Open Access Article
Open Access ArticleNanoparticles in an antibiotic-loaded nanomesh for drug delivery†
Melanie A.
Fuller
abc,
Ashley
Carey
a,
Harriet
Whiley
b,
Rio
Kurimoto
c,
Mitsuhiro
Ebara
 c and
Ingo
Köper
c and
Ingo
Köper
 *a
*a
aInstitute for NanoScale Science and Technology, Flinders University, Bedford Park, South Australia 5042, Australia. E-mail: Ingo.koeper@flinders.edu.au
bCollege of Science and Engineering, Flinders University, Bedford Park, South Australia 5042, Australia
cInternational Center for Materials Nanoarchitectonics, National Institute for Materials Science, Tsukuba, Ibaraki 305-0047, Japan
First published on 24th September 2019
Abstract
Antibiotic loaded nanomeshes were fabricated by electrospinning polycaprolactone, a biocompatible polymer, with 12.5% w/w Colistin, 1.4% w/w Vancomycin and either cationic or anionic gold nanoparticles in varying combinations. The resulting nanomeshes had different antibiotic release profiles, with citrate capped gold nanoparticles combined with Colistin having the highest sustained release over 14 days for a 4 mg, 1.5 cm2 nanomesh. The electrospinning parameters were optimised to ensure the spinning of a homogenous mesh and the addition of antibiotics was confirmed through 1H NMR and ATR-FTIR. This research, as a proof of concept, suggests an opportunity for fabricating nanomeshes which contain gold nanoparticles as a drug release mechanism for antibiotics.
1 Introduction
With the over-prescription of antibiotics worldwide, bacterial resistance is becoming a significant threat to public health.1 When bacteria are resistant to three or more types of antibiotic classes, they are labelled as multi-drug resistant and there is a limited number of antibiotics available to be used in a final attempt to treat the infection.2,3 Two examples of these last line drugs are the polypeptide antibiotics, Colistin and Vancomycin.4 Colistin, which has previously been limited in its use due to the incidence of adverse effects including nephrotoxicity and neurotoxicity, is resurfacing more frequently for treatment.5,6 It is often used to treat multi-resistant Gram-negative bacteria, where other antibiotics are no longer effective. Colistin's mechanism relies on the electrostatic interaction between the negatively charged phosphate groups of the lipopolysaccharide (LPS) bacterial outer cell membrane and the positively charged amino groups of Colistin.7,8 When the positively charged Colistin comes into contact with the bacterial cell membrane, it causes defects in the membrane, leading to leakage of the cell contents and ultimately cell death.9 It also binds and neutralizes the lipid A portion of the LPS, which is the endotoxin of Gram-negative bacteria.8 Often, multiple antibiotics are used in combination to treat resistant bacteria, and Vancomycin is one of the more popular choices to treat Gram-positive Methicillin-Resistant Staphylococcus aureus (MRSA) infections.10,11 Vancomycin's antibacterial action is different from that of Colistin, where it inhibits the cell wall synthesis of susceptible organisms.12 It does this by inhibiting the peptidoglycan synthesis in late stage bacterial cell wall formation.13Delivering the antibiotics directly to the infection site rather than via an oral dosage is beneficial as the oral ingested dosage is distributed not only at the infection site but also non-specifically around the body. The non-specificity means a high dosage needs to be given to ensure the concentration at the site of infection is significant enough to effectively treat it. The dosage can be lowered if delivered to the infection site directly, which can reduce side effects and complications. Although the dosage is ‘reduced’ compared to the oral dosage, the concentration of antibiotics delivered to the infection site can still be higher, ensuring the bacteria cannot survive and cause resistance.
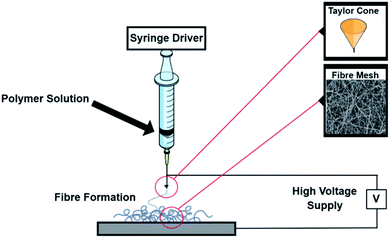
In order to deliver the antibiotics to a specific site, the antibiotics need to be immobilised on a scaffold, and in this case they have been embedded into a fibrous mesh produced using a technique called electrospinning. Electrospinning has gained considerable interest in the biomedical community as it offers promise in many applications,14 including wound management,15 drug delivery16 and antibiotic coatings.17 Other emerging applications for electrospun meshes include air filtration18–22 and oil–water emulsion separation.23,24 In electrospinning, a syringe is loaded with a polymer that has been dissolved in a volatile solvent. A high voltage is then applied between the needle connected to the syringe, and the collector plate (Fig. 1). The voltage causes the polymer solution to form a Taylor cone as it leaves the syringe, at which point the electrostatic forces induce a jet of liquid, as it overcomes the surface tension.25,26
Electrospinning can produce controlled micro or nano-sized fibres that are deposited onto a substrate. The properties of the fibres can be adjusted by varying parameters such as the viscosity of the polymer solution, choice of solvent, the voltage supplied, needle gauge as well as the needle to collector plate distance.27,28
The production process allows for various additives to be included into the fibres, as long as they can be mixed with the original polymer solution. Here, two antibiotics, Colistin (Col) and Vancomycin (Van) were added. Additionally, 5 nm diameter gold nanoparticles with either a negatively charged citrate capping Au(−) or a positively charged polydiallyldimethylammonium chloride (PDADMAC) coating Au(+), were added to determine if small, charged particles within the polymer matrix affect the antibiotic release.
