Open Access Article
Open Access ArticlePotassium 2-methoxy-4-vinylphenolate: a novel hit exhibiting quorum-sensing inhibition in Pseudomonas aeruginosa via LasIR/RhlIR circuitry†
Mayank D. Shaha,
Prashant S. Kharkar b,
Niteshkumar U. Sahub,
Zoya Peerzadaa and
Krutika B. Desai
b,
Niteshkumar U. Sahub,
Zoya Peerzadaa and
Krutika B. Desai *c
*c
aSunandan Divatia School of Science, SVKM's NMIMS (Deemed to be University), Mumbai, 400056, India
bInstitute of Chemical Technology, Category I Deemed to be University (MHRD/UGC), Mumbai-400019, India
cMithibai College of Arts & Science & Amrutben Jivanlal College of Commerce & Economics, Mumbai, 400056, India. E-mail: krutika.desai@mithibai.ac.in
First published on 4th December 2019
Abstract
The emergence of multidrug-resistant (MDR) bacterial strains in the last decade is astonishingly alarming. Many of the widely used antibiotics have failed to exhibit clinical efficacy against such strains. Eventually we will exhaust all the resources in our antibiotic armamentarium. As a need of the hour, novel strategies are desperately required not only to curb, but also to reverse, the development of resistance in these pathogens, thereby maintaining their sensitivity towards current antibiotics. Intervention of bacterial virulence, rather than killing them, by inhibiting specific pathways/targets has emerged as a novel approach to tackle the drug resistance problem. The bacterial virulence is regulated via quorum-sensing, a cell–cell communication process precisely controlled by autoinducer molecules such as acyl homoserine lactone (AHL). The present study aimed at identifying promising quorum-sensing inhibitors in Pseudomonas aeruginosa, an opportunistic human pathogen especially associated with nosocomial infections, yielding four potential hits. Out of these, potassium 2-methoxy-4-vinylphenolate was the most potent quorum-sensing inhibitor targeting P. aeruginosa LasIR/RhlIR circuitry. It also inhibited biofilm formation, various virulence factors like LasA protease, LasB elastase and pyocyanin, and motility of bacteria like swarming and twitching.
1. Introduction
The past decade has witnessed a surge in antibiotic resistance. Several, otherwise antibiotic-sensitive microbes, have become resistant, leading to utter therapeutic failure. The World Health Organization (WHO) went to the extent of calling the 21st century as the ‘Post-antibiotic era’.1 The WHO fact sheet on antimicrobial resistance (released on February 15, 2018) is concerning.2 The overall statistics indicate the gravity of the situation. Moreover, the emergence of multidrug- and pan-drug-resistant pathogens, particularly the Gram-negative ones, has made the matter worse.3 Among these pathogens, mostly responsible for the nosocomial infections, Klebsiella pneumoniae, Acinetobacter baumannii, and Pseudomonas aeruginosa are the front-runners. In particular, Pseudomonas aeruginosa is most dangerous. It is an aerobic, Gram-negative, opportunistic human pathogen majorly responsible for healthcare-associated infections (the sixth most common nosocomial pathogen) in general and ventilator-associated pneumonia (VAP) (the second most common pathogen), in particular.4,5 The emergence of multidrug-resistant (MDR) strains of Pseudomonas aeruginosa have created havoc in the not-so-distant past.6,7 Several conventional and emerging strategies have been and are being utilized to combat the disastrous situation.8–11 One such strategy is to inhibit a phenomenon named quorum sensing in these deadly pathogens.12Quorum sensing is a bacterial cell-to-cell communication mechanism mediated by a set of chemicals known as autoinducers.13 At the core of quorum sensing process is the production, release, accumulation and population-wide detection of the autoinducers. The Gram-negative bacteria use small-molecules while Gram-positive bacteria use oligopeptides as autoinducers. These molecules accumulate in the environment as the bacterial population density increases, and this information is monitored by the bacteria to track variations in their cell numbers and ultimately to control the gene expression. Such coordinated activities brought about via quorum sensing include bioluminescence, secondary metabolites such as antibiotics production, biofilm formation, and virulence factor secretion, to name a few.14,15
In Pseudomonas aeruginosa, there are two circuits as integral parts of quorum sensing mechanism which are LasI/R and RhlI/R (Fig. 1), resembling LuxI/R system in Vibrio fischeri.16 The LasI/R and RhlI/R produce small-molecule autoinducers, N-3-oxo-dodecanoyl homoserine lactone (3OC12-HSL) and C4-homoserine lactone (C4-HSL), respectively.17,18 These molecules bind to their cognate receptors forming the autoinducer–receptor complex, which ultimately regulates various downstream molecules/processes including virulence factors like LasA protease, LasB elastase, pyocyanin (PCN), swarming and twitching activity and biofilm formation. Given the importance of quorum sensing in the bacterial virulence and biofilm formation, its inhibition has emerged as an important strategy to tackle the menace of MDR and pan-drug-resistant (PDR) bacterial pathogens.19,20 The concept is very simple. By interfering with their virulence factors and not killing them via inhibition of quorum sensing, we are in a way slowing down the emergence of MDR and PDR strain. Thanks to the weaker selective pressure exerted on bacteria by these mechanisms!
