Open Access Article
Open Access ArticleTo beat the heat – engineering of the most thermostable pyruvate decarboxylase to date†
Samuel Sutionoa,
Katharina Satzinger‡
a,
André Picka,
Jörg Carstenab and
Volker Sieber*abcd
aChair of Chemistry of Biogenic Resources, Campus Straubing for Biotechnology and Sustainability, Technical University of Munich, Schulgasse 16, 94315 Straubing, Germany. E-mail: sieber@tum.de
bCatalytic Research Center, Technical University of Munich, Ernst-Otto-Fischer-Straße 1, 85748 Garching, Germany
cStraubing Branch BioCat Fraunhofer IGB, Schulgasse 11a, 94315 Straubing, Germany
dSchool of Chemistry and Molecular Biosciences, The University of Queensland, 68 Copper Road, St. Lucia 4072, Australia
First published on 20th September 2019
Abstract
Pyruvate decarboxylase (PDC) is a key enzyme for the production of ethanol at high temperatures and for cell-free butanol synthesis. Thermostable, organic solvent stable PDC was evolved from bacterial PDCs. The new variant shows >1500-fold-improved half-life at 75 °C and >5000-fold-increased half-life in the presence of 9 vol% butanol at 50 °C.
For many years, bioethanol has been used widely as an additive in gasoline or as a fuel by itself.1 To avoid competition with food, production of ethanol from plant-derived starch or sugar (first generation) has shifted to lignocellulosic biomass (second generation). To date, most of the bioethanol produced industrially still utilises yeasts as the main fermentation organism.1–3 However, in recent years, several efforts have been made to use thermophilic microorganisms as the new framework.1,3 The main motivating forces behind this are the volatility of ethanol at higher temperatures, which facilitates easy product removal, reduced risk of contamination and feasibility of consolidated bioprocessing (CBP). In this process, cellulolytic ethanologenic thermophilic microorganisms are applied, thus reducing the energy consumption required for heat exchange between biomass pre-treatment, fermentation and product separation and to avoid cellulase inhibition by accumulating glucose.3,4
There are two major pathways for producing ethanol from pyruvate, the central metabolite in glycolysis: non-oxidative decarboxylation via pyruvate decarboxylase (PDC) and oxidative decarboxylation via pyruvate dehydrogenase (PDH) or pyruvate ferredoxin oxidoreductase (PFOR). Non-oxidative ethanol productions are commonly found in yeasts and other ethanologenic bacteria, while PDH and PFOR routes are described in thermophilic microorganisms.3,4 PDC mode may offer advantages compared with PDH and PFOR, as it is thermodynamically more favourable and will not lead to any other organic acid by-products.3–5 In fact, all industrially relevant ethanol producing organisms use PDC. Thermostable PDCs are, therefore, suggested to play an important role in creating homo-ethanologenic, thermophilic microorganisms.5,6
Until recently, there were only limited efforts described in literature for improving the thermostability of PDC. Ancestral sequence reconstruction, supposedly a powerful method of resurrecting ancient thermostable enzymes, did not yield more thermostable PDC.7 A computational Rosetta-based design resulted in several variants with improved stability, determined by differential interference contrast (DIC) microscopy; unfortunately, the work was not corroborated by kinetic activity and stability data.8 Another effort to redirect the specificity of a thermostable, acetolactate synthase to perform a PDC-like reaction by conventional directed evolution did not yield variants either, with activity similar to that of bacterial PDC.9 Thus, creating thermostable PDC variants remains as the main challenge towards second generation ethanol production by thermophilic microorganisms.