Gold nanoparticles were chosen in addition to the antibiotics as positively charged nanoparticles have shown in literature to cause damage to the bacterial membranes.29,30 In a recent study, a strong correlation was found between poor bacterial viability and the attachment of positively charged gold nanoparticles on Gram-negative bacteria.31 Thus both anionic and cationic gold nanoparticles were included in the mesh to determine if they have any effect when paired with antibiotics within the nanomesh.
2 Materials and methods
2.1 Mesh formation
4, 7, 8, 9 and 10% w/w ε-polycaprolactone (PCL) solutions were prepared by dissolving PCL (average Mn 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000) (Sigma Aldrich, Kaohsiung, Taiwan) in 1,1,1,3,3,3-hexafluoroisopropanol (HFIP) (Sigma Aldrich, Kaohsiung, Taiwan) and left overnight at ∼40 °C. The PCL/HFIP solution was then loaded into a 5 mL syringe with a 22-gauge needle and electrospun with an applied voltage of 20 kV (Nanon-01A, MECC Co. Ltd, Fukuoka, Japan), with a 20 cm working distance and 20 cm horizontal needle movement for 3 h at flow rates of 0.5 mL h−1 and 1 mL h−1. The fibres were spun directly onto aluminium foil on a stationary collector plate. The fibre mesh was removed from the aluminium foil prior to further investigation. After spinning was completed, the mesh was dried in a vacuum to remove any excess HFIP and stored at −20 °C until its use.
000) (Sigma Aldrich, Kaohsiung, Taiwan) in 1,1,1,3,3,3-hexafluoroisopropanol (HFIP) (Sigma Aldrich, Kaohsiung, Taiwan) and left overnight at ∼40 °C. The PCL/HFIP solution was then loaded into a 5 mL syringe with a 22-gauge needle and electrospun with an applied voltage of 20 kV (Nanon-01A, MECC Co. Ltd, Fukuoka, Japan), with a 20 cm working distance and 20 cm horizontal needle movement for 3 h at flow rates of 0.5 mL h−1 and 1 mL h−1. The fibres were spun directly onto aluminium foil on a stationary collector plate. The fibre mesh was removed from the aluminium foil prior to further investigation. After spinning was completed, the mesh was dried in a vacuum to remove any excess HFIP and stored at −20 °C until its use.
2.2 Mesh imaging
A 0.5 cm2 piece of nanomesh was sputter coated with a thin layer of gold before being imaged via SEM (NEO-Scope JCM-5000 table top SEM, JEOL, Japan) at an accelerating voltage of 10 kV. The software ImageJ was used to analyse the diameter of the fibres, where the mean diameter of a minimum 50 fibres was used to determine fibre thickness.2.3 Antibiotic addition to nanomesh
60 mg of Colistin sulphate salt (≥15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 U mg−1) (Sigma Aldrich, Castle Hill, Australia) was dissolved in 1 mL HFIP (for a total 12.5 w/w loading of Colistin) before being vortexed with 7% w/w PCL just prior to electrospinning. All meshes were electrospun with the same spinning parameters as described in the PCL mesh formation, except for the flow rate which was kept constant at 1 mL h−1. For the addition of Vancomycin, 100 μL of 50 mg mL−1 Vancomycin hydrochloride in DMSO (Sigma Aldrich, Tokyo, Japan) was mixed with 0.5 mL HFIP and then vortexed before being added to 7% w/w PCL in HFIP for a total w/w Vancomycin loading of 1.4%. In the case of the meshes containing both antibiotics, the antibiotics were prepared as above and mixed together prior to being added to the PCL solution. For the addition of the negatively charged gold nanoparticles, 500 μL of 5 nm diameter citrate capped gold nanoparticles (1013 particles per mL) (Nanocomposix, San Diego, USA) were added to the drug solution prior to addition to the PCL solution. For the addition of positively charged gold nanoparticles, 5 nm diameter PDADMAC coated gold nanoparticles were fabricated using a previously published method.32 500 μL of the fabricated nanoparticles (1013 particles per mL) were then added as per the citrate capped gold nanoparticle method.