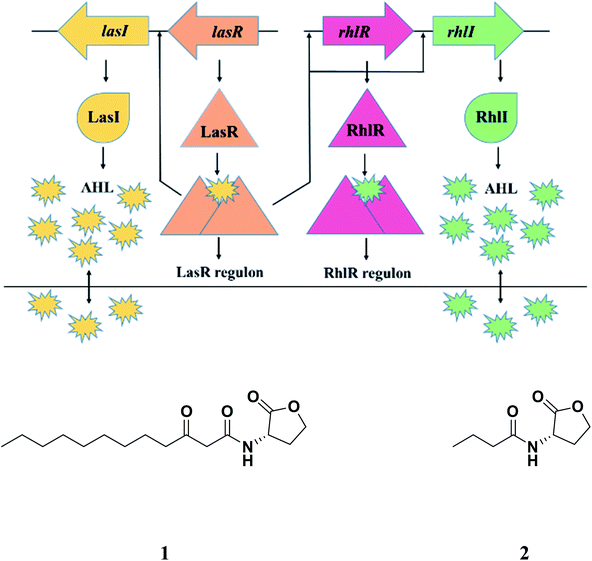 | ||
| Fig. 1 Quorum sensing in Pseudomonas aeruginosa; AHL: acyl homoserine lactone; 1: N-3-oxo-dodecanoyl homoserine lactone (3OC12-HSL); 2: N-butanoyl-L-homoserine lactone (C4-HSL). | ||
Due to the promise offered by quorum sensing inhibition, it is not surprising that a variety of small-molecules (synthetic as well as natural products) with significant antiquorum sensing and antibiofilm activities in Gram-negative and Gram-positive bacteria have been reported.21–24 Literature is full of reports particularly on quorum sensing inhibition in Pseudomonas aeruginosa.25–36
Carvacrol (essential oil) was reported to inhibit biofilm formation in S. aureus and P. aeruginosa and pyocyanin production in P. aeruginosa significantly.37 Baicalein (flavonoid from the roots of Scutellaria baicalensis) was demonstrated to inhibit several virulence factors and biofilm formation.38 Another interesting study revealed that [6]-gingerol, a pungent active from fresh ginger which is structurally analogous to 3OC12-HSL, reduced biofilm formation, virulence factors such as pyocyanin, exoprotease and rhamnolipid and mice mortality.39 The in silico studies confirmed that [6]-gingerol bound specifically to LasR receptor. The QS genes involved in production of virulence factors were specially repressed as seen from the transcriptome analysis.
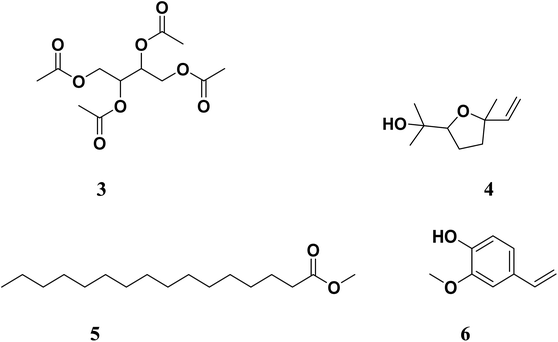
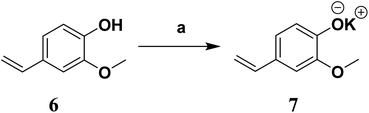
Working on the similar lines, Gala et al. (author's lab) identified active components from Tinospora cordifolia (family: Menispermaceae, common name: Guduchi) following isolation and identification using column chromatography, gas chromatography-mass spectrometry (GC-MS) and in silico molecular studies as potential anti-quorum sensing inhibitors.40 Four molecules, namely 2,3,4-triacetoxybutyl acetate (3), 2-(5-ethenyl-5-methyloxolan-2-yl)propan-2-ol (4), methyl hexadecanoate (5) and 2-methoxy-4-vinylphenol (6) (Fig. 2) were postulated to inhibit QS via either LasI or LasR.41 We were the first to report these molecules as potential QS inhibitors. Interestingly, the partial structural similarity of 5 and 6 with previously reported [6]-gingerol was intriguing, which further strengthened our belief that these leads would provide us newer chemotypes for QS inhibitors. In the present study, we report further profiling of these leads (3–6, Fig. 2) by quorum quenching studies against P. aeruginosa. Although the major issue with compound 6 was its significantly lesser aqueous solubility. We were determined to improve its aqueous solubility by suitable structural and/or chemical modification. Herein, we describe the complete profiling of modified compound 6 as a potential QS inhibitor in a battery of assays validating the original observations and the hypothesis.
2. Material and methods
2.1. General
All the chemicals such as 2-(5-ethenyl-5-methyloxolan-2-yl)propan-2-ol (4, Fig. 2), erythritol, 2-methoxy-4-vinyl phenol and elastin congo red (ECR), propidium iodide, solvents and reagents were purchased from the approved vendors such as Acros Organics (Geel, Belgium), Alfa Aesar (Karlsruhe, Germany), Spectrochem (Mumbai, India), Sigma-Aldrich (Steinheim, Germany, St. Louis, MO, USA) or Merck (Darmstadt, Germany) and used without further purification unless otherwise indicated. SYTO-9 dye was purchased from Thermo Fisher, Waltham, MA. Reactions were carried out under dry N2 atmosphere and thin-layer chromatography (TLC), wherever required, was performed using an aluminum plate coated with silica gel 60 F254 (Merck Millipore, Billerica, MA, USA). 2-Methoxy-4-vinyl phenol was purchased from Alfa Aesar by Thermo Fisher Scientific (Tewksbury, MA). Melting points were recorded using conventional Thiele tube and are uncorrected. The NMR spectra were recorded in either in CDCl3 or DMSO-d6 with tetramethylsilane (TMS) as internal standard. FT-IR spectra were recorded on PerkinElmer RX1 instrument (Waltham, MA). 1H-NMR spectra were obtained on Bruker Advance 400 (400 MHz) spectrometer. Mass spectra (MS) were recorded either on a Shimadzu 8040 LC-MS/MS system (Japan), or Agilent Infinity 1290 series (Agilent Technologies, USA) instrument coupled with quadrupole time of flight (iFunnel Q-TOF LC-MS 6550) and equipped with electrospray ionization (ESI) mode.Pseudomonas aeruginosa PAO1 strain was used as a standard strain for evaluation of all antiquorum, antibiofilm and antivirulence activity studies. Staphylococcus aureus 737 was used for LasA protease study. Chromobacterium violaceum ATCC 12472 and Escherichia coli MG4/pKDT17 were used as a reporter strain in variety of assays. Luria Bertani (LB) broth and agar powder were purchased from Himedia Laboratories, Mumbai, India. o-Nitrophenyl-4-D-galactopyranoside (ONPG) was purchased from Sisco Research Laboratories (SRL), Mumbai, India. Rest of the chemicals were purchased from Qualigens Fine Chemicals, Mumbai, India. E. coli MG4/pKDT17 was grown in LB medium containing 100 μg ampicillin. All other bacterial strains were incubated overnight at 37 °C, wherever required.
