Open Access Article
Open Access ArticleSparganin A alleviates blood stasis syndrome and its key targets by molecular docking†
Minghua Xian‡
 abc,
Sulong Ji‡abc,
Chen Chenabc,
Shengwang Liang*abc and
Shumei Wang*bcd
abc,
Sulong Ji‡abc,
Chen Chenabc,
Shengwang Liang*abc and
Shumei Wang*bcd
aSchool of Traditional Chinese Medicine, Guangdong Pharmaceutical University, Guangzhou 510006, China. E-mail: swliang371@163.com; Tel: +86-20-39352177
bKey Laboratory of Digital Quality Evaluation of Chinese Materia Medica of State Administration of TCM, Guangdong Pharmaceutical University, Guangzhou 510006, China. E-mail: gdpuwsm@126.com; Tel: +86-20-39352177
cEngineering & Technology Research Center for Chinese Materia Medica Quality of the Universities of Guangdong Province, Guangdong Pharmaceutical University, Guangzhou 510006, China
dSchool of Pharmacy, Guangdong Pharmaceutical University, Guangzhou 510006, China
First published on 21st November 2019
Abstract
Blood stasis syndrome is implicated in the development of chronic conditions, including cardio- and cerebrovascular diseases. Cyclo-(Tyr-Leu), named Sparganin A (SA), is a compound isolated from the ethanol extract of Rhizoma Sparganii. Here, the successful extraction of SA from Rhizoma Sparganii was verified by extensive spectral analysis using 1H NMR and 13C NMR. To determine the biological effects of SA, a mouse model of acute blood stasis was established by subcutaneous injection of adrenaline hydrochloride and placing the animals in an ice water bath. In this model, the concentration of TXB2, PAI-1, FIB, ET-1 was measured by ELISA, and thymus index (TI), hepatic index (HI), and spleen index (SI) were calculated. Molecular docking by SYBYL and functional analysis of the putative targets by STRING and Cytoscape were employed to identify the key targets of SA. The accumulated results documented that SA exhibits anticoagulative activity, and its key targets are VEGFA and SERPINE1. SA may be involved in the pathological process of complement and coagulation cascades. This study demonstrates that SA may be a promising drug to control coagulation in blood stasis syndrome.
Introduction
Blood stasis syndrome is one of the most common clinical-pathological concepts in traditional Chinese medicine (TCM). Blood stasis is implicated in the development of chronic conditions, including cardio- and cerebrovascular diseases,1,2 since it increases the risk of microcirculatory disturbances and thrombus formation.3 Pathologic evidence demonstrates that blood stasis is typically accompanied by the accumulation of free radicals, inflammation, and hematological disorders such as hemorrhage, congestion, and thrombosis.Patients suffering from thrombotic occlusions induced by blood stasis are at a high risk of cardiovascular and cerebrovascular diseases such as acute myocardial infarction and ischemic stroke.4,5 Thus, the need to optimize anti-thrombotic therapy is apparent.6 Several approved anti-thrombotic drugs, such as warfarin and rivaroxaban, are targeting the complex biological process of thrombogenesis.7 However, the use of these drugs is associated with significant adverse effects, such as gastrointestinal bleeding.8,9 Therefore, new oral antithrombotic drugs need to be designed to prevent or treat the development of blood stasis, with the objective of improving safety over the older, natural anticoagulants, such as heparin. Achieving the goal of treating blood stasis requires understanding the mechanisms underlying this condition.
Rhizoma Sparganii originates from the rhizome of Sparganium stoloniferum Buch.–Ham. It is a traditional Chinese medicine frequently used for the treatment of clinical blood stasis. The composition of Rhizoma Sparganii is complex, although some studies on its chemical composition have been performed.10 The essential oil of Rhizoma Sparganii contains thirty-three components, including 3,5,6,7,8,8a-hexahydro-4,8a-dimethyl-6-(11-methylethenyl)-2(1H) naphthalenone.10 However, which ingredients of Rhizoma Sparganii represent active compounds is not clear, and the current knowledge regarding their pharmacological effects is limited. Therefore, further research in this area is of great significance for the development of new drugs and guidelines for their clinical use.
Based on the previous work, our laboratory has separated one of the components of Rhizoma Sparganii, named Sparganin A (cyclo-(Tyr-Leu), SA).11 The present work is focused on the content of SA in Rhizoma Sparganii and its efficacy. Since the mechanisms by which SA relieves the blood stasis syndrome remain elusive, the comprehensive effects of SA and its key target were analyzed in a mouse model of blood stasis. The overarching objective was to fully understand the effects of SA and provide a foundation for future studies of the pharmacological mechanism.