000 U mg−1) (Sigma Aldrich, Castle Hill, Australia) was dissolved in 1 mL HFIP (for a total 12.5 w/w loading of Colistin) before being vortexed with 7% w/w PCL just prior to electrospinning. All meshes were electrospun with the same spinning parameters as described in the PCL mesh formation, except for the flow rate which was kept constant at 1 mL h−1. For the addition of Vancomycin, 100 μL of 50 mg mL−1 Vancomycin hydrochloride in DMSO (Sigma Aldrich, Tokyo, Japan) was mixed with 0.5 mL HFIP and then vortexed before being added to 7% w/w PCL in HFIP for a total w/w Vancomycin loading of 1.4%. In the case of the meshes containing both antibiotics, the antibiotics were prepared as above and mixed together prior to being added to the PCL solution. For the addition of the negatively charged gold nanoparticles, 500 μL of 5 nm diameter citrate capped gold nanoparticles (1013 particles per mL) (Nanocomposix, San Diego, USA) were added to the drug solution prior to addition to the PCL solution. For the addition of positively charged gold nanoparticles, 5 nm diameter PDADMAC coated gold nanoparticles were fabricated using a previously published method.32 500 μL of the fabricated nanoparticles (1013 particles per mL) were then added as per the citrate capped gold nanoparticle method.
2.4 Characterisation of antibiotic addition into the mesh
A 1 cm2 piece of mesh was cut and placed on the crystal of an Attenuated Total Reflection Fourier Transform Infra-Red (ATR-FTIR) spectrophotometer (FTIR-8400S; Shimadzu Co., Ltd, Kyoto, Japan) and was used to confirm Vancomycin was present in the mesh. To confirm the addition of Colistin, experiments were performed using 1H NMR (Bruker 600 MHz Avance III, Australia) where 10 mg of mesh was dissolved in a mix of 95% DMSO-d6 and 5% D2O. A 5 mm BBFO probe was used and excitation sculpting was conducted to remove the D2O peak. The parameters used for the 1H NMR experiments were an acquisition time of 1.36 s, a relaxation delay of 1 s and line broadening of 0.3.2.5 In vitro antibiotic release
Drug release studies were conducted by cutting 1.5 cm2 pieces of mesh, weighing them and placing them in vials with 2 mL Dulbecco's Phosphate Buffered Saline (D-PBS) (Sigma Aldrich, Tokyo, Japan). The mesh pieces were then placed in a water bath with shaking at 37 °C to mimic physiological conditions. At different time points the 2 mL DPBS was removed and replaced under sink conditions. After being removed, the aliquot was immediately frozen and stored at −20 °C until ready to be measured, to prevent loss of antibiotic action. Removal of the aliquot at the various time points was completed in triplicate for all meshes measured. The concentration of the two antibiotics were determined by UV-vis spectrophotometry (JASCO V-650 Spectrophotometer, Japan) at a wavelength of 214 nm for Colistin and 280 nm for Vancomycin using a quartz cuvette. The data was normalised to 4 mg mesh weights for consistency. The cumulative percentage release was calculated using eqn (1).31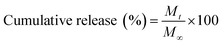 | (1) |
2.6 Broth dilution assay
1 mL of DH5α Escherichia coli (E. coli) bacteria (Invitrogen, Japan) was added to 1 mL of lysogeny broth (LB) medium (Sigma Aldrich, Japan) and incubated for 18 h at 37 °C. After 18 h, 1 mL of the bacteria was added to each of the 2 mL aliquots taken from the in vitro drug release study and then incubated at 37 °C for 24 h. Turbidity was used as an indicator of cell growth in the presence of antibiotics released from in vitro studies. The absorbance was measured using UV-vis Spectrophotometry (JASCO V-650 spectrophotometer, Japan) at 600 nm optical density. Experiments were conducted in triplicate and the average absorbance was determined. Vancomycin mesh release was used as a positive control.2.7 Zone of inhibition (ZOI) assay
E. coli American Type Culture Collection (ATCC) 700891 bacterial lawns were grown on nutrient agar plates (Sigma-Aldrich, Australia) for 24 h at 37 °C. 8 mm diameter disks were cut from various meshes and placed under UV-light to sterilise for 20 minutes. The disks were then transferred onto the agar plates and incubated for 48 h at 37 °C. After 48 h, the diameter of the zone of inhibition was recorded. Each mesh type was tested in triplicate. PCL with no antibiotics was used as a negative control and the Vancomycin only mesh was used as a positive control.3 Results and discussion
