic curve did not fit to a single exponential function because of the presence of catalytically inactive conformations that required refolding for becoming catalytic through a stepwise process.37 On the other hand, the substrate cleavage under the multiple-turnover condition conducted with 10 mM MgCl2 at 37 °C was extremely slow, taking 90 hours to show the turnover number of about 6 (a cleaved fraction of ≈0.3) including a burst phase during the first turnover (a cleaved fraction of ≈0.05) (Fig. 1B). The cleavage proceeded linearly with time, and the kinetic analysis determined the turnover frequency to be 0.050 h−1 (molecules of cleaved substrate per molecule of ribozyme, per hour) and the turnover rate to be 0.083 fmol min−1. The low activity is attributed to the substrate recognition through a long sequence whereas much faster turnover rates were observed for short hammerhead ribozyme constructs (Fig. S2, ESI†), indicating the trade-off between the extension of the substrate recognition sequence and the low turnover activity, as reported for HDV-like ribozymes.15
When the pH was dropped from 7.0 to 6.0, the single-turnover rate of HH10 fell to 0.38-fold of the value at pH 7.0 whereas the multiple-turnover rate did not greatly change. The results suggest that the substrate cleavage through an acid–base mechanism is the rate-limiting step under the single-turnover condition but not under the multiple-turnover condition.
Effects of conventional additives
It has been reported that an addition of PEG, conventionally used as a molecular crowding agent, alters the catalytic activity of several types of ribozymes, including hammerhead ribozymes.38,39 We observed here that PEGs with average molecular weights of 200 (PEG200) and 8000 (PEG8000) in a concentration range from 10% to 30% by weight increased the turnover rate of HH10 by 4.3–6.4-fold (Fig. 1C and D). Urea, which is widely used as a nucleic acid denaturant, in a concentration range from 1.0 to 4.0 M also increased the turnover rate by 3.7–5.8-fold (Fig. 1E). Therefore, these conventionally used additives, which affect the strength of RNA interactions by various factors, such as changes in the solution properties (e.g., water activity) and disruption of the helical conformation of RNA,40,41 can enhance the turnover of the native hammerhead ribozyme that forms long base pairing with its RNA substrate.Effects of tetraalkylammonium ions
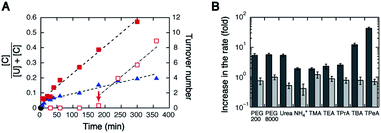
Tetraalkylammonium ions are monovalent cations that modulate the stability of Watson–Crick base pairs in a size-dependent manner. Small cations stabilize base-paired RNA structures by preferential electrostatic binding to densely packed structures. In contrast, bulky cations, such as tetrabutylammonium (TBA) and tetrapentylammonium (TPeA) ions, have the ability to destabilize base pairing.42–44 This unique property of bulky cations is caused by steric effects that reduce the accessibility to base-paired regions but can electrostatically interact with unpaired regions such as single strands.45 It is also remarkable that the large tetraalkylammonium ion affects metal ion binding to RNA, resulting in a change in metal ion requirements for catalysis by several types of ribozymes and deoxyribozymes.46 Here, we tested different-sized tetraalkylammonium ions containing methyl, ethyl, propyl, butyl, or pentyl groups for the turnover of HH10. Tetrahexylammonium ion could not be tested due to low aqueous solubility.The addition of tetramethylammonium (TMA), tetraethylammonium (TEA), or tetrapropylammonium (TPrA) at a concentration of 100 mM only slightly increased the turnover rate, as observed using NH4+. Remarkably, TBA substantially increased the rate by 12-fold (the turnover frequency was 0.61 h−1), and TPeA greatly increased the rate by 43-fold (frequency = 2.1 h−1) (Fig. 2 and S3, ESI†), indicating that the bulky cations play the same function as PEGs and urea in enhancement of the catalytic turnover, with a much lower quantity. A rapid increase in the cleavage yield by the addition of TPeA was also observed 3 h after the addition of MgCl2 (Fig. 2A). The turnover frequency in the presence of TPeA did not change significantly (1.7 h−1), when the ribozyme concentration decreased from 5.0 to 1.0 nM. In contrast, these tetraalkylammonium ions, including TBA and TPeA, produced only small effects in the single-turnover rate (Fig. 2B) and did not promote the cleavage in the absence of Mg2+, indicating that the cations themselves possess no intrinsic RNA cleavage activity. It is mentioned that our previous study showed TBA-induced reductions in the single-turnover rates of lead-dependent ribozyme and 17E deoxyribozyme especially under low metal ion concentrations.46 Because these nucleic acid enzymes consisted of short sequences, their catalytically active conformations were likely to be easily disrupted by TBA. In contrast, the catalytically active conformation of HH10 is relatively stable, as indicated by the observation of efficient substrate cleavage even in the presence of large amounts of urea.