Materials and methods
Plant materials
Rhizoma Sparganii was purchased from Guangzhou Zhixin Pharmaceutical Co., Ltd., and was identified as the black-triangular Sparganium stoloniferum Buch.-Ham. by an expert in Chinese medicine identification, Professor Li Shuyuan (Guangdong Pharmaceutical University).Chemicals and reagents
Adrenaline hydrochloride injections were purchased from Grand Pharmaceutical (China) Co., Ltd. (batch no.: 20170214), aspirin enteric-coated tablets were from Bayer HealthCare Co., Ltd. (batch no.: BJ32400), mouse thromboxane B2 (TXB2), endothelin 1 (ET-1), plasminogen activator inhibitor 1 (PAI-1), and fibrinogen (FIB) enzyme-linked immunosorbent assay kit were from Tianjin Annuruikang Biotechnology Co., Ltd. All other reagents were of analytical grade. Cyclo-(Tyr-Leu) was synthesized by ChinaPeptides Biotech Co., Ltd.Animals
Female Kunming mice, weighing 20–30 g, were obtained from the Guangdong Medical Laboratory Animal Center (license no.: SCXK-Guangdong-2013-0002). The animal studies adhered to the principles of the Institutional Animal Care and Use Committee Guidebook. All animal experiments were approved by the Animal Ethics Committee of Guangdong Pharmaceutical University.Mouse model of acute blood stasis12
Female mice were randomly divided into 6 groups according to body weight: the normal group, the model group, the aspirin control group (ASP group), and three SA-treated groups (SA1, 0.12 mg kg−1; SA2, 0.24 mg kg−1; SA3, 0.48 mg kg−1). Except for the normal group, animals in other groups were given subcutaneous adrenaline hydrochloride injection (1 ml kg−1). After 2 hours, the mice were placed in 0 °C ice water for 5 minutes, and then subcutaneously injected with 1 ml kg−1 epinephrine hydrochloride. These steps were repeated 2 hours later. This protocol resulted in acute blood stasis. After the second ice water bath, mice have fasted for 12 hours and only water was given. Subsequently, after 12 h interval, blood was collected in tubes containing heparin and the blood rheology index was measured. The remaining plasma was centrifuged, and the supernatant was used to detect the concentration of TXB2, ET-1, PAI-1, and FIB by ELISA. The thymus, spleen, and liver were collected and weighted to calculate the thymus index (TI), spleen index (SI), and liver index (HI).SA extraction procedure
The extraction protocol is illustrated in Fig. 1. Briefly, 14 kg of the dried powdered plant was extracted three times with 60% (v/v) ethanol for two hours and filtered. The filtrates were combined and the ethanol was removed to yield the total extract. The total extract was suspended in water and extracted three times with petroleum ether, chloroform, ethyl acetate, and n-butanol. The extracts were combined and recovered under reduced pressure to obtain 26 g of petroleum ether extract, 34 g of chloroform extract, 11.5 g of ethyl acetate fraction extract, 42 g of n-butanol fraction extract.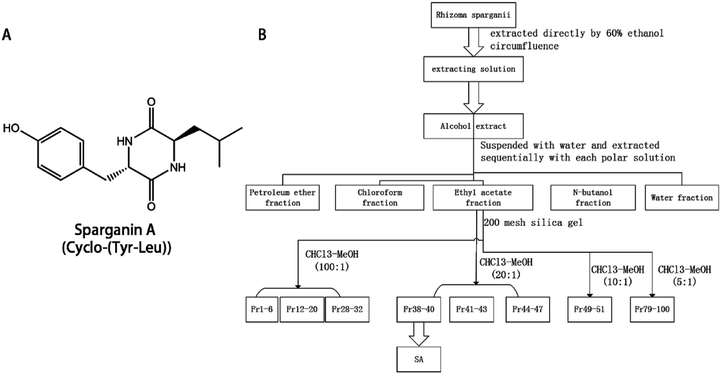 | ||