3.1 Optimisation of parameters for mesh formation
Meshes were produced with various flow rates and % w/w of PCL, while the working distance, voltage, deposition time and needle size were kept constant. An ideal mesh for drug release would have homogenous fibres with no visible defects.33 In order to develop an ideal mesh, the concentrations of PCL as well as the flow rates were altered and the meshes were then imaged to determine their homogeneity and fibre thickness (Fig. 3). 4% w/w PCL led to significant beading defects regardless of the flow rate. Beading leads to inhomogeneous fibres and can result from two different processes. The low viscosity of the solution can cause the solution to fall through the syringe too quickly, leading to a droplet. If the droplet falls, a new Taylor cone needs to be formed which leads to distinct and separated beads. The second reason for beading being observed is when the polymer solution is too viscous, as the solution will take too long to fall through the syringe which causes breaks in the electrospinning.34 Again, a new Taylor cone will need to be formed causing beading defects. The 1 mL h−1 was observed to have a greater number of defects and this may be due to the faster flow rate, where the low viscosity means the polymer solution is falling through as well as being pushed faster than the 0.5 mL h−1. To remove the beading, a higher polymer weight was used.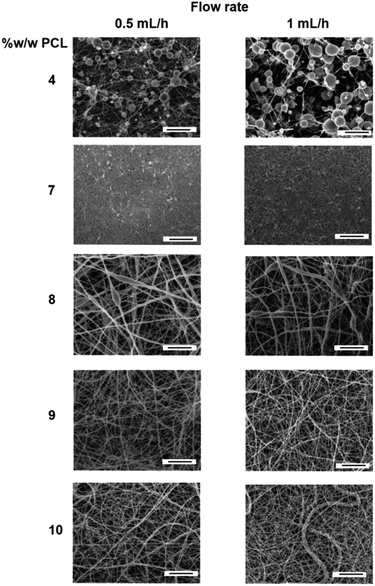 | ||
| Fig. 3 SEM images of different %w/w of PCL in HFIP showing the changes in morphology at a flow rate of 0.5 mL h−1 and 1 mL h−1. All scale bars are 10 μm. | ||
Polymer percentages of 7, 8, 9 and 10% w/w resulted in meshes with various fibre diameters. For 7% w/w PCL, the fibres had a diameter of 246 ± 76 nm with homogenous fibre diameters and no apparent beading defects. The 8% and 9% w/w PCL had no observable difference between the flow rates, however minor defects and various fibre diameters can be observed. These defects are different from the beading in the 4% w/w PCL as they are elongated within the fibre, causing differences in thicknesses along the length of the fibre. This is likely due to the viscosity being too high, leading to elongated beading within the fibre itself. Finally, the 10% w/w PCL mesh showed a variety of fibre diameters, which is not ideal in a drug delivery application (Table S1†). Thus for the electrospinning conditions used, the optimal polymeric solution concentration was determined to be 7% w/w as it produced the thinnest, most homogenous fibres compared to the other percentages tested. 7% w/w PCL was used in the formation of all the meshes to assess the addition of antibiotics and nanoparticles for drug delivery.
3.2 Addition of antibiotics into the nanomesh
Confirmation of the addition of Colistin and Vancomycin was achieved through a series of characterisation techniques. For NMR, peaks corresponding to the introduction of Colistin occurred at approximately 4.5, 2.9 and 1.85 ppm and were not observed in the PCL sample (Fig. 4).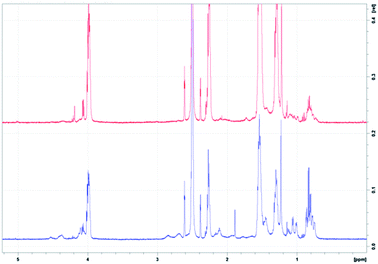 | ||
| Fig. 4 (Top) 1H NMR of 10 mg PCL in 95% DMSO, 5% D2O. (Bottom) 1H NMR of 10 mg PCL with Colistin in 95% DMSO, 5% D2O. | ||
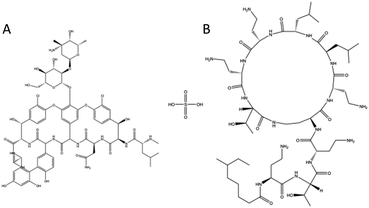
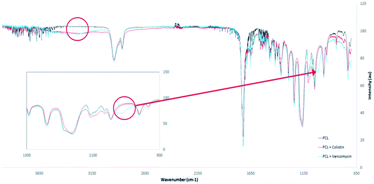
ATR-FTIR confirmed the addition of Vancomycin into the nanomesh. The chemical structures of both antibiotics (Fig. 2) have many of the same functional groups, with the exception of an ether. The FTIR spectrum showed a characteristic ether peak at ∼1030 cm−1 which was only present in the sample containing Vancomycin (Fig. 5). Similarly, both antibiotics contain an amine group within their structure whereas PCL does not. The signal at ∼3300 cm−1 corresponding to the amine group further indicates the presence of both Colistin and Vancomycin in the mesh.