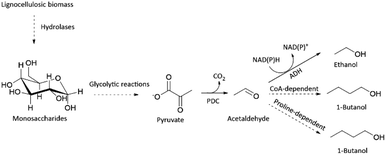
Besides acting as an important enzyme for ethanol production, PDC has also been applied in the in vitro production of n-butanol (Scheme 1).10,11 Butanol together with other longer-chain alcohols, such as 1-propanol, isobutanol and isopentanol, is regarded as the next-generation biofuel, due to its closer resemblance to traditional gasoline.12 Thus, there is an increasing demand to produce longer-chain alcohols sustainably.12 Industrially, butanol is still produced from petroleum-derived propene utilizing Co-catalyst, H2, and CO via oxo synthesis and Reppe process.13 Albeit its efficiency, production of butanol from petroleum is not sustainable.
In a more sustainable way, butanol can be produced via an acetone–butanol–ethanol (ABE) process, utilising Clostridium acetobutylicum.12,14 Recent work on metabolic engineering has demonstrated the possibility of producing butanol in other microorganisms.15,16 However, one of the major challenges in butanol fermentation is still the toxicity of butanol to many microorganisms at relatively low concentrations (<2 vol%).17,18 This effect, however, is less pronounced for the enzymes catalysing butanol production in vitro.10,19 Many enzymes can already tolerate higher organic solvent concentrations than the corresponding microorganisms.20 Furthermore, engineering enzyme stability towards organic solvents is less challenging than evolving microorganisms to withstand the same concentration of organic solvents.17,21 Therefore, in vitro butanol production emerges as a very promising alternative to traditional butanol fermentation. Thus, in this work, we focused on the engineering of PDC towards higher thermal stability for prospective ethanol production in thermophilic microorganisms and improved stability in butanol for applications in in vitro, artificial butanol synthesis.
Prior to engineering a PDC, we characterised two new yeast PDCs from Candida glabrata (CgPDC) and Zygosaccharomyces rouxii (ZrPDC), as well as other PDCs reported in the literature, to determine the most suitable template. As presented in Table 1, the two new PDCs from the yeast are not thermostable and showed typical substrate cooperativity, as described for other yeast PDCs.22 Characterisation of bacterial PDCs showed similar findings to the previous report, with PDC from Acetobacter pasteurianus (ApPDC) demonstrating the highest kinetic stability (T501 h) and melting temperature (Tm).22 T501 h is defined as the temperature at which 50% of the enzymes remained active after 1 h incubation, while Tm is defined as the temperature at which 50% of the enzymes are in an unfolded state.23 In general, bacterial PDCs showed higher activity at 25 °C and overall stability (T501 h and Tm) than newly characterised CgPDC and ZrPDC.
| Enzyme | Vmax (U mg−1) | Km (mM) | T501 ha (°C) | Tmb (°C) |
|---|---|---|---|---|
a T501 h is defined as the temperature at which the enzyme gives 50% remaining activity after 1 h incubation (Fig. S1).23b Melting temperature was determined using Thermofluor assay (Fig. S1).c The Hill coefficient was determined according to the equation: 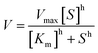 . Cg is Candida glabrata, Zr is Zygosaccharomyces rouxii, Ap is Acetobacter pasteurianus, Zm is Zymomonas mobilis and Zp is Zymobacter palmae. . Cg is Candida glabrata, Zr is Zygosaccharomyces rouxii, Ap is Acetobacter pasteurianus, Zm is Zymomonas mobilis and Zp is Zymobacter palmae. |
||||
| CgPDC | 22.2 ± 0.4 | 9.1 ± 0.2 h: 2.02c | 49.6 | 55.5 |
| ZrPDC | 16.7 ± 0.4 | 11.6 ± 0.4 h: 2.39c | 42.9 | 50.5 |
| ApPDC | 80.3 ± 0.6 | 1.9 ± 0.1 | 64.9 | 70.0 |
| ZmPDC | 92.5 ± 0.7 | 1.3 ± 0.1 | 62.4 | 66.5 |
| ZpPDC | 79.7 ± 0.4 | 0.8 ± 0.1 | 61.2 | 65.0 |