| Fig. 1 The structure and the extraction route map of SA. The structure and the extraction route map of SA. (A) the structure of SA; (B) the extraction route map of SA. | ||
Separation and purification of components
The ethyl acetate fraction (11.5 g) was dissolved in 50 ml of ethyl acetate, and subjected to silica gel (200–300 mesh) column chromatography; gradient elution was performed using a chloroform–methanol solvent system. A flow chart depicting the crude fraction of the ethyl acetate moiety is shown in Fig. 2. Eluted fractions 38–40 were combined, concentrated on a rotary evaporator, dissolved in methanol, and allowed crystallize at room temperature to form solid SA.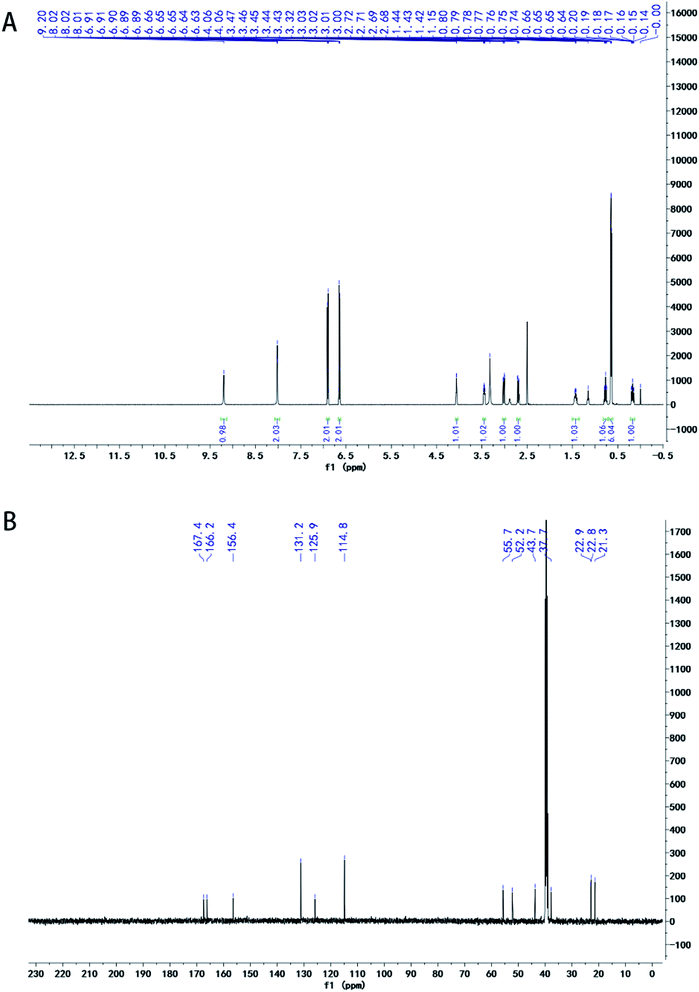 | ||
| Fig. 2 1H-NMR and 13C NMR spectrum of Sparganin A. 1H-NMR and 13C NMR spectrum of Sparganin A. (A) 1H-NMR; (B) 13C NMR spectrum. | ||
Identification of SA structure
The structure of SA was resolved by extensive spectral analysis, 1H NMR, 13C NMR. The data was analyzed using MestreNova software (Mestrelab Research S.L., version: 11.0.3-18688).Molecular docking of SA
To compile a list of blood stasis syndrome-associated genes, genes on coagulation, hemorrhage, and thrombus formation were extracted from the DisGeNET (v6.0) resource (http://www.disgenet.org/),13 and DrugBank (https://www.drugbank.ca/).14 Molecular docking was analyzed using the SYBYL-X 2.1.1 software (Certara, L. P.). The scoring function total-score equal to 5 was used as a threshold to evaluate the interaction between ingredients and targets. The structure of disease targets employed in the analysis of docking was obtained from the Protein Data Bank (PDB, http://www.rcsb.org).15 The co-crystalized ligand and water molecules were removed from the structure, while H atoms were added and side chains were fixed during protein preparation. The Surflex-Dock (SFXC) docking mode was used, and the procedure was conducted as previously described.16 Total Surflex-Dock scores represent binding affinities.Functional analysis of the putative targets by STRING
The STRING database (https://string-db.org/) was used to determine protein–protein interactions (PPI). This resource includes data on interacting proteins or genes in humans.17 Interactions among the putative targets were identified using a threshold score of 0.7. To mine the critical targets related to SA, a target-function network of SA was constructed, with the target-function relation based on the network topological analysis by Cytoscape 3.7.1.18Results
Extraction and identification of SA from Rhizoma Sparganii