3.3 Zone of inhibition assay
Zone of inhibition assays were used to test the antibiotic activity of Colistin released from the mesh (Fig. 6). E. coli lawns were grown on nutrient agar and small circular pieces of mesh were cut out and placed on the bacterial lawns. As E. coli is Gram-negative, only Colistin meshes were tested as E. coli is not susceptible to Vancomycin. All meshes containing Colistin produced a ZOI with varying diameters. The control meshes of PCL and PCL with both cationic and anionic nanoparticles showed no inhibition of the bacteria. The ZOI assay was used as a qualitative method only to test for antibiotic action as Colistin is known to diffuse slowly and poorly though agar and often leads to unreliable and inaccurate diameters.35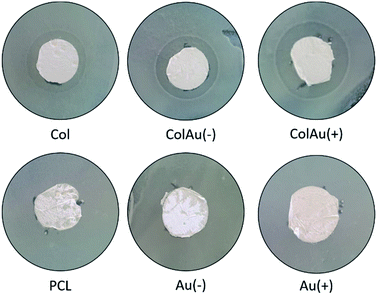 | ||
| Fig. 6 Zone of inhibition assays after 48 h for meshes tested on E. coli lawns with 8 mm mesh disks. | ||
3.4 Drug release studies
With confirmation of antibiotic loading through FTIR and 1H NMR, a drug release study was conducted to determine the amount of antibiotic being released from the mesh over time (Fig. 7). Drug concentrations were determined by UV/vis spectroscopy.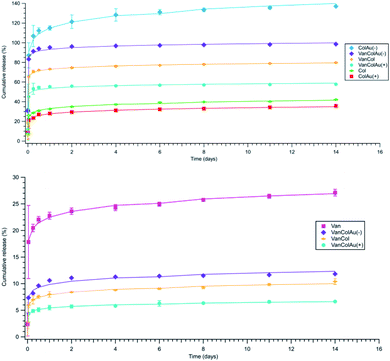 | ||
| Fig. 7 (Top) Colistin cumulative release, (Bottom) Vancomycin cumulative release. Both show Korsmeyer–Peppas fitting models. | ||
The drug release behaviour for 7% w/w PCL with varying antibiotic and gold nanoparticle combinations was monitored to determine the most efficient drug release profile. All samples exhibited a burst release in the first few hours before reaching a plateau around day 5. This burst release is due to diffusion, where the drug diffuses out from the mesh within the first few hours of being submerged in DPBS.36,37
The total amount of drug released varied significantly between different drug combinations. As all the mesh combinations had the same amount of Colistin added during the formation of the mesh, the differences observed are most likely due to different release mechanisms. It is noted the cumulative percentage is above 100% however this is due to the theoretical maximum amount of Colistin being calculated for the total weight of the mesh and then divided by the average 1.5 cm2 piece of mesh weight.
The ColAu(−) sample had the greatest release of Colistin compared to the other combinations (Fig. 7). The addition of positively charged gold nanoparticles saw a similar release to the Colistin mesh alone. This suggests that nanoparticles of the opposite charge to the drug can increase the release from the fibres. This altered release has been previously documented for changes in fibre texture, the pH differences of core and shell fibres and fibre shell thickness however to the author's knowledge, changes to release rates due to charged nanoparticles within the mesh has not been previously observed.38,39 However, it has been shown that in core–shell electrospinning, positively charged drugs are found to migrate to the surface of the fibres compared to neutral drugs which remain in the core.40 This is due to the charge generation of the surface of the polymer during electrospinning. The positively charged drugs are repelled from the inner needle surface and are drawn toward the grounded collector plate.41 Although in this case there is no core–shell, the negatively charged nanoparticles are likely interacting with the Colistin, in effect neutralising its cationic charge and encapsulating it further into the fibre. Whereas for the meshes with positively charged nanoparticles the particles and drug repel each other with minimal interaction, allowing the highly cationic Colistin (+5 net charge) to migrate to the fibre surface.42 Thus, the addition of anions when using a cationic drug should increase the release time of the drug from fibres.