2.2. Synthesis of test compounds
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching), 1242, 1220 (C–O–C stretching).
O stretching), 1242, 1220 (C–O–C stretching).![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching), 1171, 1198 (C–O–C stretching).
O stretching), 1171, 1198 (C–O–C stretching).![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C stretching), 1124 (C–O–C stretch).
C stretching), 1124 (C–O–C stretch).2.3. Anti-quorum sensing activity
The test compounds, dissolved in 50% DMSO at specified concentration, i.e., 4 mg mL−1, were screened for short acyl-HSL and long acyl-HSL inhibition according to the protocols given below. The corresponding concentrations of the test compounds were 13.7 mM (3), 14.7 mM (4), 23.4 mM (5) and 21.4 mM (7).![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) EtOH 1
EtOH 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 as solvent) was used as positive control and PBS
1 as solvent) was used as positive control and PBS![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) EtOH (1
EtOH (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
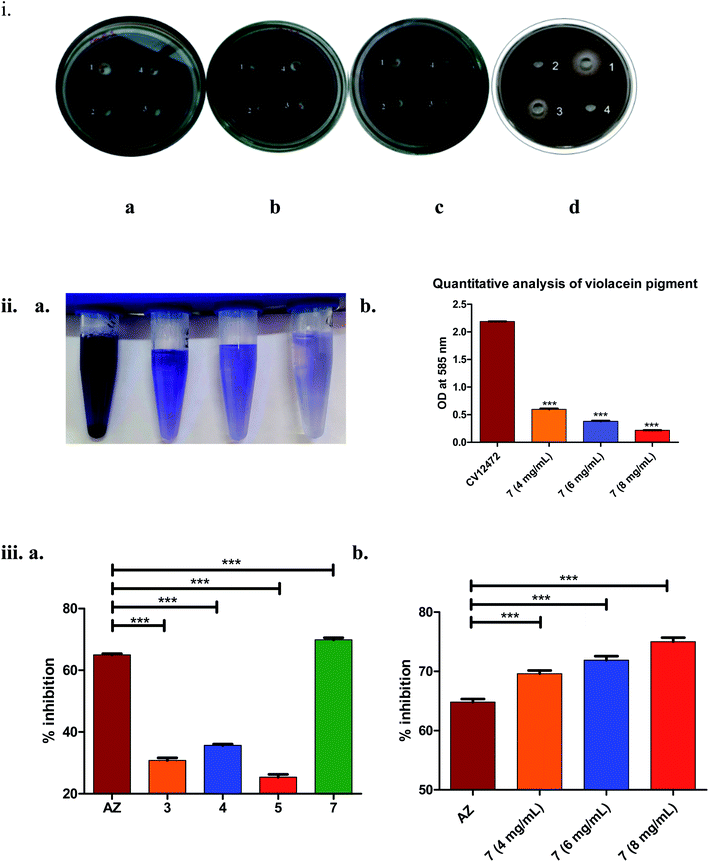
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) as its negative control. Production of pigment inhibition was checked after 24 h incubation.40 To support this assay quantitative analysis of violacein pigment inhibition was done. Growth curve analysis was done to confirm that compound 7 possess anti-quorum sensing activity not anti-bacterial. Bacterial growth curve was observed by measuring cell density at 600 nm at various time point until it reached to stationary phase.
1) as its negative control. Production of pigment inhibition was checked after 24 h incubation.40 To support this assay quantitative analysis of violacein pigment inhibition was done. Growth curve analysis was done to confirm that compound 7 possess anti-quorum sensing activity not anti-bacterial. Bacterial growth curve was observed by measuring cell density at 600 nm at various time point until it reached to stationary phase.2.4. Effect of the compound on P. aeruginosa biofilm formation as analyzed by crystal violet assay
Biofilm formation was checked using 96-well flat-bottom microtiter plate, as per the modified method reported by Mathur et al. (2006).43 Overnight culture of P. aeruginosa was diluted with fresh LB broth supplemented with 0.5% glucose, up to 0.2 OD600. Then control/test compound was added. The positive control, azithromycin dihydrate, was dissolved in PBS![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) EtOH (1
EtOH (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), pH 6.0, and 50% DMSO/PBS
1), pH 6.0, and 50% DMSO/PBS![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) EtOH (1
EtOH (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) was used as negative control. Microtiter wells were filled with 200 μL aliquots of the diluted cultures as described above, uninoculated broth and uninoculated broth containing the compound. The plates were incubated for 24 h, at 37 °C. The wells were aspirated, washed with deionized water to remove the planktonic bacteria, and dried. The biofilm, formed in microtiter plate wells, was fixed with 200 μL MeOH for 15 min. Then the solvent was removed and the plates were dried. Next, the biofilm was stained using 200 μL of 1% crystal violet dye for 15 min. Excess stain was removed and the plate washed with tap water and dried. The bound dye was resolubilized in 95% EtOH (180 μL) for 5 min and the absorbance at 490 nm recorded using BioTek Microplate reader Epoch™ 2.40
1) was used as negative control. Microtiter wells were filled with 200 μL aliquots of the diluted cultures as described above, uninoculated broth and uninoculated broth containing the compound. The plates were incubated for 24 h, at 37 °C. The wells were aspirated, washed with deionized water to remove the planktonic bacteria, and dried. The biofilm, formed in microtiter plate wells, was fixed with 200 μL MeOH for 15 min. Then the solvent was removed and the plates were dried. Next, the biofilm was stained using 200 μL of 1% crystal violet dye for 15 min. Excess stain was removed and the plate washed with tap water and dried. The bound dye was resolubilized in 95% EtOH (180 μL) for 5 min and the absorbance at 490 nm recorded using BioTek Microplate reader Epoch™ 2.40
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 concentration. The mixture of dyes (5 μL) was added onto the slide and coverslip was fixed with transparent nail paint. Samples were observed under Spinning disc confocal laser microscope (CLSM, Zeiss LSM 800, Carl Zeiss, Jena) at 40X.
