DOI:
10.1039/C9RA06761B
(Paper)
RSC Adv., 2019,
9, 40855-40862
Exploring the antifouling effect of elastic deformation by DEM–CFD coupling simulation†
Received
27th August 2019
, Accepted 25th November 2019
First published on 10th December 2019
Abstract
The influence of elastic deformation and elastic modulus on the release of adhered bacteria was investigated in this paper. Four silicone elastomers (SE) with different elastic moduli and one rigid polystyrene sheet were prepared to verify the antifouling effect of elastic deformation. The SE film has an elastic deformation effect under the stimulus of fluid medium, which makes the surface unstable. That could reduce the adhesion of fouling organisms and provide a foul-release basis. Distinct anti-adhesion properties were observed in our study in that cells more easily adhered to the rigid surface than the elastic surfaces under hydrodynamic conditions. However, the bacterial attachment test showed a similar antifouling performance of SE and the rigid surface under static conditions. To investigate the anti-adhesion ability of the elastic surface and rigid surface, the bacterial adhesive kinetics were studied by Discrete Element Method (DEM)–Computational Fluid Dynamics (CFD) coupling simulation. Results indicated the number of bacteria adhering on the elastic wall was significantly lower than on the rigid wall. And as the elastic modulus increased, the bacterial adhesion increased accordingly within a certain range. This work should not only enhance understanding of elastomer-based antifouling materials, but also facilitate the design and construction of other types non-toxic foul-release materials.
Introduction
Antifouling coatings are widely used on the hulls of ships to prevent the adhesion of marine organisms. Marine biofouling is a widespread problem in the maritime industry, and it has serious impacts, such as increased navigation resistance, higher fuel consumption, and decreased navigation speed.1–4 In the recent past, several environmentally benign strategies have been proposed increasingly to control biofouling, such as foul-release coatings.5–7 Foul-release coatings provide a very smooth, low-friction surface and have an adjusted elasticity, which reduces the strength of adhesion of fouling.8–10 The most promising coatings are generally based on silicone elastomers (SE), which have been considered environmentally benign.11,12 SE has a number of properties, such as low surface energy, low microroughness and low modulus, which are necessary to reduce chemical and mechanical locking of fouling organisms.13,14 Although all these properties are benefit to foul-release, but the surface instability, which induced by the elastic deformation under the stimulus of a fluid medium, could inhibit the incipient bacterial adhesion. Understanding the antifouling mechanisms is conducive to the development of environmentally friendly fouling-resistant technologies.
Biofouling occurs via the formation of biofilm, which triggered by the adhesion of primary colonizing bacteria.15 In the early stages of biofilm formation, physical interactions are the first forces for bacteria adhesion, which frequently influenced by the physical properties of interaction surface.16 As previous reports, the micron-scale deformations are presented on elastic surface, and it is benefit to antifouling.17 However, the relationship between bacterial adhesion and the elastic deformation was not completely clear. This work describes an easy and novel method to investigate the influence of elastic deformation on antifouling effects. In this paper, four SE films with adjusted elasticity was prepared by varying the graphene concentration. This surface of graphene-SE (GSE) had elastic deformation effect under the stimulus of fluid medium (Scheme 1), which could inhibit the adhesion of biofouling and provide foul release basis. Traditional ideas describing the mechanisms of foul-release coatings is that mechanical factors and surface chemistry are major determinants of adhesion strength, whereas settled cells of macro-fouling species appear to be more sensitive to surface chemistry.13,18 However, a different relationship was observed in our study that cells were more easily adhere to the polystyrene (PS) sheet (rigid material) than the elastic surfaces (Scheme 1).
 |
| | Scheme 1 Schematic illustration showing the different antifouling response of elastic film and rigid film under marine environment. | |
A novel method was explored to investigate the bacterial deposition corresponding to flowing particles being captured by the elastic and rigid membrane. The bacterial adhesive kinetics, including bacterial motions and collisions, were simulated by coupling the Discrete Element Method (DEM) and Computational Fluid Dynamics (CFD) methods for the first time.15,19 The simulations of particulates as described in this work can predict initial deposition patterns of both elastic membrane and rigid membrane. It evaluates the role of elastic deformation, which could influence the bacteria adhesion. Simulations considered 1 μm diameter as the bacterial particle size, which is about the same size as common marine microorganisms, such as Paracoccus pantotrophus (P. pantotrophus). Results indicated that there is a great difference of bacterial adhesion between elastic wall and rigid wall. The number of bacteria adhering on elastic wall is significantly lower than that of the rigid wall, indicating that the elastic wall is more effective to inhibit bacterial adhesion under the effect of water flow. The bacteria morphology will influence the adhesion rate for elastic wall, but for rigid surface, the influence of bacteria morphology is ignorable. The purpose of this study was to investigate the initial attachment kinetics of bacteria on elastic surface and rigid surface, which should provide insight into the functions of elastic deformation. Moreover, this study demonstrated the usefulness of DEM–CFD technique for the investigation of particulate fouling, and that could enhance the understanding of the adhesion behavior of bacteria on different surfaces, facilitating the construction of eco-friendly coatings.