For the Vancomycin release, when on its own, Vancomycin releases approximately 25% of the initial loading which is relatively low compared to the release of Colistin. This is due to the differences in their solubility in the electrospinning solvent HFIP. However, in comparison to the other combinations of the Vancomycin meshes, the Vancomycin only mesh release is high, with the VanCol, VanColAu(+) and VanColAu(−) only releasing between 6–12%. The addition of the positively charged nanoparticles to the Vancomycin mesh had no significant effect, which mirrored what was observed in the Colistin mesh. The addition of the negatively charged nanoparticles also had no significant effect which is expected as Vancomycin is amphoteric with only a slight positive charge when dissolved within HFIP.43 This further supports the idea that the charge of a drug and the addition of charged particles within a nanomesh system affects the drug positioning within a fibre, which ultimately determines the release profile.
To analyse the release kinetics, all meshes containing Colistin were fitted to zero-order, first-order, Higuchi, Hixson–Crowell and Korsmeyer–Peppas models (Table S2†). The fitting was evaluated by the correlation coefficient (R2). These models were chosen as they model different release mechanisms including diffusion and erosion.
The kinetics did not correlate with zero order, which compares the cumulative amount of drug released versus time. The plots shows a curvilinear profile for all meshes and the regression values were low indicating the release is not zero-order. The first order model, which compares the log of cumulative percentage of drug remaining versus time had a similar profile to zero order, where the data shows to be curvilinear. Again, the regression value was smaller than other models for all meshes tested.
The Korsmeyer–Peppas model (log cumulative percentage of drug released compared to log time) had the best fit for the release of Colistin for all the meshes, characterised by the highest R2 values (Table S2†). The analysis of the Korsmeyer–Peppas model provides insight into the mechanisms of drug release, being both erosion and diffusion based.44 This is determined by the values of the drug release exponent, n, which in this case is the slope of the Korsmeyer–Peppas model plot (Table S2†). When n is equal to or less than 0.45, it is an indication that the release mechanism is Fickian diffusion.45 Fickian diffusion occurs when the polymer's relative relaxation time is considerably shorter than the diffusion time of water transport, which is controlled by the concentration gradient. If the n value is between 0.45 and 0.89 it indicates non-Fickian diffusion also known as analogous transport.46 This type of transport has both erosion and diffusion as part of the release mechanism. When n is equal to or greater than 0.89 it indicates class II transport where the mechanism is erosion based.47 In all meshes, n is less than 0.45 for Colistin release which is an indication of Fickian diffusion.
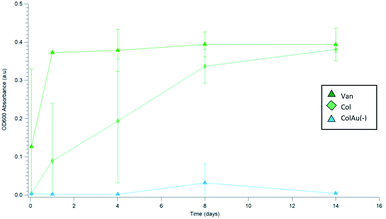
In order to examine how the release would affect bacteria over a 14 day time period, an in vitro bacterial study was conducted where E. coli was added to the aliquots of DPBS that was removed at various time points during the Colistin release study. The absorbance was then used to determine bacterial growth over time. The in vitro study confirmed the initial results showing that ColAu(−) produced the most efficient nanomesh. For the ColAu(−) nanomesh, the bacterial growth was severely hindered over 14 days whereas all other meshes had bacterial growth observed through their absorbance at 600 nm (Fig. 8). This data was in agreement with the results from the cumulative Colistin release, showing that the addition of small charged particles can alter the release profile.
4 Conclusion
This research described the fabrication of Colistin and Vancomycin loaded PCL nanomesh utilising an electrospinning approach for sustained drug release. Homogenous fibres were formed through optimisation of the electrospinning process, with 7% w/w PCL at 1 mL h−1 producing the most uniform nanomesh. 12.5% w/w of Colistin and 1.4% w/w of Vancomycin were introduced into the polymer matrix prior to spinning. Both Colistin and Vancomycin were released from the nanomesh over a 14 day period, with Colistin releasing at a higher cumulative percentage than Vancomycin. The addition of small charged nanoparticles altered the release of the antibiotics from the nanomesh. The addition of citrate capped gold nanoparticles likely neutralised Colistin's charge, causing the antibiotic to migrate toward the centre of the fibre, prolonging its release profile. However, the addition of positively charged gold nanoparticles did not significantly alter the release of Colistin from the mesh compared to Colistin alone. Further investigation is needed to determine if other small charged particles affect the release of drugs from single spun fibres and how it affects the release over time. As it is a pharmaceutical application, the stability of the mesh under different storage conditions as well as the toxicological properties also need to be evaluated.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors would like to thank the Australian Institute of Nuclear Science and Engineering (AINSE Ltd) (Award – PGRA) for the top-up scholarship, the National Institute for Materials Science (NIMS) for the internship and the Australian Government Research Training Program for providing financial assistance. The authors would also like to acknowledge the assistance of Martin Johnston from Flinders University for his assistance with running the 1H NMR.Notes and references
- C. Llor and L. Bjerrum, Ther. Adv. Drug Saf., 2014, 5, 229–241 CrossRef.