| 5NPU7 | 20.7 ± 0.8 | 4.8 ± 0.4 h: 1.28c | 51.9 | 55.5 |
| 5TMA8 | 87.0 ± 0.7 | 1.8 ± 0.1 | 54.7 | 59.5 |
| PDC-Var. 1 | 91.7 ± 0.7 | 2.1 ± 0.1 | 73.1 | 77.0 |
| PDC-Var. 2 | 71.6 ± 0.7 | 1.5 ± 0.1 | 78.5 | 82.0 |
Another PDC that was reported to have improved stability, called 5TMA (Rosetta-designed ZmPDC), was also characterised in this study to fill the gap remaining due to the lack of data on activity and kinetic stability in the original study.8 It was claimed that 5TMA did not show any loss of molar ellipticity, measured by circular dichroism and that it was stable at 60 °C, observed by DIC microscopy. However, our kinetic stability studies based on activity suggested otherwise. We observed that 5TMA was less stable than wild-type (WT) ZmPDC, the parental PDC, in respect to T501 h and Tm (Table 1). We could not explain this major discrepancy, except that we used a buffer of pH 6.5, while the previous work used pH 7.5.8 It is worth mentioning that during our T501 h experiment, we observed that 5TMA did not show any aggregation in contrast to other WT PDCs. This could explain why the authors did not observe a phase transition in DIC of 5TMA, but did for WT ZmPDC.
Our previous work on evolving thermostable, branched-chain α-keto acid decarboxylase from Lactococcus lactis (LlKdcA) showed that improved thermostability was also translated to increased stability against isobutanol.24 The direct relationship between thermostability and organic solvent stability has also been described in other enzyme classes.21,25,26 Therefore, to save time and expense in finding PDC variants with increased thermostability and butanol-stability, we developed a simple screening platform to screen only for improved thermostability. For library development, staggered extensions process (StEP) was utilised.27 StEP has been shown in previous work to be a robust and relatively easy approach for increasing thermostability.27 Three PDCs from Zymomonas mobilis (ZmPDC), Zymobacter palmae (ZpPDC) and ApPDC with relatively similar stability and activity, were chosen as parental templates.
After screening about 800 colonies, we obtained several variants that showed improved thermostability in comparison with our control (WT ApPDC). The most thermostable variant (renamed PDC-Var. 1) was selected, and the mutations were determined by DNA sequencing. PDC-Var. 1 showed the highest sequence similarity to ApPDC (98%), bearing 13 substitutions (Fig. S2 and S3†). In comparison with ApPDC, the most thermostable PDC reported to date, PDC-Var. 1 exhibited increases of 8.2 °C and 7.0 °C in T501 h and Tm, respectively. Surprisingly, PDC-Var. 1 also showed improved activity in comparison with WT ApPDC, without significant change in Km (Table 1). This indicated that increased thermostability is not always accompanied by a decrease in activity.28 We later used this new variant as a new template to further increase its thermostability.
In a previous work with LlKdcA, we successfully improved its thermal and isobutanol-stability by introducing seven amino acid substitutions (Var. 7M.D).24 As ApPDC and LlKdcA belong to the same α-keto acid decarboxylase family, we intended to transfer the seven beneficial substitutions of Var. 7M.D to our new PDC-Var. 1. A similar screening procedure was applied. From the seven libraries, only one variant showed improved stability. We named the new variant PDC-Var. 2, which displays a change at position 441 of PDC-Var. 1 from isoleucine to valine (Fig. S3†).24 PDC-Var. 2 exhibited T501 h and Tm of 78.5 °C and 82.0 °C, respectively, meaning an improvement of 5.4 °C and 5.0 °C compared with PDC-Var. 1 or an increase of 13.6 °C and 12.0 °C compared with WT ApPDC.