Experimental section
Materials
Room temperature vulcanized silicone rubber (RTV-2) was purchased from Guangdong Bo Rui Co., Ltd. (Guangdong, China). Bacterial strain P. pantotrophus ATCC 35512 was obtained from Chuanxiang Biotechnology, Ltd. (Shanghai, China). Graphene were obtained from Nanjing SCF Nanotech, Ltd. Dimethyloctadecyl[3-(trimethoxysilyl)propyl] ammonium chloride (C26H58ClNO3Si) were purchased from Sigma-Aldrich. Silane coupling agent KH-550 was purchased Nanjing Aocheng Chemical Co., Ltd. Ethanol, tetrahydrofuran and acetone were obtained from Beijing Chemical Industry Group Co., Ltd. All aqueous solutions were prepared using ultra-pure water (18.2 MΩ, Milli-Q, Millipore). All the chemicals were used as received without further purification.
Preparation of GSE antifouling films
Firstly, graphene was added in anhydrous ethanol, then KH-550 was added and mechanical stirring for 2 h. Then, the graphene mixture was placed in a vacuum drying chamber for 12 h at 80 °C, and obtaining the pre-treated graphene dispersions. Secondly, RTV-2 silicon rubber and pre-treated graphene were mixed and added in tetrahydrofuran, and mechanical stirring for hours at bath temperature 55 °C until the tetrahydrofuran evaporated completely. Then, the mixture of graphene and RTV-2 was placed in a vacuum chamber for 10 min at 60 °C until no bubbles overflowed. Finally, the mixture of graphene and RTV-2 was poured into an acrylic mold, and then it was cured at room temperature for 24 h. Four kinds of GSE with different graphene concentration were prepared, and the content of graphene was 0 wt%, 0.18 wt%, 0.36 wt% and 0.72 wt%, respectively.
Measurements and characterizations
The samples were characterized by scanning electron microscope (SEM) images using a HITACHI S-4500 instrument. Raman spectra were obtained using a micro-Raman system (JY-Horiba, LabRam300) which combined a 50× objective lens (Olympus, NA 0.75) with a 633 nm CW He–Ne laser. FT-IR characterization was carried out on a BRUKE Vertex 70 FTIR spectrometer.
Measurement of elastic modulus
Tensile tests were used to investigate the elastic modulus and tensile properties of the pristine silicone elastomers (SE) and GSE composites. The tensile tests were measured using a rubber electronic tensile testing machine (UTM5305, Youhong Measurement and Control Technology Co., LTD, Shanghai, China). These samples were cut into type I dumbbell-shape according to China National Standard GB/T 528-2009 (ISO 37:2005, IDT).20 The Transcell BSS-500 kg (0.01 N) force transducer was used. The frequency of sampling was 250 ms, and the tensile speed was 500 mm min−1.
Measurement of contact angle
Static contact angle measurements with the sessile drop method were recorded and analyzed at room temperature (DSA225, KRUSS, Germany). Deionized water, ethanediol, formamide were used in this experiment, each reported contact angles were measured three times to minimize experimental error.
Calculation of the surface energy
The surface energy is calculated by the followed methods based on previous reports.20 The correlation of contact angle and surface energy is as follows:
where θ stands for contact angle, γS and γL represents surface energy of solid and liquid, respectively. β is a constant with the value 1.057 × 10−4 m2 mJ−1. The surface energy of deionized water (γS) is 72.8 mN m−1.21
Bacterial attachment experiments
Monocolonies of P. pantotrophus on the solid ATCC 1396 medium agar plate were transferred to 20 mL of liquid culture medium respectively and grown at 37 °C for 12 h under 180 rpm rotation. Then the bacteria were diluted with broth to 106 cfu mL−1.22 In all experiments, the concentrations of bacteria were determined by measuring the optical density at 600 nm (OD600nm). Firstly, bacteria attachment was tested under quasi-static condition. The PS sheet and GSE films were cut into 1 cm × 1 cm and incubated with bacteria for 24 h. Then, the amount of adhesion bacteria was measured after ultrasonic. Secondly, the anti-fouling ability was studied at simulated marine environment. We designed an instrument to simulate the marine environment as boat sail in the flowing medium. These anti-fouling surfaces were settled at the bottom of test area, and the flowing speed was about 1.0–2.5 m s−1. Then, as-prepared bacteria solution (500 μL) was mixed with artificial seawater and incubated with these anti-fouling surfaces in this instrument. After 60 hours or 120 hours, washed with PBS buffer under aseptic condition to eliminate medium and unbound bacteria.22 The generated biofilm was measured by crystal violet staining method,19 selecting the area of 1 cm × 1 cm in these samples to quantify the biofilms.