The structure of SA was elucidated by extensive spectral analysis using 1H NMR and 13C NMR (Fig. 2). 1H-NMR (500 MHz, DMSO) spectrum: δ 9.20 (s, 1H), 8.02 (t, J = 3.3 Hz, 2H), 6.93–6.88 (m, 2H), 6.67–6.62 (m, 2H), 4.08–4.03 (m, 1H), 3.48–3.42 (m, 1H), 3.02 (dd, J = 13.6, 3.8 Hz, 1H), 2.70 (dd, J = 13.6, 4.8 Hz, 1H), 1.50–1.36 (m, 1H), 0.77 (ddd, J = 13.7, 9.1, 4.8 Hz, 1H), 0.65 (dd, J = 6.6, 5.1 Hz, 6H), 0.17 (ddd, J = 14.0, 9.3, 5.2 Hz, 1H). 13C NMR (125 MHz, DMSO-d6) spectrum: δ 167.4, 166.2, 156.4, 131.2 (2C), 125.8, 114.8 (2C), 55.6, 52.3, 43.7, 37.7, 22.9, 22.8, 21.3.The anticoagulation effect of SA
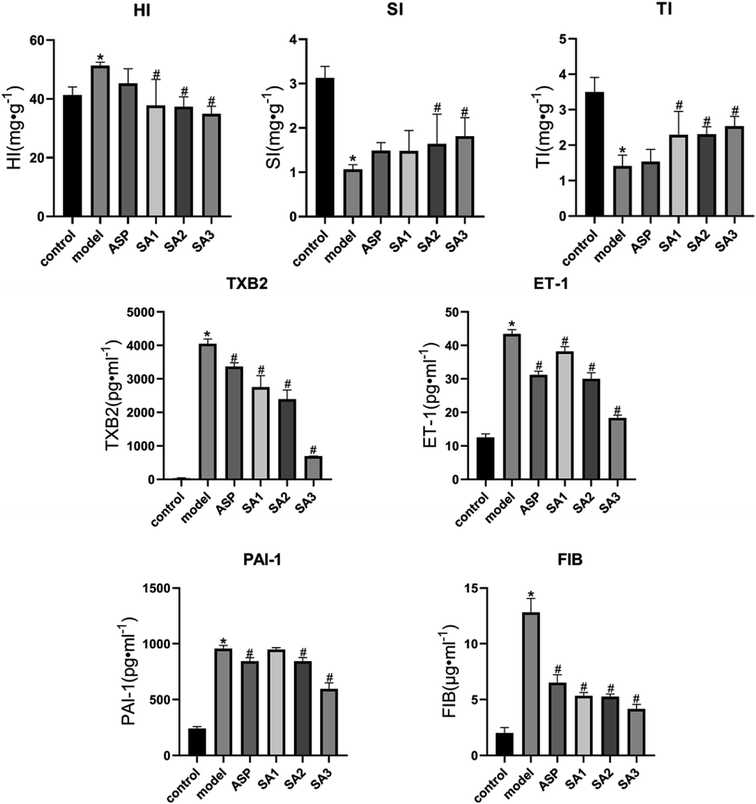
Since animal experiments require a large amount of the compound, SA was obtained by synthesis and its identity was verified by HPLC and MS spectrum (ESI A†). SA significantly reduced the level of TXB2 (SA1 group: 2757.64 ± 341.50 pg ml−1; SA2 group: 2395.76 ± 271.21 pg ml−1; SA3 group: 687.67 ± 10.25 pg ml−1; model group: 4058.09 ± 137.50 pg ml−1), PAI-1 (SA1 group: 947.84 ± 17.92 pg ml−1; SA2 group: 843.27 ± 33.89 pg ml−1; SA3 group: 595.56 ± 55.50 pg ml−1; model group: 955.87 ± 30.19 pg ml−1), ET-1 (SA1 group: 38.18 ± 1.47 pg ml−1; SA2 group: 30.01 ± 1.84 pg ml−1; SA3 group: 18.39 ± 0.81 pg ml−1; model group: 43.43 ± 1.30 pg ml−1), and FIB (SA1 group: 5.33 ± 0.31 μg ml−1; SA2 group: 5.26 ± 0.24 μg ml−1; SA3 group: 4.15 ± 0.43 μg ml−1; model group: 12.83 ± 1.24 μg ml−1) (P < 0.01) in the mouse model of acute blood stasis (as shown in Fig. 3). SA can increase the thymus index (SA1 group: 2.29 ± 0.66 mg g−1; SA2 group: 2.31 ± 0.21 mg g−1; SA3 group: 2.53 ± 0.28 mg g−1; model group: 1.41 ± 0.31 mg g−1), spleen index (SA1 group: 1.48 ± 0.46 mg g−1; SA2 group: 1.64 ± 0.67 mg g−1; SA3 group: 1.81 ± 0.42 mg g−1; model group: 1.06 ± 0.11 mg g−1), and reduce the hepatic index (SA1 group: 37.77 ± 8.92 mg g−1; SA2 group: 37.34 ± 3.37 mg g−1; SA3 group: 34.93 ± 2.58 mg g−1; model group: 51.37 ± 1.15 mg g−1) (as shown in Fig. 3). Thus, SA may effectively counteracts blood stasis.Molecular docking of SA
To identify proteins associated with the blood stasis syndrome, 88 targets related to coagulation hemorrhage and thrombus were screened using the DisGeNET (v6.0) and DRUGBANK resources, and their 3D crystal structure and the structures of their active ligands were obtained from PDB. There were 61 targets with a total score greater than 5 indicating relatively strong interaction with SA (ESI B†). The top 10 results of molecular docking scores obtained using the SYBYL-X 2.1.1 software are shown in Table 1 at the size of grid box 6 Å. Moreover, we exhibit the zoomed images of the interaction sites mentioning the name of the residues with proper binding energy due to H bonding (ESI C†). These targets are likely to have a critical function in the regulation of coagulation by SA.| GENE | PDB ID | Total score | Hydrogen-bonding residues |