1 concentration. The mixture of dyes (5 μL) was added onto the slide and coverslip was fixed with transparent nail paint. Samples were observed under Spinning disc confocal laser microscope (CLSM, Zeiss LSM 800, Carl Zeiss, Jena) at 40X.2.5. Anti-virulence assays
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) EtOH (1
EtOH (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1)]. The mixtures were poured onto plates and allowed to solidify. P. aeruginosa (10 μL) was point inoculated in the middle of the plate, which were then incubated at 37 °C for 48 h. The degree of swarming was determined by measuring the diameter of the swarm and compared with control.44
1)]. The mixtures were poured onto plates and allowed to solidify. P. aeruginosa (10 μL) was point inoculated in the middle of the plate, which were then incubated at 37 °C for 48 h. The degree of swarming was determined by measuring the diameter of the swarm and compared with control.44![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) EtOH 1
EtOH 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1)/50% DMSO/PBS
1)/50% DMSO/PBS![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) EtOH (1
EtOH (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) was added to the diluted culture (4.9 mL) and further incubated at 37 °C for 24 h. The culture was centrifuged at 10
1) was added to the diluted culture (4.9 mL) and further incubated at 37 °C for 24 h. The culture was centrifuged at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm under cold (4 °C) condition for 15 min. The culture supernatant (0.5 mL) was mixed with 1.5 mL of 15 min boiled suspension of Staphylococcus aureus in 0.02 M Tris–HCl buffer (pH 8.5). The OD600 was measured at 0 and 3 h with UV-Vis spectrophotometer (PerkinElmer Lambda 25) post incubation at 37 °C.45,46
000 rpm under cold (4 °C) condition for 15 min. The culture supernatant (0.5 mL) was mixed with 1.5 mL of 15 min boiled suspension of Staphylococcus aureus in 0.02 M Tris–HCl buffer (pH 8.5). The OD600 was measured at 0 and 3 h with UV-Vis spectrophotometer (PerkinElmer Lambda 25) post incubation at 37 °C.45,46![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) EtOH 1
EtOH 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1)/50% DMSO/PBS
1)/50% DMSO/PBS![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) EtOH (1
EtOH (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) was added to the diluted culture (4.9 mL) and incubated at 37 °C for 24 h. The culture was centrifuged at 10
1) was added to the diluted culture (4.9 mL) and incubated at 37 °C for 24 h. The culture was centrifuged at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm (4 °C). Buffer (0.1 mL, 0.1 M Tris, 1 mM CaCl2, and pH 7.5) containing ECR substrate (10 mg) was mixed with culture supernatant (0.5 mL), incubated at 37 °C for 24 h. The tubes were centrifuged at 10
000 rpm (4 °C). Buffer (0.1 mL, 0.1 M Tris, 1 mM CaCl2, and pH 7.5) containing ECR substrate (10 mg) was mixed with culture supernatant (0.5 mL), incubated at 37 °C for 24 h. The tubes were centrifuged at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm (4 °C) for 10 min, and OD495 of the supernatant was measured.47
000 rpm (4 °C) for 10 min, and OD495 of the supernatant was measured.47![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) EtOH 1
EtOH 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1)/50% DMSO/PBS
1)/50% DMSO/PBS![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) EtOH (1
EtOH (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) were added to the diluted culture (4.9 mL) and incubated for 48 h at 37 °C. The culture was centrifuged at 10
1) were added to the diluted culture (4.9 mL) and incubated for 48 h at 37 °C. The culture was centrifuged at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm (4 °C) for 15 min. To the culture supernatant (5 mL), CHCl3 (3 mL) was added, vortexed to extract blue pyocyanin in CHCl3, followed by absorbance measurement at 690 nm. Pyocyanin concentration was calculated as mg mL−1 using the formula, OD690/16.40,48
000 rpm (4 °C) for 15 min. To the culture supernatant (5 mL), CHCl3 (3 mL) was added, vortexed to extract blue pyocyanin in CHCl3, followed by absorbance measurement at 690 nm. Pyocyanin concentration was calculated as mg mL−1 using the formula, OD690/16.40,48![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) EtOH 1
EtOH 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) (positive control)/negative control (50% DMSO/PBS
1) (positive control)/negative control (50% DMSO/PBS![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) EtOH (1
EtOH (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1)) was mixed with LB agar. The mixtures were poured into plates and allowed to solidify. The Pseudomonas aeruginosa culture (PAO1) was stabbed to bottom in the middle of the plate. Plates were then incubated at 37 °C for 24 h. The extent of twitching was determined by measuring the diameter of growth zone (in mm), followed by comparison with the control.49