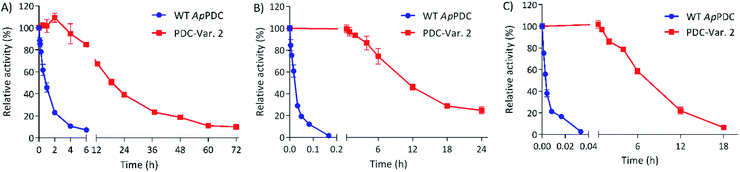
To check the stability of PDC-Var. 2 in comparison with WT ApPDC for ethanol fermentation applications via thermophilic microorganisms, both enzymes were incubated at 65 °C, 70 °C and 75 °C (Fig. 1). At 65 °C, WT ApPDC showed a first-order deactivation kinetics, with a half-life of 57 min, while PDC-Var. 2 showed a half-life of 18 h. This improvement represented an almost 19-fold increase. At higher temperatures, the stability effect of PDC-Var. 2 became even more apparent. At 70 °C, WT ApPDC completely lost its activity after 10 min of incubation, while PDC-Var. 2 retained 25% of initial activity after 24 h incubation. At 75 °C, WT ApPDC was completely deactivated after 2 min, while PDC-Var. 2 retained more than 20% of initial activity after 12 h incubation. Based on a first-order kinetic deactivation, WT ApPDC exhibited half-lives of 1.2 min and 10.8 s at 70 °C and 75 °C, respectively. For PDC-Var. 2, the half-lives at 70 °C and 75 °C are 10.7 h and 7.3 h, respectively. As such, the improved stability of PDC-Var. 2 corresponded to a 522-fold and a 2438-fold increased half-life at 70 °C and 75 °C (Fig. 1), respectively. PDC-Var. 2 also demonstrated improved stability towards ethanol by tolerating ethanol up to 40 vol% at 50 °C in comparison with WT ApPDC that showed stability up to only 22 vol% (Fig. S4†). We stress that this new improved characteristic is important for ethanol production in vitro.19
Biotechnological production of butanol is challenging, mainly due to its high toxicity for microbial cells. Until now, no microorganism has been found in nature that can grow at butanol concentrations higher than 4 vol%.29 Recently, production schemes were proposed that utilise the solubility limit of butanol in water (9 vol% at 50 °C).10,11 Using solvent-stable enzymes and hands-on in vitro enzyme cascade, cell-free butanol production within a two-phase water/butanol system can be envisioned. Some enzymes from thermophilic microorganisms can already tolerate higher alcohol concentrations, such as aldolase from Saccharolobus solfataricus (previously known as Sulfolobus solfataricus), which was shown to retain full activity in a two-phase system.19
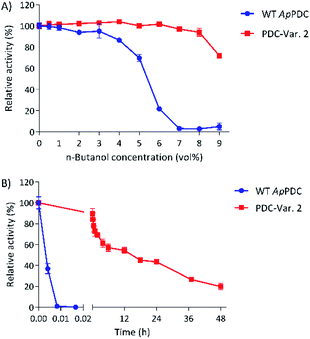
At 50 °C, the most thermostable WT PDC, ApPDC, showed stability up to only 5.6 vol% butanol after incubation for 1 h (Fig. 2A). In 7 vol% butanol, WT ApPDC was completely deactivated. As expected, PDC-Var. 2 demonstrated exceptional stability, by retaining more than 70% of its initial activity in 9 vol% butanol after 1 h incubation at 50 °C (Fig. 2A). Further kinetic stability studies revealed that WT ApPDC was completely deactivated after 30 s incubation at 9 vol% butanol at 50 °C (Fig. 2B). PDC-Var. 2, however, maintained more than 40% of its initial activity after 24 h incubation and 15% of its initial activity after 48 h incubation (Fig. 2B). This corresponds to a half-life of 15 h and thus an over 5000-fold increase in half-life in comparison with WT ApPDC.
When a higher temperature is desired, e.g. 60 °C, as indicated by previous work, WT ApPDC could retain 50% of its activity only up to 2 vol% of butanol at 60 °C, while PDC-Var. 2 started to lose 50% of initial activity in the presence of 8 vol% butanol (Fig. S5†).10,11 Hence, with this new improved process stability and high activity (Table 1), PDC-Var. 2 rises as a better candidate than any PDCs reported to date and holds promise for the application of in vitro butanol production at its solubility limit.