- M. T. Sweeney, B. V. Lubbers, S. Schwarz and J. L. Watts, J. Antimicrob. Chemother., 2018, 73, 1460–1463 CrossRef CAS.
- J. Li, C. R. Rayner, R. L. Nation, R. J. Owen, D. Spelman, K. E. Tan and L. Liolios, Antimicrob. Agents Chemother., 2006, 50, 2946–2950 CrossRef CAS.
- H. Garbis, M. R. van Tonningen and M. Reuvers, in Drugs During Pregnancy and Lactation, ed. C. Schaefer, P. Peters and R. K. Miller, Academic Press, Oxford, 2 nd edn, 2007, pp. 123–177 Search PubMed.
- N. Markou, H. Apostolakos, C. Koumoudiou, M. Athanasiou, A. Koutsoukou, I. Alamanos and L. Gregorakos, Crit. Care, 2003, 7, R78–R83 CrossRef.
- N. E. Edrees, A. A. A. Galal, A. R. Abdel Monaem, R. R. Beheiry and M. M. M. Metwally, Chem. Biol. Interact., 2018, 294, 56–64 CrossRef CAS.
- M. Gurjar, J. Intensive Care Med., 2015, 3, 3 CrossRef.
- A. Z. Bialvaei and H. Samadi Kafil, Curr. Med. Res. Opin., 2015, 31, 707–721 CrossRef CAS.
- N. Martis, S. Leroy and V. Blanc, J. Infect., 2014, 69, 1–12 CrossRef.
- A. Hassoun, P. K. Linden and B. Friedman, Crit. Care, 2017, 21, 211 CrossRef.
- T. L. Holland, C. Arnold and V. G. Fowler, J. Am. Med. Assoc., 2014, 312, 1330–1341 CrossRef.
- C. Watanakunakorn, J. Antimicrob. Chemother., 1984, 14, 7–18 CrossRef CAS.
- P. E. Reynolds, Eur. J. Clin. Microbiol. Infect. Dis., 1989, 8, 943–950 CrossRef CAS.
- S. Agarwal, J. H. Wendorff and A. Greiner, Polymer, 2008, 49, 5603–5621 CrossRef CAS.
- M. Gizaw, J. Thompson, A. Faglie, S.-Y. Lee, P. Neuenschwander and S.-F. Chou, Bioengineering, 2018, 5, 9 CrossRef.
- E. J. Torres-Martinez, J. M. Cornejo Bravo, A. Serrano Medina, G. L. Pérez González and L. J. Villarreal Gómez, Curr. Drug Delivery, 2018, 15, 1360–1374 CrossRef CAS PubMed.
- J. L. Castro-Mayorga, M. J. Fabra, L. Cabedo and J. M. Lagaron, Nanomaterials, 2016, 7, 4 CrossRef.
- D. Lv, R. Wang, G. Tang, Z. Mou, J. Lei, J. Han, S. De Smedt, R. Xiong and C. Huang, ACS Appl. Mater. Interfaces, 2019, 11, 12880–12889 CrossRef CAS.
- M. Zhu, J. Han, F. Wang, W. Shao, R. Xiong, Q. Zhang, H. Pan, Y. Yang, S. K. Samal, F. Zhang and C. Huang, Macromol. Mater. Eng., 2017, 302, 1600353 CrossRef.
- D. Lv, M. Zhu, Z. Jiang, S. Jiang, Q. Zhang, R. Xiong and C. Huang, Macromol. Mater. Eng., 2018, 303, 1800336 CrossRef.
- M. Zhu, R. Xiong and C. Huang, Carbohydr. Polym., 2019, 205, 55–62 CrossRef CAS.
- M. Zhu, D. Hua, M. Zhong, L. Zhang, F. Wang, B. Gao, R. Xiong and C. Huang, Colloid Interface Sci. Commun., 2018, 23, 52–58 CrossRef CAS.