Results and discussion
Prepared and characterizations of elastic films and PS sheet
To revealed the influence of elastic deformation on antifouling effects, four elastic surfaces of GSE film with different graphene concentration were prepared, and the content of graphene was 0 wt%, 0.18 wt%, 0.36 wt% and 0.72 wt%, respectively (Fig. S1†). Meanwhile, PS sheet (rigid surface) was prepared as a control. The surface morphology was characterized by SEM, as revealed in Fig. 1. The pristine surface of SE was smooth (Fig. 1a), and the surface of GSE became crimpling after graphene was mixed in this film (Fig. 1b and c). However, the graphene was aggregation in the film when the content of graphene was 0.72 wt%. That might be induced by the high concentration of graphene nanosheets which made the compatibility decreasing between graphene and silicone rubber (Fig. 1d). The wrinkles in these surfaces are benefit to elastic deformation, and it should make the surface unstable and drive bacteria departing from these surfaces. Meanwhile, the elastic modulus measurement was conducted and the elastic modulus curves for these anti-fouling films were shown in Fig. 2a. We could conclude that the addition of graphene (GSE) decreased the elastic modulus compared to the pristine SE films (0 wt%), and the elastic modulus of 0.36 wt% GSE was the lowest than other GSE films. As previous reports, a low modulus is favorable for foul-release coatings.23,24 When the concentration of graphene was 0.72 wt%, the elastic modulus was higher than 0.36 wt% GSE, that is corresponding with the result of SEM images. Therefore, either the property of elastic modulus or the ability of elastic deformation of 0.36 wt% GSE was more proper for fouling-release than others. The presence of graphene in the composite film was further confirmed by Raman spectroscopy. The Raman spectrum of GSE film displays two prominent peaks (Fig. 2b). One is G-band at 1588 cm−1 and another is relatively broad 2D-band of GSE film at around 2680 cm−1.25 Surface energy is one of the important factors which could affect bacterial adhesion and biofilm formation. So, the contact angle of elastic surface (GSE film) and rigid surface (PS sheet) were measured, and the surface energy of these materials were determined by previous reported method.20 As revealed in Fig. S2,† it is observed that the mean surface energy values for the PS sheet and GSE films (0–0.72 wt%) were 28.9, 19.6, 19.2, 20.6 and 19.1 mN m−1, respectively. According to Baier curve, the surface energy of PS sheet and GSE film are all close to foul-release zone.26,27
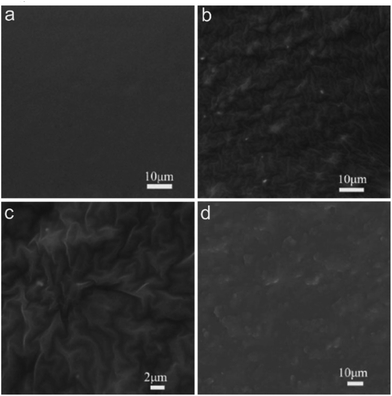 |
| | Fig. 1 The elastic film of (a) pristine SE film (0 wt%), and GSE film with different graphene content, the content of graphene was (b) 0.18 wt%, (c) 0.36 wt% and (d) 0.72 wt%, respectively. | |
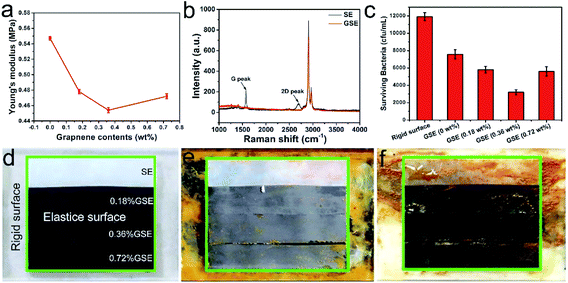 |
| | Fig. 2 (a) The elastic modulus of pristine SE film and GSE film with different graphene content. (b) Raman spectra of the pristine SE and GSE films. (c) The surviving bacteria of P. pantotrophus incubated on rigid surface and antifouling surfaces. Representative digital images showed the rigid surface (outside the green border) and elastic surface (inside the green border) after incubated with P. pantotrophus for (d) 0 h, (e) 60 h and (f) 120 h in simulated marine environment. The graphene concentration of elastic surface is 0 wt%, 0.18 wt%, 0.36 wt% and 0.72 wt%, respectively. | |
Inhibitory effects of elastic film and rigid film on bacteria adhesion and biofilm formation