1)) was mixed with LB agar. The mixtures were poured into plates and allowed to solidify. The Pseudomonas aeruginosa culture (PAO1) was stabbed to bottom in the middle of the plate. Plates were then incubated at 37 °C for 24 h. The extent of twitching was determined by measuring the diameter of growth zone (in mm), followed by comparison with the control.492.6. Gene regulation
Overnight culture of P. aeruginosa PAO1 (OD600: 0.2) was inoculated with LB medium supplemented with different concentration of compound 7 (2 mg mL−1 to 8 mg mL−1) and incubated at 37 °C. Total RNA was extracted using TriZol and concentration and purity were determined by UV absorption (260/280 nm) using Take 3 plate in Epoc2 (BioTek). cDNA was synthesized from purified mRNA sample using PrimeScript reagent kit (TaKaRa). Real Time PCR was carried out using SYBR green master mix (Thermo scientific, USA) with StepOne Applied Biosystems (California, USA). The reaction procedure as follows: 95 °C for 30 s, 40 cycles of 95 °C for 5 s, and 60 °C for 5 s, and a final melting curve analysis from 65 °C to 95 °C, with increments of 0.5 °C every 5 s. Real-time PCR amplifications were conducted in triplicate. Primer sequences (Sigma-Aldrich, USA) for P. aeruginosa QS genes were used as described in Table S1.† 50 The ribosomal gene rpsL was chosen as a housekeeping gene to normalize the qRT-PCR data and to calculate the relative fold changes in gene expression. The fold change of target genes for each group with respect to the control group was calculated using the ΔΔCt method.3. Results and discussion
In our previous report, we demonstrated the anti-QS and anti-biofilm activities of the cold EtOAc extract of Tinospora cordifolia stem along with probable chemical constituents of the extract.40 We selected four of these constituents (3–6, Fig. 2) based the molecular docking studies. The compounds were either synthesized (3 or 5) or procured from the vendors (4 and 6). While working with compound 6, we observed solubility issues (precipitation) at the concentrations tested. To circumvent these issues, the potassium salt of 6 (7, Scheme 1) was subsequently prepared, thoroughly characterized and then used in further studies.3.1. Evaluation of anti-quorum sensing activity
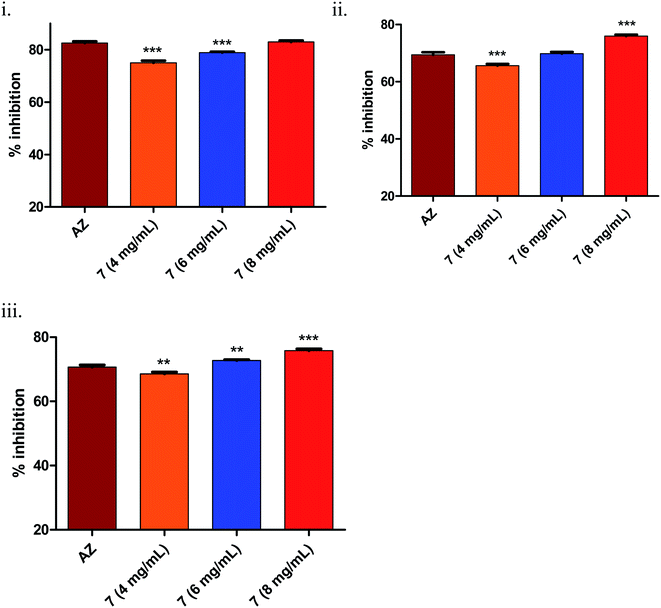
As seen from Fig. 3(iii)(a), it was clearly evident that compound 7, compared to 3–5 and the positive control AZ (65 ± 0.43%), showed remarkable inhibition (70 ± 0.62%) of long-chain AHL at the concentration tested, i.e., 4 mg mL−1. The observed activity, i.e., inhibition of long-chain AHL in P. aeruginosa, was indicative of the compound effect on the las QS circuitry. Further, the dose–response studies of compound 7 at 4, 6 and 8 mg mL−1 were carried out. The results precisely indicated the dose-dependent increase in % inhibition as 69 ± 0.52% (4 mg mL−1), 72 ± 0.68% (6 mg mL−1) and 75 ± 0.69% (8 mg mL−1). Overall, the results were encouraging.
3.2. Evaluation of anti-biofilm forming activity
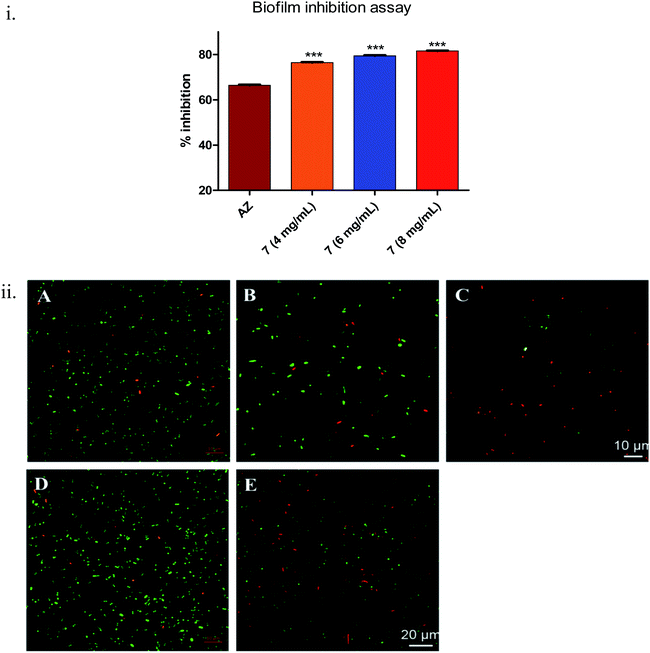
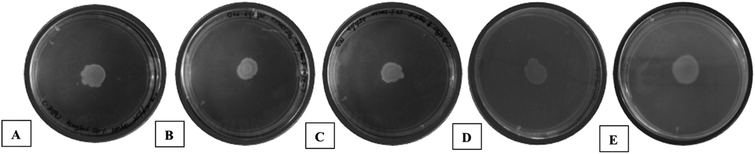
During biofilm formation, bacteria are covered in a matrix of extracellular polymeric substances (EPS) that hold microbial cells together onto a surface. Biofilm formation cycle of P. aeruginosa PAO1 can be divided into five major phenotypic steps. The progression commences by the reversible adhesion of planktonic bacteria onto the surface suitable for growth (Stage I), followed by irreversible attachment of bacteria, which thereafter form microcolonies in the EPS matrix (Stage II). Gradually, these microcolonies expand and their confluence leads to a more structured phenotype with noncolonized space (Stage III). Further, the noncolonized spaces are filled with bacteria, which finally cover the entire surface (Stage IV). Finally, the bacteria disperse from the sessile structure and re-enter in the planktonic state to spread and colonize other surfaces (Stage V).52Compound 7 demonstrated statistically significant (p < 0.001) dose-dependent increase in the anti-biofilm forming activity (as assessed by crystal violet staining) in 24 h treated culture of Pseudomonas aeruginosa PAO1 (Fig. 4(i)) compared to the positive control, AZ. The corresponding values were AZ (66 ± 0.3%), 7 – 4 mg mL−1 (76 ± 0.24%), 6 mg mL−1 (79 ± 0.28%) and 8 mg mL−1 (82 ± 0.2%). The results were in line with earlier anti-QS activity outcome (Fig. 3). The spinning-disc confocal microscopy studies corroborated the results from anti-QS and anti-biofilm forming activity studies (Fig. 4(ii)). As precisely seen in Fig. 4(ii)(A–C), the number of dead cells (red stained) increased while the viable cell (green stained) number decreased in dose-dependent manner compared to the untreated control (Fig. 4(ii)(D)), which was attributed to the increased cell-killing (in the absence of antibacterial activity of 7) due to biofilm inhibition. Azithromycin, at the concentration tested, also showed inhibition of biofilm formation, albeit to lesser extent as assessed qualitatively from Fig. 4(ii)(E). The higher number of red stained (dead) cells signifies a greater number of static cells present onto the surface, which in turn, denote the reduced biofilm formation.