|---|---|---|---|
| FLT3 | 6il3 | 7.5293 | GLU573/ARG834 |
| TNF | 5yoy | 7.2241 | VAL16/TYR59 |
| PTGS2 | 5kir | 7.1925 | ALA378 |
| SLC23A1 | 5vnn | 6.9739 | ASN365/GLU535/ARG609 |
| KLKB1 | 6O1S | 6.9247 | SER578/ASP572 |
| CYP2C9 | 5X23 | 6.9195 | AGR108/SER365/THR364 |
| EPO | 1eer | 6.8649 | SER152/THR151/ARG10/GLU13 |
| SERPINC1 | 4EB1 | 6.8426 | GLU255/SER235/TYR253/LYS265 |
| SPP1 | 3cxd | 6.8083 | PRO170/THR111 |
| GUCY1A1 | 4ni2 | 6.8074 | THR602/GLY598/ASP477/ASN431 |
Key targets of the SA
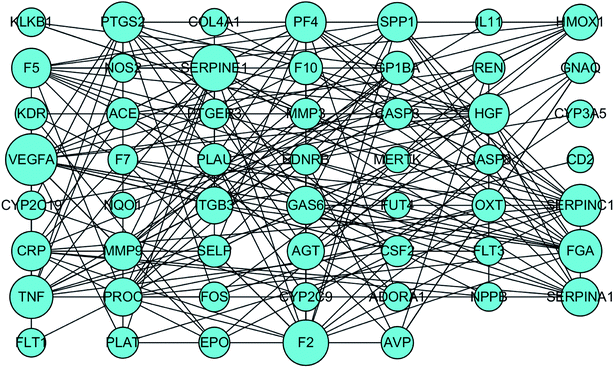
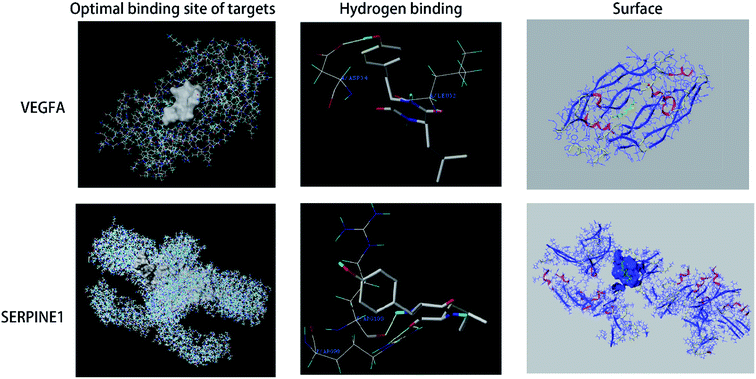
To identify the key targets of SA, the 61 targets with a total score greater than 5 were imported into STRING to find out the protein–protein interactions (PPI). The results of this search are shown in Fig. 4. The enrichment of functional pathway was found out that the top 10 of pathway, such as the complement and coagulation cascade pathways, is illustrated in Fig. 5. The network topological analysis by Cytoscape demonstrated that targets with the top five scores of degree were vascular endothelial growth factor A (VEGF-A) and serine proteinase inhibitor family member 1 (SERPINE1) (Table 2). The key pathway of SA includes the complement and coagulation cascades pathway. As depicted in Fig. 6, SA is bound to residues outside the active pocket of VEGF-A (key residues including ASP34/LEU32) and SERPINE1 (key residues including ARG100/ARG98) by hydrogen bonds. The overall spatial structure indicates that the VEGF-A/SA and SERPINE1/SA complexes are stable, indicating the interactions of SA with their targets may have an active functional role.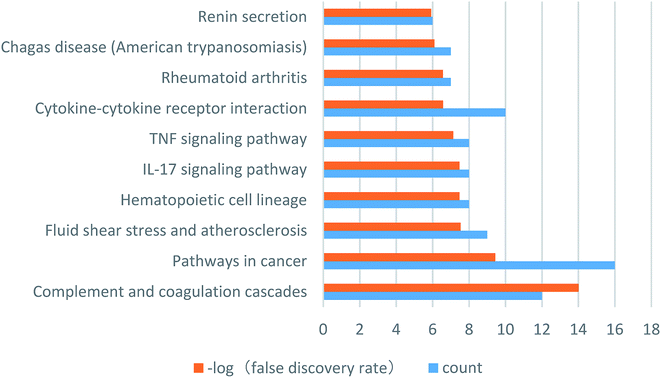 | ||
| Fig. 5 The enrichment of functional pathway of SA. Orange pillars represent the false discovery rate, blue pillars represent the count of targets. | ||
| Name | Degree | Betweenness Centrality | Closeness Centrality |
|---|---|---|---|
| VEGFA | 22 | 0.18984028 | 0.6 |
| SERPINE1 | 18 | 0.0843947 | 0.57303371 |
| F2 | 17 | 0.10466867 | 0.5 |
| TNF | 15 | 0.13513865 | 0.54255319 |
| FGA | 15 | 0.03441782 | 0.5257732 |
| SERPINC1 | 14 | 0.03725382 | 0.46788991 |
| PTGS2 | 13 | 0.17484413 | 0.5257732 |
| PF4 | 13 | 0.0575878 | 0.51 |
| HGF | 13 | 0.04974032 | 0.5049505 |
| CRP | 12 | 0.06029181 | 0.51515152 |
Discussion
The major finding of the present study is the demonstration that SA has an anticoagulative activity in the mouse model of blood stasis syndrome. Analysis of molecular docking indicated that the key targets of SA are VEGF-A and SERPINE1.Blood stasis syndrome is one of the common clinical conditions and may be caused by several factors. According to the theory of traditional Chinese medicine, blood stasis is induced by the “cold” and “worry”. In the protocol employed in the current work, the subcutaneous