Bacterial biofilms are important initiators for the successful settlement of marine organism. Therefore, we compared the anti-adhesion ability between the rigid surface and GSE surface under different conditions, including static condition and simulated marine environment. Firstly, the PS sheet and elastic films were incubated with bacteria for 24 h in quasi-static condition, and the results were determined by optical density (OD) measurements. As Fig. S3† revealed, the quantities of bacteria adhering to rigid surface and elastic surface were almost same. That might be attributed to the similar surface energy of GSE films and PS sheet. Secondly, we designed an instrument to simulate the marine environment that boat sail in fluid medium, as described in Fig. S4.† These coatings were settled at the bottom of test area in this instrument, and the flowing speed was set as about 0.2–0.5 m s−1. We systematically tested the adhesion efficiency of P. pantotrophus, which was chosen as a model to evaluate the anti-adhesion performance and mechanism. Fig. 2d–f showed the biofilm formation of P. pantotrophus on these surfaces, respectively. Compared with the rigid surface, a remarkable difference was observed in the elastic surface of GSE after 60 h and 120 h incubation. Clear biofilm bands were observed on the rigid surface after 60 h and 120 h incubation. In contrast, no evident biofilms were observed on the surface of 0.36 wt% GSE, indicating that most bacteria were expelled from the surface under the fluid medium. However, some of the biofilm removed from the rigid surface after 120 h incubation, that might because the life cycle and aging of biofilm, which resulted in the release of planktonic cells and biofilm dispersal.28,29 Further tests of the biofilm formation were conducted by observing the number of colony-forming units on agar plate (Fig. S5†), and the numbers of bacteria were quantified (Fig. 2c). The results demonstrated that after 120 h culture, elastic GSE exhibited stronger anti-adhesion activity than rigid surface, and the anti-adhesion ability of 0.36 wt% GSE was the best. Meanwhile, the biofilm mass was quantified by the crystal violet staining method as well.19 The results revealed that the 0.36 wt% GSE film were effective for biofilm inhibition (Fig. S6†), and it confirmed that 0.36 wt% GSE film exhibited the best inhibition ability toward bacteria adhesion and biofilm formation.
Exploring the influence and mechanism of elastic deformation on antifouling performance by DEM–CFD coupling simulation
The non-bactericidal film of GSE can expel bacteria through elastic deformation and make the surface unstable. The pressure pulsations of turbulent flow give rise to various fluid/solid interactions. The elastic modulus is defined as:
According to this equation, under the same F (pressure pulsations), elastic materials, which have a lower elastic modulus than rigid materials, will achieve a larger deformation. In the ocean, the intensity of turbulence covers a wide-range, so that materials with a low elastic modulus are helpful for interacting with a wider range of flow. To further explore the anti-adhesion mechanism of GSE and study the influence of elastic modulus, DEM–CFD coupling simulation was used in this paper.
DEM Mechanistic principles are implemented via the EDEM 2.3 software, which allows the consideration of particle contact through elastic Hertz models, and enables coupling with CFD data.25 The coupling method used in this paper is one-way DEM–CFD coupling. It permits the fluid to influence particles only, while particles loading has no influence on the fluid. DEMs have been used to simulate the flow of granular materials in various applications.30,31 In the process of DEM calculation, the contact model between particles is defined as soft-particle contact models, and a single particle is regarded as a calculation unit. From this simulation, we can calculate the relative displacement and the interaction force between the particles. The commercial software FLUENT 12.1 was used to solve the steady-state Navier–Stokes (N–S) equations under laminar flow hydrodynamics,32,33 using the SIMPLEC (SIMPLE-consistent) algorithm, this expressed as:
| |
 | (1) |
where
ug is speed,
t is time,
ρg is density of liquids,
εg is void fraction of liquids;
| |
 | (2) |
where
g is gravity,
ug is dynamic viscosity of liquids,
S is momentum sink and
P is liquid pressure.
S represent the summation of the drag force, and it is achieved through the calculation of the drag force produced by the relative motion between two phases. The momentum sink S is calculated by
| |
 | (3) |
where
FD is the drag force in mesh cell,
V is the volume of the CFD mesh cell.