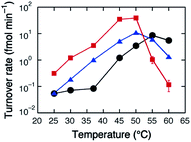
The stability of the catalytically active conformation of HH10 was studied by measuring the catalytic activity at different temperatures. The turnover rate in the absence of additives increased as the temperature was raised to ≈55 °C but decreased at higher temperatures by thermal denaturation. The temperature at which the rate began to decrease was reduced by 5–10 °C in the presence of TBA or TPeA (Fig. 3), indicating destabilization of the catalytically active conformation, but still higher than the temperature used in the above experiments, 37 °C. The destabilization has disadvantages in forming the catalytically active conformation but has advantages in cleaved product dissociation for catalytic turnover.
The large tetraalkylammonium ions have the properties that are useful for increasing the turnover rate. The large cations destabilize RNA base pairing by preferential interactions with single strands and affect metal ion binding to RNA.43 Thus, it is probable that the mechanism for increasing the turnover rate is different from those of PEGs and urea: the large tetraalkylammonium ions can affect the substrate cleavage in several ways, including destabilization of the complex between the ribozyme and the cleaved products, destabilization of interfering RNA structures, and changes in the property of Mg2+ binding to RNA. Further research is necessary to understand the mechanism for increasing the turnover rate in the presence of the bulky cations, such as kinetic analysis of the rate of base pair dissociation.
Search for optimum salt conditions for the multiple-turnover cleavage
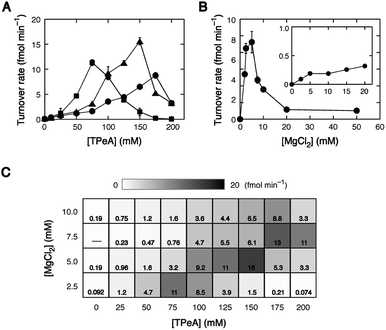
The addition of 100 mM TPeA provided a 43-fold increase in the turnover rate with 10 mM MgCl2. Then, we searched for optimum concentrations of MgCl2 and TPeA for the turnover of HH10. At a constant MgCl2 concentration of 10 mM, the magnitude of the increase in the turnover rate changed depending on TPeA concentration, reaching 105-fold at 175 mM, but decreasing at higher concentrations due to increased destabilization of the catalytically active conformation. The TPeA concentration that produced the highest rate decreased as MgCl2 concentration decreased: 150 mM at 5.0 mM MgCl2 and 75 mM at 2.5 mM MgCl2 (Fig. 4A). On the other hand, the magnitude of the increase at a constant TPeA concentration of 100 mM varied with MgCl2 concentration, with the greatest increase, reaching 110-fold at 5.0 mM MgCl2, but decreasing at higher concentrations (Fig. 4B). These observations suggest competitive binding of TPeA and Mg2+ ions. It is striking that the optimal conditions for multiple-turnover cleavage lay on the diagonal of the MgCl2–TPeA concentration matrix, as shown in Fig. 4C. The maximum increase in the turnover rate was greater than 180-fold (9.1 h−1 or 16 fmol min−1 at 5.0 mM MgCl2 and 150 mM TPeA). These results show that large tetraalkylammonium ions promote the turnover much more effectively than PEG and urea and that the rate increase under optimized salt conditions can be higher than those reported using other additives. We suggest that bulky cations provide a new method to enhance the catalytic performance of the native hammerhead ribozyme, although it will be important to optimize salt conditions for applying to other nucleic acid enzymes that have different structural stability and roles of metal ions in catalysis.Conclusions
This study showed that large tetraalkylammonium ions have an ability to greatly enhance the catalytic turnover of the native hammerhead ribozyme, like RNA chaperones. Because the large tetraalkylammonium ions interact with RNAs through electrostatic forces and steric hindrance, our results provide physical insights into interactions of ribozymes with large cationic molecules, such as basic proteins. The study using tetraalkylammonium ions may be useful for understanding influences of RNA-binding proteins on the turnover of intracellular ribozymes.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This research was supported by in part by Grants-in-Aid for Scientific Research, Japan Society for the Promotion of Science (JSPS KAKENHI grant numbers, 15K05575 and 18K05325).Notes and references
- V. Bourdeau, G. Ferbeyre, M. Pageau, B. Paquin and R. Cedergren, Nucleic Acids Res., 1999, 27, 4457–4467 CrossRef CAS.