injection of epinephrine hydrochloride and ice water bath mimics human anger. As a result, the adrenal glands secrete a large amount of adrenaline which, together with the cold, engenders an acute blood stasis. The application of bioinformatics to the KEGG pathway analysis revealed that SA may be involved in the pathological process of complement and coagulation cascades. Animal experiments have demonstrated that SA significantly reduces the levels of TXB2, PAI-1, ET-1, and FIB in the mouse model of acute blood stasis model mouse. Together, the results of the current work documented the critical role of SA in blood stasis syndrome.
Molecular docking technology is a computational method for predicting the three-dimensional structure of complexes formed by small molecule drugs and proteins.19 The computer-aided method screens the drugs by analyzing their binding to proteins by geometric matching and energy matching. The present investigation utilized network topological analysis by Cytoscape and demonstrated that the SA targets with the top scores of the degree of matching are VEGF-A and SERPINE1.
Different subtypes of VEGF, including VEGF-A and VEGF-B, are expressed in many tissues and have multiple biological functions. The expression of VEGF can be induced by constitutive production and thrombin in vascular smooth muscle cells, endothelial cells, megakaryocytes, and platelets.20,21 VEGF controls wound healing and hemostasis by modifying the hemostatic properties and proliferation of endothelial cells.22 In particular, VEGF regulates the coagulation cascade that affects bleeding and clotting times.23 In current work, molecular docking approach demonstrated that SA can bind the VEGF-A protein. This finding implies that SA controls the coagulation cascade by regulating the activity of VEGF-A.
SERPINE1, also known as the plasminogen activator inhibitor 1 (PAI-1), is encoded by the SERPINE1 gene, is associated with thrombophilia.24 SERPINE1 is a primary inhibitor of tissue-type plasminogen activator (PLAT) and urokinase-type plasminogen activator (PLAU), which are required for the downregulation of fibrinolysis and controlled degradation of blood clots.25,26 SERPINE1 promotes the procoagulatory state and clot formation in the cerebral microvasculature, where it functions as an important endogenous inhibitor of the fibrinolytic pathway.27 SERPINE1 can also block Factor XIa (FXIa) and induce its clearance and degradation by forming a complex with FXIa on endothelial cells.28 Moreover, the suppression of SERPINE1 activity can ameliorate lipopolysaccharide-induced thrombosis in rats.29 The present work demonstrated by molecular docking that SA can bind to the SERPINE1 protein of SERPINE1 by molecular docking, indicating that interaction of SA and SERPINE1 can regulate the coagulation cascade.