The interphase force between seawater and bacterial particles is mainly drag force, so we choose Ergun and Wen & Yu (Gidaspow) as drag models in the EDEM–FLUENT coupling modules,34,35 which expressed as:
| |
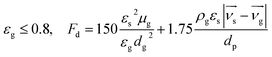 | (4) |
| |
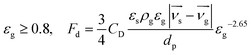 | (5) |
| |
 | (6) |
| |
 | (7) |
where
εs is the volume fraction of particles,
εg is the volume fraction of liquid,
Fd is the drag force of single particle,
CD is the drag coefficient,
dp is particle diameter,
ρg is the density of liquid,
μg is the viscosity coefficient of liquid,
νg is the liquid velocity, and
νs is the particle velocity.
To establish the bacterial particles model, the morphology of P. pantotrophus was studied by SEM. The SEM images indicated these bacteria are spherical or ellipsoidal, and the diameter is 1–2 μm. In order to make the model similar to the real bacterial morphology, three types of bacterial model were established in EDEM particle modeling module, as described in Fig. 3. The simulation parameters common to all tested configurations are presented in Table S1.†
 |
| | Fig. 3 SEM images of P. pantotrophus (a and b) and three types of bacterial models, including coccoid bacteria (c), dividing bacteria (d), and combined spheroidal (e) models. | |
Consider the viscous contact between bacterial particles, the Hertz–Mindlin with JKR Cohesion was chosen as particle contact models. The contact force for bacterial particles was represented by the Hertz–Mindlin elastic contact model.36,37 Except for the contact repulsive force, the cohesive nature of bacterial particles leads to an attractive force. Herein, we used the JKR cohesion model,38 which was originally implemented to incorporate the van der Waals forces in the contact domain:
| |
 | (8) |
| |
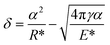 | (9) |
where
R* is equivalent radius, and
E* is equivalent Young's modulus. In this model, the cohesion force is mainly determined by the contact overlap (
δ), interaction parameter, and the surface energy (
γ).
For this simulation experiment, the reasonable parameter setting can improve the accuracy of the results. Before the simulation calculation, it is necessary to determine the properties of each material and the relevant parameters. In order to better distinguish the properties of elastic surface and rigid surface, silicone rubber and stainless steel were selected as the model of elastic surface and rigid surface. And the corresponding parameters were presented in Table 1. In this model, these simulation parameters are used common to all tested configurations.
Table 1 Parameters of materials
| Material parameters |
Elastic surface |
Rigid surface |
| Poisson's ratio |
0.48 |
0.30 |
| Density (kg m−3) |
1.03 × 103 |
8.03 × 1011 |
| Shear modulus (Pa) |
1.5 × 105 |
1.9 × 1011 |
| Elastic modulus (Pa) |
4.6 × 105 |
5.6 × 1011 |
Parameter setting of bacterial particle factory
Bacterial particles were randomly created one by one following a uniform law over the control surface (Fig. 4), meaning that the probability of particles creation was equal and uniform for each point of the inlet window at each time step. In order to allow a high enough concentration of particles in the computational volume and favour particle-to-particle collisions and cluster formation, the bacterial particle creation rate was set to 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 particles per s and the generation method was spray form. The initial orientation is 60° from the particle formation surface and the initial average velocity of particles is set to 0.001 m s−1.
000 particles per s and the generation method was spray form. The initial orientation is 60° from the particle formation surface and the initial average velocity of particles is set to 0.001 m s−1.
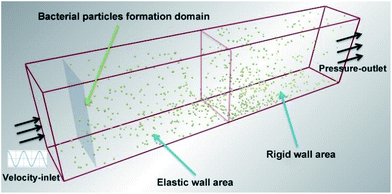 |
| | Fig. 4 Schematic diagram of EDEM–FLUENT coupling calculation model. The specific size of the calculation domain is length, width, height is 200 mm × 30 mm × 60 mm. | |
The simulation parameters are usually set as empirical values, and the corresponding specific parameters of this paper are shown in Table S2.† The calculation time-step is set in accordance with EDEM setting principle, which is 18% of Rayleigh time-step. Grid size is 3Rmin and the total simulation time is 0.075 s. After the above operation completed, the CFD coupling program can be started by opening the EDEM coupling connection. In the whole EDEM–FLUENT coupling simulation, the calculation of EDEM is completely controlled by FLUENT.
FLUENT simulation
First of all, the structured hexahedron mesh is generated by the Hypermesh software, which is shown in Fig. S7.† Boundary conditions for the model include inlet-velocity and pressure-outlet, as shown in Fig. 4. The relationship between inlet-velocity and flow-time no-slip was described in Fig. S8.† The remaining walls were treated as wall boundary conditions.