- M. Z. Barciszewska, M. Szymanski, E. Wyszko, J. Pas, L. Rychlewski and J. Barciszewski, Mutat. Res., 2005, 589, 103–110 CAS.
- K. Salehi-Ashtiani, A. Luptak, A. Litovchick and J. W. Szostak, Science, 2006, 313, 1788–1792 CrossRef CAS.
- M. Martick, L. H. Horan, H. F. Noller and W. G. Scott, Nature, 2008, 454, 899–902 CrossRef CAS.
- M. de la Peña and I. Garcia-Robles, EMBO Rep., 2010, 11, 711–716 CrossRef.
- C. H. Webb and A. Luptak, RNA Biol., 2011, 8, 719–727 CrossRef CAS.
- C. Hammann, A. Luptak, J. Perreault and M. de la Peña, RNA, 2012, 18, 871–885 CrossRef CAS.
- M. de la Peña and A. Cervera, RNA Biol., 2017, 14, 985–991 CrossRef.
- R. Yamagami, J. L. Bingaman, E. A. Frankel and P. C. Bevilacqua, Nat. Commun., 2018, 9, 2149 CrossRef.
- R. Yamagami, R. Huang and P. C. Bevilacqua, Biochemistry, 2019, 58, 3971–3979 CrossRef CAS.
- L. Citti and G. Rainaldi, Curr. Gene Ther., 2005, 5, 11–24 CrossRef CAS.
- D. Rueda and N. G. Walter, Methods Mol. Biol., 2006, 335, 289–310 CAS.
- S. Li, M. Nosrati and M. Kashani-Sabet, Methods Mol. Biol., 2008, 405, 113–131 CrossRef.
- E. Bertrand, R. Pictet and T. Grange, Nucleic Acids Res., 1994, 22, 293–300 CrossRef CAS.
- C. T. Webb and A. Luptak, Biochemistry, 2018, 57, 1440–1450 CrossRef CAS.
- J. C. Penedo, T. J. Wilson, S. D. Jayasena, A. Khvorova and D. M. Lilley, RNA, 2004, 10, 880–888 CrossRef CAS.
- M. Martick, T. S. Lee, D. M. York and W. G. Scott, Chem. Biol., 2008, 15, 332–342 CrossRef CAS.
- T. S. Lee, G. M. Giambasu, C. P. Sosa, M. Martick, W. G. Scott and D. M. York, J. Mol. Biol., 2009, 388, 195–206 CrossRef CAS.
- E. A. Frankel, C. A. Strulson, C. D. Keating and P. C. Bevilacqua, Biochemistry, 2017, 56, 2537–2548 CrossRef CAS.
- H. Chen, T. J. Giese, B. L. Golden and D. M. York, Biochemistry, 2017, 56, 2985–2994 CrossRef CAS.
- A. L. Feig, W. G. Scott and O. C. Uhlenbeck, Science, 1998, 279, 81–84 CrossRef CAS.
- A. Mir and B. L. Golden, Biochemistry, 2016, 55, 633–636 CrossRef CAS.