In summary, SA alleviates the blood stasis syndrome by targeting VEGF-A and SERPINE1, as demonstrated by molecular docking. Given that the effect of SA as an anticoagulant is similar that of Rhizoma Sparganii, and the determination of the composition of Rhizoma Sparganii is currently not available in the Chinese Pharmacopoeia, the measurement of SA can serve as the content index of Rhizoma Sparganii, facilitating its quality control. In addition, since SA may be involved in the pathological process of complement and coagulation cascades, it may represent a promising molecule to be used in the treatment of the blood stasis syndrome.
Conclusion
This study identified the anticoagulative activity of SA mediated by its binding to VEGF-A and SERPINE1. SA may be involved in the pathological process of complement and coagulation cascades. The work demonstrates that SA may be a promising drug to control coagulation in the blood stasis syndrome.Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
We thank Haibin Luo from Sun Yat-sen University for technical assistance in SYBYL. This work was supported by Technology Funds obtained from the Key Unit of Chinese Medicine Digitalization Quality Evaluation of State Administration of Traditional Chinese Medicine.References
- S.-J. Yue, L.-T. Xin, Y.-C. Fan, S.-J. Li, Y.-P. Tang, J.-A. Duan, H.-S. Guan and C.-Y. Wang, Sci. Rep., 2017, 7, 40318–40332 CrossRef CAS PubMed.
- P. Liu, E. X. Shang, Y. Zhu, J. G. Yu, D. W. Qian and J. A. Duan, Front. Pharmacol., 2017, 8, 677–691 CrossRef PubMed.
- Z. J. Zou, Z. H. Liu, M. J. Gong, B. Han, S. M. Wang and S. W. Liang, Phytomedicine, 2015, 22, 333–343 CrossRef CAS PubMed.
- S. K. De, A. Myat, J. Cotton, S. James, A. Gershlick and G. W. Stone, Heart, 2017, 103, 546–562 CrossRef PubMed.
- K. S. Wong, L. R. Caplan and J. S. Kim, Front. Neurol. Neurosci., 2016, 40, 58–71 Search PubMed.
- R. Bauersachs, C. Espinola-Klein, H. Lawall, M. Storck, T. Zeller and S. Debus, Dtsch. Med. Wochenschr., 2019, 144, 683–689 CrossRef PubMed.
- D. Sibbing and M. Spannagl, Hämostaseologie, 2014, 34, 78–84 CrossRef CAS PubMed.
- T. Hirai, Y. Hamada, Y. Geka, S. Kuwana, K. Hirai, M. Ishibashi, Y. Fukaya and T. Kimura, Eur. J. Clin. Pharmacol., 2017, 73, 1491–1497 CrossRef PubMed.
- G. Y. H. Lip, L. Mauri, G. Montalescot, M. Ozkor, P. Vardas, P. G. Steg, D. L. Bhatt, S. H. Hohnloser, C. Miede, M. Nordaby, M. Brueckmann, J. Kreuzer, T. Kimura, J. Oldgren, J. M. Ten Berg and C. P. Cannon, Am. Heart J., 2019, 212, 13–22 CrossRef CAS PubMed.
- F. M. Zhu, D. U. Bin, L. I. Jun, H. S. Gao and C. J. Liu, Nat. Prod. Res. Dev., 2010, 22, 253–256 CAS.
- L. Bei, W. Shu-mei, W. Bai-lin, X. Ya-tao, Y. Er-lei, H. Xu-guan and L. sheng-wang, Chin. Tradit. Pat. Med., 2015, 37, 34–39 Search PubMed.
- L. Zhang, B. J. Zhao, J. R. Yuan, C. F. Wang, D. Zhao, L. Feng and X. B. Jia, Chinese Traditional & Herbal Drugs, 2016, pp. 493–497 Search PubMed.
- J. Pinero, A. Bravo, N. Queralt-Rosinach, A. Gutierrez-Sacristan, J. Deu-Pons, E. Centeno, J. Garcia-Garcia, F. Sanz and L. I. Furlong, Nucleic Acids Res., 2017, 45, D833–D839 CrossRef CAS PubMed.