In consonance with our previous report,40 where we postulated the hits (compound 6) would bind to LasR as supported by the molecular docking analysis, it made perfect sense that 7 would inhibit biofilm formation in Pseudomonas aeruginosa by interfering one of the QS circuitries (las, rhl or pqs), preferably las. In summary, these preliminary results with 7 encouraged us to further evaluate it in the anti-virulence assays.
To gain further insights into structural requirements for biofilm inhibition, we studied literature reports detailing the structure–activity relationship (SAR) around 2-methoxyphenols bearing 4-substituent [H (Guaiacol), ethyl (4-ethylguaiacol), allyl (eugenol), 2-methylvinyl (isoeugenol) and propyl (2-methoxy-4-propylphenol)] which can easily be compared with compound 7 (Fig. S2†). The report52 discussed the inhibition of biofilm formation in one of the Gram-negative microorganisms, E. coli O157:H7 by essential oils (containing eugenol-like compounds). It was found that the presence of double bond as part of 4-substituent, clearly imparted significant biofilm inhibitory activity to the studied compounds. The order of activity with respect to biofilm inhibition was – eugenol > isoeugenol > 2-methoxy-4-propylphenol > 4-ethylguaiacol > guaiacol. The hydrophobic substitution at position 4 on the 2-methoxyphenol led to increased activity. Further potentiation of activity was observed when the substituent with unsaturation was used. The study53 provided important clues with respect to structural requirement for biofilm inhibitory activity in Gram-negative microbes. The presence of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C bond at position 4 on the aromatic ring, keeping other substituents constant, made the major impact on biofilm formation. Given the strict structural requirements at position 4, it can be postulated that the 4-substituent played an important role in modulating the biofilm inhibition possibly by interacting with appropriate target(s) in the QS circuitry in the Gram-negative microbes.
C bond at position 4 on the aromatic ring, keeping other substituents constant, made the major impact on biofilm formation. Given the strict structural requirements at position 4, it can be postulated that the 4-substituent played an important role in modulating the biofilm inhibition possibly by interacting with appropriate target(s) in the QS circuitry in the Gram-negative microbes.
3.3. Anti-virulence assays
Hentzer et al. synthesized halogenated furanones (produced by Australian red macro-alga Delisea pulchra) and checked anti-quorum sensing and anti-virulence activity in P. aeruginosa. As a result of that study, furanones showed inhibition in quorum sensing and virulence factors like LasA protease and elastase.55
3.4. Gene regulation
Studies on down regulation of LasIR and RhlIR circuitry have been done using real time PCR. Phenotypic assays showed inhibition of biofilm and virulence factors in P. aeruginosa. To confirm the same at genetic level, real time PCR was performed and observed significant down regulation (in terms of fold change) of all four genes viz lasI, rhlI, lasR and rhlR as concentration of compound 7 increase (2 mg mL−1, 4 mg mL−1, 6 mg mL−1 and 8 mg mL−1) in the presence of housekeeping gene (rpsL) (Fig. S3†). Result confirms the inhibition of both circuitry LasIR and RhlIR in P. aeruginosa PAO1. In fact, down regulation of these genes confirms the inhibition of biofilm and other virulence factors. The lasI–lasR and rhlI–rhlR quorum sensing systems regulate expression of various virulence genes and play an important role in biofilm formation. Luo et al. reported that baicalin (5,6,7-trihydroxyflavone, flavonoid monomer purified from Scutellaria baicalensis) significantly repressed all four gene in Pseudomonas aeruginosa strain.59 Similar study was conducted by Hossain et al. where methyl gallate (phenolic compound) significantly down regulated all four genes.604. Conclusion
Inhibition of QS in Pseudomonas aeruginosa remains an elusive target for tackling the menace of its MDR strains and the subsequent complications. Continuing on the same path, in the present study, our group extensively investigated the potential of previously identified small-molecule QS inhibitors (from Tinospora cordifolia stem EtOAc extract) in the QS, biofilm-formation, and virulence inhibition assays. Of the initial four hits, compound 7 was a clear winner. It exhibited significant anti-QS, anti-biofilm forming and anti-virulence activities in relevant assays in dose-dependent manner. Compound 7 also down regulated all four genes responsible for quorum sensing in P. aeruginosa in dose dependent manner. The activities were comparable or even better than the positive control AZ at the doses tested. Given the tiny structure of 7, there is so much scope for further structural modifications and functional studies centered on it. These results were in line with other reports with respect to the structural requirements for biofilm inhibitory activity by 4-substituted 2-methoxyphenols. The discovery of hit 7 is just the beginning of the story. The results reported in the present study are likely to motivate researchers in the field to further explore it.Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.Conflicts of interest
We do not have any conflicts of interest to declare.Acknowledgements
The authors deeply acknowledge Sophisticated Analytical Instrument Facility (SAIF), Indian Institute of Technology Bombay (IIT-B), Mumbai, India for confocal analysis. MS thanks SVKM's NMIMS (Deemed to be University), Mumbai, India for providing the necessary facilities. We also thanks Mr Mitesh Joshi, SVKM's NMIMS, Sunandan Divatia School of Science for his help with the artwork.References
- A. G. Tedros, Prioritization of pathogens to guide discovery, research and development of new antibiotics for drug-resistant bacterial infections, including tuberculosis, World Health Organisation, Geneva, December, 2017 Search PubMed.
- Antimicrobial resistance, World Health Organization, 1998, 1–6 Search PubMed.
- A. Y. Bhagirath, Y. Li, R. Patidar, K. Yerex, X. Ma, A. Kumar and K. Duan, Int. J. Mol. Sci., 2019, 20, 1781 CrossRef.
- D. M. Sievert, P. Ricks, A. J. Kallen, J. R. Edwards, A. Schneider, J. Patel, A. Srinivasan, B. Limbago and S. Fridkin, Infect. Control Hosp. Epidemiol., 2013, 14, 1–14 CrossRef PubMed.