- W. Ma, M. Zhang, Z. Liu, C. Huang and G. Fu, Environ. Sci.: Nano, 2018, 5, 2909–2920 RSC.
- W. Ma, J. Zhao, O. Oderinde, J. Han, Z. Liu, B. Gao, R. Xiong, Q. Zhang, S. Jiang and C. Huang, J. Colloid Interface Sci., 2018, 532, 12–23 CrossRef CAS.
- I. Kurtz and J. Schiffman, Materials, 2018, 11, 1059 CrossRef.
- J. K. Y. Lee, N. Chen, S. Peng, L. Li, L. Tian, N. Thakor and S. Ramakrishna, Prog. Polym. Sci., 2018, 86, 40–84 CrossRef CAS.
- D. Li and Y. Xia, Adv. Mater., 2004, 16, 1151–1170 CrossRef CAS.
- M. Mirjalili and S. Zohoori, J. Nanostruct. Chem., 2016, 6, 207–213 CrossRef CAS.
- X. Li, S. M. Robinson, A. Gupta, K. Saha, Z. Jiang, D. F. Moyano, A. Sahar, M. A. Riley and V. M. Rotello, ACS Nano, 2014, 8, 10682–10686 CrossRef CAS.
- Y. Zhou, Y. Kong, S. Kundu, J. D. Cirillo and H. Liang, J. Nanobiotechnol., 2012, 10, 19 CrossRef CAS.
- Z. V. Feng, I. L. Gunsolus, T. A. Qiu, K. R. Hurley, L. H. Nyberg, H. Frew, K. P. Johnson, A. M. Vartanian, L. M. Jacob, S. E. Lohse, M. D. Torelli, R. J. Hamers, C. J. Murphy and C. L. Haynes, Chem. Sci., 2015, 6, 5186–5196 RSC.
- M. Fuller and I. Köper, Polymers, 2018, 10, 1336 CrossRef.
- P. Bala Balakrishnan, L. Gardella, M. Forouharshad, T. Pellegrino and O. Monticelli, Colloids Surf., B, 2018, 161, 488–496 CrossRef.
- A. Haider, S. Haider and I.-K. Kang, Arabian J. Chem., 2015, 1165–1188 Search PubMed.
- L. Poirel, A. Jayol and P. Nordmann, Clin. Microbiol. Rev., 2017, 30, 557–596 CrossRef CAS.
- S. Misra, H. Pandey, S. Patil, P. Ramteke and A. Pandey, Fibers, 2017, 5, 41 CrossRef.
- W.-C. Lin, I.-T. Yeh, E. Niyama, W.-R. Huang, M. Ebara and C.-S. Wu, Polymers, 2018, 10, 231 CrossRef.
- Y. Ding, W. Li, F. Zhang, Z. Liu, N. Zanjanizadeh Ezazi, D. Liu and H. A. Santos, Adv. Funct. Mater., 2019, 29, 1802852 CrossRef.
- K. T. Shalumon, G.-J. Lai, C.-H. Chen and J.-P. Chen, ACS Appl. Mater. Interfaces, 2015, 7, 21170–21181 CrossRef CAS.
- U. Angkawinitwong, S. Awwad, P. T. Khaw, S. Brocchini and G. R. Williams, Acta Biomater., 2017, 64, 126–136 CrossRef CAS.
- B. Pourdeyhimi, A. L. Yarin and S. Ramakrishna, Fundamentals and Applications of Micro- and Nanofibres, Cambridge University Press, Cambridge, 1st edn, 2014 Search PubMed.
- P. J. Bergen, J. Li, C. R. Rayner and R. L. Nation, Antimicrob. Agents Chemother., 2006, 50, 1953–1958 CrossRef CAS.
- S. Jamzad and R. Fassihi, AAPS PharmSciTech, 2006, 7, E33 CrossRef.
- P. Kumar, A. L. Ganure, B. B. Subudhi and S. Shukla, Int. J. Pharm. Res., 2015, 14, 677–691 CAS.
- M. V. Cabañas, J. Peña, J. Román and M. Vallet-Regí, Eur. J. Pharm. Sci., 2009, 37, 249–256 CrossRef.
- T. Hayashi, H. Kanbe, M. Okada, M. Suzuki, Y. Ikeda, Y. Onuki, T. Kaneko and T. Sonobe, Int. J. Pharm., 2005, 304, 91–101 CrossRef CAS.
- N. Amit, L. Bibek and S. Kalyan, Acta Pharm., 2011, 61, 25–36 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9ra06398f |
| This journal is © The Royal Society of Chemistry 2019 |