- D. S. Wishart, Y. D. Feunang, A. C. Guo, E. J. Lo, A. Marcu, J. R. Grant, T. Sajed, D. Johnson, C. Li, Z. Sayeeda, N. Assempour, I. Iynkkaran, Y. Liu, A. Maciejewski, N. Gale, A. Wilson, L. Chin, R. Cummings, D. Le, A. Pon, C. Knox and M. Wilson, Nucleic Acids Res., 2018, 46, D1074–D1082 CrossRef CAS PubMed.
- S. K. Burley, H. M. Berman, C. Bhikadiya, C. Bi, L. Chen, L. Di Costanzo, C. Christie, K. Dalenberg, J. M. Duarte and S. Dutta, Nucleic Acids Res., 2019, 47, D464–D474 CrossRef CAS PubMed.
- G.-F. Jorge, M. Enrique, M.-R. Wilson, O.-Z. Carlos, O.-V. Jesus and P.-M. Elsie, J. Mol. Graphics Modell., 2017, 75, 250–265 CrossRef PubMed.
- D. Szklarczyk, A. L. Gable, D. Lyon, A. Junge, S. Wyder, J. Huerta-Cepas, M. Simonovic, N. T. Doncheva, J. H. Morris, P. Bork, L. J. Jensen and C. V. Mering, Nucleic Acids Res., 2019, 47, D607–D613 CrossRef CAS PubMed.
- P. Shannon, A. Markiel, O. Ozier, N. S. Baliga, J. T. Wang, D. Ramage, N. Amin, B. Schwikowski and T. Ideker, Genome Res., 2003, 13, 2498–2504 CrossRef CAS PubMed.
- P. Deb, B. Chandrasekaran, R. Mailavaram, R. Tekade and A. Jaber, Drug discovery today, 2019, 24, 1854–1864 CrossRef CAS PubMed.
- S. Bassus, O. Herkert, N. Kronemann, A. Gorlach, D. Bremerich, C. M. Kirchmaier, R. Busse and V. B. Schini-Kerth, Arterioscler., Thromb., Vasc. Biol., 2001, 21, 1550–1555 CrossRef CAS PubMed.
- R. Mohle, D. Green, M. A. S. Moore, R. L. Nachman and S. Rafii, Proc. Natl. Acad. Sci. U. S. A., 1997, 94, 663–668 CrossRef CAS PubMed.
- K. Tezono, K. P. Sarker, H. Kikuchi, M. Nasu, I. Kitajima and I. Maruyama, Haemostasis, 2001, 31, 71–79 CAS.
- X. Liu, L. Hao, S. Zhang, Y. Ji, Y. Zhang, X. Lu, B. Shi, H. Pei, Y. Wang, D. Chen, X. Guan and Y. Zheng, IUBMB Life, 2010, 62, 819–824 CrossRef CAS PubMed.
- P. Hendrix, P. Foreman, M. Harrigan, W. Fisher, N. Vyas, R. Lipsky, M. Lin, B. Walters, R. Tubbs, M. Shoja, J. Pittet, M. Mathru and C. Griessenauer, World Neurosurg., 2017, 105, 672–677 CrossRef PubMed.
- M. H. Lee, E. Vosburgh, K. Anderson and J. McDonagh, Blood, 1993, 81, 2357–2362 CrossRef CAS PubMed.
- W. P. Fay, A. C. Parker, L. R. Condrey and A. D. Shapiro, Blood, 1997, 90, 204–208 CrossRef CAS PubMed.
- E. Griemert, S. Schwarzmaier, R. Hummel, C. Gölz, D. Yang, W. Neuhaus, M. Burek, C. Förster, I. Petkovic, R. Trabold, N. Plesnila, K. Engelhard, M. Schäfer and S. Thal, Ann. Neurol., 2019, 85, 667–680 CrossRef CAS PubMed.
- C. Puy, A. Ngo, J. Pang, R. Keshari, M. Hagen, M. Hinds, D. Gailani, A. Gruber, F. Lupu and O. McCarty, Arterioscler. Thromb. Vasc. Biol., 2019, 39, 1390–1401 CrossRef CAS PubMed.
- M. Tsantarliotou, S. Lavrentiadou, D. Psalla, I. Margaritis, M. Kritsepi, I. Zervos, M. Latsari, V. Sapanidou, I. Taitzoglou and Z. Sinakos, Food Chem. Toxicol., 2019, 125, 190–197 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9ra06329c |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2019 |

![[x with combining macron]](https://www.rsc.org/images/entities/i_char_0078_0304.gif)