- N. Lynn, G. Joshua, G. Katherine and M. Conan, ContagionLive, Infectious Diseases Today, 2018, 20, 1–6 Search PubMed.
- S. B. Vickery, D. McClain and K. A. Wargo, Pharmacotherapy, 2016, 36, e154–e159 CrossRef CAS PubMed.
- I. Agnese, M. Haenni and J. Y. Madec, Microbiol. Spectrum, 2018, 6, 1–16 Search PubMed.
- M. S. Mulani, E. E. Kamble, S. N. Kumkar, M. S. Tawre and K. R. Pardesi, Front. Microbiol., 2019, 10, 1–12 CrossRef PubMed.
- T. L. Tagliaferri, M. Jansen and H.-P. Horz, Front. Cell. Infect. Microbiol., 2019, 9, 1–13 CrossRef PubMed.
- G. G. Zhanel, A. R. Golden, S. Zelenitsky, K. Wiebe, C. K. Lawrence, H. J. Adam, T. Idowu, R. Domalaon, F. Schweizer, M. A. Zhanel, P. R. S. Lagacé-Wiens, A. J. Walkty, A. Noreddin, J. P. Lynch and J. A. Karlowsky, Drugs, 2019, 79, 271–289 CrossRef CAS PubMed.
- D. R. Giacobbe, M. Mikulska and C. Viscoli, Expert Rev. Clin. Pharmacol., 2018, 11, 1219–1236 CrossRef CAS PubMed.
- M. Shaaban, A. Elgaml and E. S. E. Habib, Microb. Pathog., 2018, 127, 138–143 CrossRef PubMed.
- S. Mukherjee and B. L. Bassler, Nat. Rev. Microbiol., 2019, 7(6), 371–382 CrossRef PubMed.
- W.-L. Ng and B. L. Bassler, Annu. Rev. Genet., 2009, 43, 197–222 CrossRef CAS PubMed.
- S. T. Rutherford and B. L. Bassler, Cold Spring Harbor Perspect. Med., 2012, 2, 1–25 Search PubMed.
- M. J. Gambello and B. H. Iglewski, J. Bacteriol., 1991, 173, 3000–3009 CrossRef CAS PubMed.
- J. P. Pearson, K. M. Gray, L. Passador, K. D. Tucker, A. Eberhard, B. H. Iglewski and E. P. Greenberg, Proc. Natl. Acad. Sci. U. S. A., 1994, 91, 197–201 CrossRef CAS PubMed.
- J. P. Pearson, L. Passador, B. H. Iglewski and E. P. Greenberg, Proc. Natl. Acad. Sci. U. S. A., 1995, 92, 1490–1494 CrossRef CAS PubMed.
- O. Fleitas Martínez, P. O. Rigueiras, Á. da S. Pires, W. F. Porto, O. N. Silva, C. de la Fuente-Nunez and O. L. Franco, Front. Cell. Infect. Microbiol., 2018, 8, 1–17 CrossRef PubMed.
- S. Haque, D. K. Yadav, S. C. Bisht, N. Yadav, V. Singh, K. K. Dubey, A. Jawed, M. Wahid and S. A. Dar, J. Chemother., 2019, 1–27 CAS.
- N. D. Paguigan, J. Rivera-Chávez, J. J. Stempin, M. Augustinović, A. I. Noras, H. A. Raja, D. A. Todd, K. D. Triplett, C. Day, M. Figueroa, P. R. Hall, N. B. Cech and N. H. Oberlies, J. Nat. Prod., 2019, 82, 550–558 CrossRef CAS PubMed.
- J. Chen, B. Wang, Y. Lu, Y. Guo, J. Sun, B. Wei, H. Zhang and H. Wang, Mar. Drugs, 2019, 17, 80 CrossRef CAS PubMed.
- X. Chen, L. Zhang, M. Zhang, H. Liu, P. Lu and K. Lin, Expert Opin. Ther. Pat., 2018, 28, 849–865 CrossRef CAS PubMed.
- S. Manner and A. Fallarero, Int. J. Mol. Sci., 2018, 19(5), 1346 CrossRef PubMed.
- S. A. Ahmed, M. Rudden, T. J. Smyth, J. S. G. Dooley, R. Marchant and I. M. Banat, Appl. Microbiol. Biotechnol., 2019, 103, 3521–3535 CrossRef CAS PubMed.
- J. Fong, K. T. Mortensen, A. Nørskov, K. Qvortrup, L. Yang, C. H. Tan, T. E. Nielsen and M. Givskov, Front. Cell. Infect. Microbiol., 2019, 8, 1–11 Search PubMed.
- B. Almohaywi, T. T. Yu, G. Iskander, D. S. H. Chan, K. K. K. Ho, S. Rice, D. S. C. Black, R. Griffith and N. Kumar, Bioorg. Med. Chem. Lett., 2019, 29(9), 1054–1059 CrossRef CAS PubMed.
- J. E. Paczkowski, A. R. McCready, J. P. Cong, Z. Li, P. D. Jeffrey, C. D. Smith, B. R. Henke, F. M. Hughson and B. L. Bassler, ACS Chem. Biol., 2019, 14, 378–389 CrossRef CAS PubMed.
- M. Kalia, V. K. Yadav, P. K. Singh, S. Dohare, D. Sharma, S. S. Narvi and V. Agarwal, 3 Biotech, 2019, 9(2), 40 CrossRef PubMed.
- M. N. Qiu, F. Wang, S. Y. Chen, P. C. Wang, Y. H. Fu, Y. Y. Liu, X. Wang, F. B. Wang, C. Wang, H. W. Yang, Y. Wu, S. Y. Zhu, H. B. Zhou, W. M. Chen, J. Lin, J. X. Zheng and P. H. Sun, Bioorg. Med. Chem. Lett., 2019, 29, 749–754 CrossRef CAS PubMed.
- M. Goswami, A. Espinasse and E. E. Carlson, Chem. Sci., 2018, 9, 7332–7337 RSC.
- M. M. Saleh, H. A. Abbas and M. M. Askoura, Microb. Pathog., 2019, 127, 31–38 CrossRef CAS PubMed.