Before solving this model, the flow field is set as incompressible liquid, and the flow state is set as turbulent. Meanwhile, boundary condition of the inlet fluid velocity is set by user-defined function, and the velocity equation is expressed as:
| |
 | (10) |
where
T is cycle,
L is channel length,
t is run time,
X is the coordinate in the entry surface. The entry speed was changed over time, as revealed in Fig. S8.
†
Pressure-based solver and second-order upwind discretization is used to reduce the numerical diffusion. The outlet is set as atmospheric pressure (1.01 × 105 Pa). The convergence criteria are set as RMS = 1e−005, and the relaxation factor is reduced to make the calculation more stable. The domain used in this coupling simulation is shown in Fig. 4, with size of 200 mm × 30 mm × 60 mm. The length of the domain was chosen to allow any inlet effects to dissipate and to ensure a developed flow towards the outlet end of the fetch.
Results and analysis of EDEM–FLUENT coupling calculation
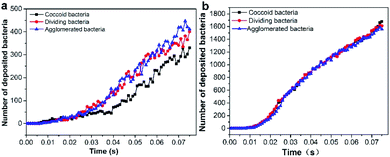
The adhesion of bacterial particles on elastic wall and rigid wall at different periods was processed by DEM–CFD coupling methods, as shown in Fig. S9† and 5. In order to distinguish the adhesion state and free state, particles were dyed according to their speed, such as red, green and blue particles. When the velocity of particles is lower than 6.6 × 10−10 m s−1, they are all dyed blue, including static particles (Fig. S9†). When t = 0.01 s, bacterial particles float in the whole flow field and some of them adhere to the bottom walls under the effect of drag and gravity, but the adhesion amount on the elastic wall or rigid wall is not obvious. And the number of green and blue bacterial particles were basically same, indicating that the particles were in a state of low-speed movement. However, when t = 0.03 s, the particles adhesion to rigid wall is much higher than that of elastic wall. This might be related to the different physical properties of rigid wall and elastic wall, including Poisson's ratio, density, shear modulus and elastic modulus. When t = 0.075 s, the bacterial adhesion on the elastic wall is distinct from the rigid wall. Meanwhile, the particles speed decreased over time and the number of blue particles were increasing gradually, even most of them adhered to the wall. And the number of bacteria adhering on rigid wall was much higher than that of the elastic wall. However, a large number of bacterial particles were observed on the boundary. That might be induced by the adhesion phenomenon between bacterial particles, which made the floating bacteria aggregated on the adjacent elastic wall. The number of bacterial particles adhering to the elastic wall increases with time, but the number of bacteria occasionally decreases at the next point. That might be due to the bacterial particles not attached to the elastic wall firmly, and separating from the elastic wall under the effect of water flow.
 |
| | Fig. 5 The number of deposited bacteria in elastic surface (a) and rigid surface (b). (c) Bacterial adhesion on elastic wall and rigid wall at t = 0.075 s. | |
The influence of bacterial morphology and elastic modulus on bacterial adhesion.
In fact, bacterial morphology is not only spherical, some of them are irregular ellipsoidal, rod-shaped, and agglomerated spheroidal, etc. Therefore, it is necessary to simulate the adhesion of bacteria on the elastic wall with different bacterial morphology. In this study, three kinds of typical bacterial models are established, which represent coccoid bacteria, dividing bacteria and agglomerated bacteria (as shown in Fig. 3), respectively. The adhesion of these bacterial morphology on the elastic wall and rigid wall was shown in Fig. 6. Results revealed that, for the elastic wall, the anti-adhesion effect for spherical particles was more effective than that of ellipsoidal and agglomerated spherical particles. But for the rigid wall, the adhesion of these bacterial particles was similar, and it was much larger than that of elastic wall, indicating that the elastic wall has the property to inhibit bacterial adhesion under the effect of water flow.
 |
| | Fig. 6 The evolution of deposited particles on elastic wall (a) and rigid wall (b) with different bacterial morphology. | |
Based on the above results, we could conclude that the elastic deformation was benefit to anti-adhesion. To further verify the effect of elastic modulus on bacterial adhesion, three models of elastic wall with different shear modulus and elastic modulus were designed, and the parameters were presented in Table 2. The deposited bacterial particles were counted and the results indicate that the adhesion of bacterial particles increased with the increase of elastic modulus at the same simulation time (Fig. 7). So, we could conclude that the smaller of the elastic modulus is, the less of bacterial particles adhering to the elastic wall. This confirmed that the elastic transform could influence the adhesion of bacteria.