- C. Hammann, R. Hormes, G. Sczakiel and M. Tabler, Nucleic Acids Res., 1997, 25, 4715–4722 CrossRef CAS.
- H. Kaddour, J. Vergne, G. Herve and M. C. Maurel, Biochim. Biophys. Acta, 2014, 1840, 1670–1675 CrossRef CAS.
- R. R. Poudyal, C. D. Keating and P. C. Bevilacqua, ACS Chem. Biol., 2019, 14, 1243–1248 CrossRef CAS.
- D. Herschlag, M. Khosla, Z. Tsuchihashi and R. L. Karpel, EMBO J., 1994, 13, 2913–2924 CrossRef CAS.
- E. Jankowsky and B. Schwenzer, Biochemistry, 1996, 35, 15313–15321 CrossRef CAS.
- E. Jankowsky and B. Schwenzer, Eur. J. Biochem., 1998, 254, 129–134 CrossRef CAS.
- J. K. Christiansen, S. Lobedanz, K. Arar, J. Wengel and B. Vester, Bioorg. Med. Chem., 2007, 15, 6135–6143 CrossRef CAS.
- S. Nakano, H. T. Karimata, Y. Kitagawa and N. Sugimoto, J. Am. Chem. Soc., 2009, 131, 16881–16888 CrossRef CAS PubMed.
- S. Nakano, M. Horita, M. Kobayashi and N. Sugimoto, Catalysis, 2017, 7, 355–368 Search PubMed.
- M. Ryckelynck, S. Baudrey, C. Rick, A. Marin, F. Coldren, E. Westhof and A. D. Griffiths, RNA, 2015, 21, 458–469 CrossRef CAS.
- J. L. O'Rear, S. Wang, A. L. Feig, L. Beigelman, O. C. Uhlenbeck and D. Herschlag, RNA, 2001, 7, 537–545 CrossRef.
- M. D. Canny, F. M. Jucker, E. Kellogg, A. Khvorova, S. D. Jayasena and A. Pardi, J. Am. Chem. Soc., 2004, 126, 10848–10849 CrossRef CAS.
- J. L. Boots, M. D. Canny, E. Azimi and A. Pardi, RNA, 2008, 14, 2212–2222 CrossRef CAS.
- J. A. Nelson and O. C. Uhlenbeck, RNA, 2008, 14, 43–54 CrossRef CAS.
- E. M. Osborne, J. E. Schaak and V. J. DeRose, RNA, 2005, 11, 187–196 CrossRef CAS.
- S. Nakano, D. Miyoshi and N. Sugimoto, Chem. Rev., 2014, 114, 2733–2758 CrossRef CAS.
- S. Nakano and N. Sugimoto, Mol. BioSyst., 2017, 13, 32–41 RSC.
- S. Nakano, L. Wu, H. Oka, H. T. Karimata, T. Kirihata, Y. Sato, S. Fujii, H. Sakai, M. Kuwahara, H. Sawai and N. Sugimoto, Mol. BioSyst., 2008, 4, 579–588 RSC.
- S. Nakano, Y. Kitagawa, D. Miyoshi and N. Sugimoto, FEBS Open Bio, 2014, 4, 643–650 CrossRef CAS.
- E. Stellwagen, J. M. Muse and N. C. Stellwagen, Biochemistry, 2011, 50, 3084–3094 CrossRef CAS PubMed.
- S. Nakano, Y. Tanino, H. Hirayama and N. Sugimoto, Biophys. J., 2016, 111, 1350–1360 CrossRef.
- N. C. Stellwagen and E. Stellwagen, J. Phys. Chem. B, 2019, 123, 3649–3657 CrossRef CAS PubMed.
- S. Nakano, T. Ayusawa, Y. Tanino and N. Sugimoto, J. Phys. Chem. B, 2019, 123, 7687–7694 CrossRef CAS PubMed.
- S. Nakano, T. Watabe and N. Sugimoto, ChemPhysChem, 2017, 18, 3614–3619 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9ra06797c |
| This journal is © The Royal Society of Chemistry 2019 |