- Y. Bin Li, J. Liu, Z. X. Huang, J. H. Yu, X. F. Xu, P. H. Sun, J. Lin and W. M. Chen, Eur. J. Med. Chem., 2018, 158, 753–766 CrossRef PubMed.
- F. D'Angelo, V. Baldelli, N. Halliday, P. Pantalone, F. Polticelli, E. Fiscarelli, P. Williams, P. Visca, L. Leoni and G. Rampioni, Antimicrob. Agents Chemother., 2018, 62(11), e01296-18 CrossRef PubMed.
- F. Soukarieh, P. Williams, M. J. Stocks and M. Cámara, J. Med. Chem., 2018, 61, 10385–10402 CrossRef CAS.
- B. Almohaywi, A. Taunk, D. S. Wenholz, S. Nizalapur, N. N. Biswas, K. K. K. Ho, S. A. Rice, G. Iskander, D. S. C. Black, R. Griffith and N. Kumar, Molecules, 2018, 23(5), 1106 CrossRef.
- M. R. Tapia-Rodriguez, A. Hernandez-Mendoza, G. A. Gonzalez-Aguilar, M. A. Martinez-Tellez, C. M. Martins and J. F. Ayala-Zavala, Food Control, 2017, 75, 255–261 CrossRef CAS.
- J. Luo, J. L. Kong, B. Y. Dong, H. Huang, K. Wang, L. H. Wu, C. C. Hou, Y. Liang, B. Li and Y. Q. Chen, Drug Des., Dev. Ther., 2016, 10, 183–203 CrossRef CAS PubMed.
- H. S. Kim, S. H. Lee, Y. Byun and H. D. Park, Sci. Rep., 2015, 5, 8656 CrossRef CAS PubMed.
- V. Gala, N. John, A. Bhagwat, A. Datar, P. Kharkar and K. Desai, Indian J. Med. Res., 2016, 144, 92 CrossRef PubMed.
- B. N. Singh, B. R. Singh, R. L. Singh, D. Prakash, B. K. Sarma and H. B. Singh, Food Chem. Toxicol., 2009, 47, 778–786 CrossRef CAS PubMed.
- J. P. Kirwan, T. A. Gould, H. P. Schweizer, S. W. Bearden, R. C. Murphy and M. E. A. Churchill, Society, 2006, 188, 784–788 CAS.
- T. Mathur, S. Singhal, S. Khan, D. Upadhyay, T. Fatma and A. Rattan, Indian J. Med. Microbiol., 2006, 24, 25–29 CrossRef CAS PubMed.
- M. S. a Khan, M. Zahin, S. Hasan, F. M. Husain and I. Ahmad, Lett. Appl. Microbiol., 2009, 49, 354–360 CrossRef PubMed.
- E. Kessler, M. Safrins, J. C. Olsonll and D. E. Ohmanll, J. Biol. Chem., 1993, 268, 7503–7508 Search PubMed.
- N. R. John, V. C. Gala and C. S. Sawant, Int. J. Pharma Bio Sci., 2013, 4, 487–495 Search PubMed.
- J. P. Pearson, E. C. Pesci and B. H. Iglewski, J. Bacteriol., 1997, 179(18), 5756–5767 CrossRef CAS PubMed.
- V. Huerta, K. Mihalik, S. H. Crixell and D. a. Vattem, Int. J. Appl. Res. Nat. Prod., 2008, 1, 9–15 Search PubMed.
- S. Datta, D. Jana, T. R. Maity, A. Samanta and R. Banerjee, 3 Biotech, 2016, 6, 1–6 CrossRef PubMed.
- H. Li, X. Li, Z. Wang, Y. Fu, Q. Ai, Y. Dong and J. Yu, BMC Microbiol., 2015, 15, 1–8 CrossRef PubMed.
- H. B. Liu, J. H. Lee, J. S. Kim and S. Park, Biotechnol. Bioeng., 2010, 106, 119–126 CAS.
- T. Rasamiravaka, Q. Labtani, P. Duez and M. El Jaziri, BioMed Res. Int., 2015, 2015, 1–17 CrossRef PubMed.
- Y. G. Kim, J. H. Lee, G. Gwon, S. Il Kim, J. G. Park and J. Lee, Sci. Rep., 2016, 6, 36377 CrossRef CAS PubMed.
- H. Oura, Y. Tashiro, M. Toyofuku, K. Ueda, T. Kiyokawa, S. Ito, Y. Takahashi, S. Lee, H. Nojiri, T. Nakajima-Kambe, H. Uchiyama, H. Futamata and N. Nomura, Appl. Environ. Microbiol., 2015, 81, 2808–2818 CrossRef CAS PubMed.
- M. Hentzer, H. Wu, J. B. Andersen, K. Riedel, T. B. Rasmussen, N. Bagge, N. Kumar, M. a. Schembri, Z. Song, P. Kristoffersen, M. Manefield, J. W. Costerton, S. Molin, L. Eberl, P. Steinberg, S. Kjelleberg, N. Høiby and M. Givskov, EMBO J., 2003, 22, 3803–3815 CrossRef CAS PubMed.
- C. T. O. Loughlin, L. C. Miller, A. Siryaporn, K. Drescher and M. F. Semmelhack, Proc. Natl. Acad. Sci. U. S. A., 2013, 110(44), 17981–17986 CrossRef PubMed.
- G. A. O'Toole and R. Kolter, Mol. Microbiol., 1998, 30, 295–304 CrossRef PubMed.
- A. Glessner, R. S. Smith, B. H. Iglewski and J. B. Robinson, J. Bacteriol., 1999, 181, 1623–1629 CAS.
- J. Luo, B. Dong, K. Wang, S. Cai, T. Liu, X. Cheng, D. Lei, Y. Chen, Y. Li, J. Kong and Y. Chen, PLoS One, 2017, 12, 1–32 Search PubMed.
- M. A. Hossain, S. J. Lee, N. H. Park, A. F. Mechesso, B. T. Birhanu, J. Kang, M. A. Reza, J. W. Suh and S. C. Park, Sci. Rep., 2017, 7, 1–16 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9ra06612h |
| This journal is © The Royal Society of Chemistry 2019 |