Table 2 Parameters settings of elastic walls for different models
| Models |
Model 1 |
Model 2 |
Model 3 |
| Shear modulus (Pa) |
1.50 × 106 |
1.50 × 105 |
1.50 × 104 |
| Elastic modulus (Pa) |
4.46 × 106 |
4.46 × 105 |
4.46 × 104 |
 |
| | Fig. 7 Evolution with time of deposited particles under different elastic modulus. | |
Conclusion
In summary, we have demonstrated the anti-adhesion activity of silicone elastomers for investigating anti-fouling mechanism based on elastic deformation. The surface of GSE had elastic deformation under the stimulus of fluid medium, so it could reduce the adhesion of biofouling and provide foul release basis. Under simulated marine environment, the GSE coatings showed excellent anti-adhesion properties than rigid surface. Moreover, to investigate the anti-adhesion mechanism of the elastic materials, we described a novel method to explore the bacterial adhesive kinetics by DEM–CFD coupling simulation. Results indicated that the number of bacteria adhering on elastic wall was significantly lower than that of the rigid wall. As the elastic modulus increased, the adhesion of bacterial particles was increased accordingly at the same time. We hope that this work has not only provided new insights into deciphering elastic material based antifouling coatings, but also will facilitate the design and construction of other types of elastic antifouling materials.
Conflicts of interest
There are no conflicts to declare.
Acknowledgements
The authors are grateful for grants received from the National Natural Science Foundation of China (Grant No. 51875240), the Pre-research Foundation of Equipment Field of China (Grant No. 61400040403), the Department of Science and Technology of Jilin Province (Grant No. 20190103114JH), the China Postdoctoral Science Foundation Funded Project (Grant No. 2018M630324), and the Changchun Science and Technology Innovation Double Ten Project (Grant No. 17SS023).
Notes and references
- Z. Chen, W. Zhao, J. Xu, M. Mo, S. Peng, Z. Zeng, X. Wu and Q. Xue, RSC Adv., 2015, 5, 36874–36881 RSC.
- I. Banerjee, R. C. Pangule and R. S. Kane, Adv. Mater., 2011, 23, 690–718 CrossRef CAS PubMed.
- L. D. Chambers, K. R. Stokes, F. C. Walsh and R. J. K. Wood, Surf. Coat. Technol., 2006, 201, 3642–3652 CrossRef CAS.
- Q. Li, S. Mahendra, D. Y. Lyon, L. Brunet, M. V. Liga, D. Li and P. J. J. Alvarez, Water Res., 2008, 42, 4591–4602 CrossRef CAS PubMed.
- D. M. Yebra, S. Kiil, K. D. Johansen and C. Weinell, Prog. Org. Coat., 2005, 53, 256–275 CrossRef CAS.
- M. S. Selim, M. A. Shenashen, S. A. El-Safty, S. A. Higazy, M. M. Selim, H. Isago and A. Elmarakbi, Prog. Mater. Sci., 2017, 87, 1–32 CrossRef CAS.
- L. A. Goetz, B. Jalvo, R. Rosal and A. P. Mathew, J. Membr. Sci., 2016, 510, 238–248 CrossRef CAS.
- M. S. Selim, M. A. Shenashen, A. Elmarakbi, N. A. Fatthallah, S. Hasegawa and S. A. El-Safty, Chem. Eng. J., 2017, 320, 653–666 CrossRef CAS.
- M. Barletta, C. Aversa, E. Pizzi, M. Puopolob and S. Vescob, Prog. Org. Coat., 2018, 123, 267–281 CrossRef CAS.
- J. Genzer and K. Efimenko, Biofouling, 2006, 22, 339–360 CrossRef CAS PubMed.
- S. B. Yeh, C. S. Chen, W. Y. Chen and C. J. Huang, Langmuir, 2014, 30, 11386–11393 CrossRef CAS PubMed.
- F. Natalio, R. Andre, A. F. Hartog, B. Stoll, K. P. Jochum, R. Wever and W. Tremel, Nat. Nanotechnol., 2012, 7, 530–535 CrossRef CAS PubMed.
- R. Holland, T. M. Dugdale, R. Wetherbee, A. B. Brennan, J. A. Finlay, J. A. Callow and M. E. Callow, Biofouling, 2004, 20, 323–329 CrossRef CAS PubMed.
- Z. Hu, J. A. Finlay, L. Chen, D. E. Betts, M. A. Hillmyer, M. E. Callow, J. A. Callow and J. M. DeSimone, Macromolecules, 2009, 42, 6999–7007 CrossRef CAS.
- F. Chaumeil and M. Crapper, J. Membr. Sci., 2013, 442, 254–263 CrossRef CAS.
- A. Rosenhahn, S. Schilp, H. J. Kreuzer and M. Grunze, Phys. Chem. Chem. Phys., 2010, 12, 4275–4286 RSC.
- H. Jin, T. Zhang, W. Bing, S. Dong and L. Tian, J. Mater. Chem. B, 2019, 7, 488–497 RSC.
- J. A. Finlay, S. M. Bennett, L. H. Brewer, A. Sokolova, G. Clay, N. Gunari, A. E. Meyer, G. C. Walker, D. E. Wendt, M. E. Callow, J. A. Callow and M. R. Detty, Biofouling, 2010, 26, 657–666 CrossRef CAS PubMed.
- T. Tsuji, K. Yabumoto and T. Tanaka, Powder Technol., 2008, 184, 132–140 CrossRef CAS.
- L. Tian, E. Jin, H. Mei, Q. Ke, Z. Li and H. Kui, Journal of Bionic Engineering, 2017, 14, 130–140 CrossRef.
- D. Y. Kwok and A. W. Neumann, Adv. Colloid Interface Sci., 1999, 81, 167–249 CrossRef CAS.
- W. Bing, Z. Chen, H. Sun, P. Shi, N. Gao, J. Ren and X. Qu, Nano Res., 2015, 8, 1648–1658 CrossRef CAS.
- K. Bazaka, M. V. Jacob, R. J. Crawford and E. P. Ivanova, Appl. Microbiol. Biotechnol., 2012, 95, 299 CrossRef CAS PubMed.
- R. F. Brady, Prog. Org. Coat., 1999, 35, 31–35 CrossRef CAS.
- Y. Song, J. Yu, L. Yua, F. E. Alama, W. Dai, C. Li and N. Jiang, Mater. Des., 2015, 88, 950–957 CrossRef CAS.
- C. M. Magin, S. P. Cooper and A. B. Brennan, Mater. Today, 2010, 13, 36–44 CrossRef CAS.
- R. E. Baier, J. Mater. Sci.: Mater. Med., 2006, 17, 1057–1062 CrossRef CAS PubMed.
- O. E. Petrova and K. Sauer, Curr. Opin. Microbiol., 2016, 30, 67–78 CrossRef CAS PubMed.
- D. McDougald, S. A. Rice, N. Barraud, P. D. Steinberg and S. Kjelleberg, Nat. Rev. Microbiol., 2012, 10, 39–50 CrossRef CAS PubMed.
- F. Bertrand, L.-A. Leclaire and G. Levecque, Chem. Eng. Sci., 2005, 60, 2517–2531 CrossRef CAS.
- J. Horabik and M. Molenda, Biosyst. Eng., 2016, 147, 206–225 CrossRef.
- R. Costa, S. Clain, G. J. Machado and R. Loubère, J. Sci. Comput., 2017, 71, 1375–1411 CrossRef.
- H. Xiao, J. L. Wu, J. X. Wang, R. Sun and C. J. Roy, J. Comput. Phys., 2016, 324, 115–136 CrossRef.
- L. Yan, Y. Cao, H. Zhou and B. He, Bioresour. Technol., 2018, 269, 384–392 CrossRef CAS PubMed.
- N. Iqbal and C. Rauh, Appl. Math. Comput., 2016, 277, 154–163 CrossRef.
- W. Yan, Y. Qian, W. Ge, S. Lin, W. K. Liu, F. Lin and G. J. Wagner, Mater. Des., 2018, 141, 210–219 CrossRef.
- M. Hiraiwa, M. A. Ghanem, S. P. Wallen, A. Khanolkar, A. A. Maznev and N. Boechler, Phys. Rev. Lett., 2016, 116, 1871–1880 Search PubMed.
- E. J. Parteli and T. Pöschel, Powder Technol., 2016, 288, 96–102 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9ra06761b |
|
| This journal is © The Royal Society of Chemistry 2019 |
Click here to see how this site uses Cookies. View our privacy policy here.  Open Access Article
Open Access Article a,
Xiaoming Wanga,
Wei Bing
a,
Xiaoming Wanga,
Wei Bing *ab,
Huichao Jin
*ab,
Huichao Jin a,
Jie Zhao
a,
Jie Zhao a and
Luquan Rena
a and
Luquan Rena





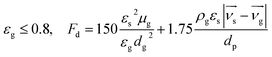
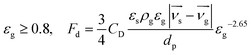




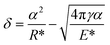
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 particles per s and the generation method was spray form. The initial orientation is 60° from the particle formation surface and the initial average velocity of particles is set to 0.001 m s−1.
000 particles per s and the generation method was spray form. The initial orientation is 60° from the particle formation surface and the initial average velocity of particles is set to 0.001 m s−1.