In conclusion, we demonstrated that organic solvent-stable PDC variants could be found only by developing a screening platform focused on improved thermostability. In general, developing a screening platform for improved thermostability is easier than for organic solvent tolerance, as some organic solvents may possess incompatibility issues with enzymatic assays, and general handling is more difficult. We expect our new thermostable PDC variant (PDC-Var. 2) to enhance the feasibility of consolidated bioprocessing approach for ethanol production in cellulolytic thermophilic organisms.3,5 Improved butanol stability of PDC-Var. 2 should drive forward the already developed in vitro butanol production, either via CoA dependent or proline-facilitated aldehyde condensation pathways (Scheme 1).10,11 Additionally, improved thermostability and stability in the presence of butanol of PDC-Var. 2 will facilitate simpler product removal at high temperatures in cell-free butanol synthesis.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the German Research Foundation (DFG) and the Technical University of Munich (TUM) in the framework of the Open Access Publishing Program. Fruitful discussions with Samed Güner and Ioannis Zachos are greatly appreciated. We thank BMBF funding (Grant No. 031L0078F) and COST action CM1303 Systems Biocatalysis.Notes and references
- D. Barnard, A. Casanueva, M. Tuffin and D. Cowan, Environ. Technol., 2010, 31, 871–888 CrossRef CAS PubMed.
- S. Yang, X. Liu and Y. Zhang, in Bioprocessing for Value-Added Products from Renewable Resources: New Technologies and Applications, ed. S. Yang, Elsevier, Amsterdam, 1st edn, 2007, pp. 73–118 Search PubMed.
- D. G. Olson, R. Sparling and L. R. Lynd, Curr. Opin. Biotechnol., 2015, 33, 130–141 CrossRef CAS PubMed.
- M. P. Taylor, K. L. Eley, S. Martin, M. I. Tuffin, S. G. Burton and D. A. Cowan, Trends Biotechnol., 2009, 27, 398–405 CrossRef CAS.
- L. Tian, S. J. Perot, S. Hon, J. Zhou, X. Liang, J. T. Bouvier, A. M. Guss, D. G. Olson and L. R. Lynd, Microb. Cell Fact., 2017, 16, 171 CrossRef.
- A. H. Thompson, D. J. Studholme, E. M. Green and D. J. Leak, Biotechnol. Lett., 2008, 30, 1359–1365 CrossRef CAS PubMed.
- L. Buddrus, E. S. V. Andrews, D. J. Leak, M. J. Danson, V. L. Arcus and S. J. Crennell, Acta Crystallogr., Sect. F: Struct. Biol. Commun., 2018, 74, 179–186 CrossRef CAS PubMed.
- D. W. Sammond, N. Kastelowitz, B. S. Donohoe, M. Alahuhta, V. V. Lunin, D. Chung, N. S. Sarai, H. Yin, A. Mittal, M. E. Himmel, A. M. Guss and Y. J. Bomble, Biotechnol. Biofuels, 2018, 11, 1–13 CrossRef.
- M. Cheng, H. Yoshiyasu, K. Okano, H. Ohtake and K. Honda, PLoS One, 2016, 1–13 Search PubMed.
- S. Reiße, M. Haack, D. Garbe, B. Sommer, F. Steffler, J. Carsten, F. Bohnen, V. Sieber and T. Brück, Front. Bioeng. Biotechnol., 2016, 4, 1–10 CrossRef.
- B. Krutsakorn, K. Honda, X. Ye, T. Imagawa, X. Bei, K. Okano and H. Ohtake, Metab. Eng., 2013, 20, 84–91 CrossRef CAS.
- M. R. Connor and J. C. Liao, Curr. Opin. Biotechnol., 2009, 20, 307–315 CrossRef CAS PubMed.
- M. Uyttebroek, W. Van Hecke and K. Vanbroekhoven, Catal. Today, 2013, 239, 7–10 CrossRef.
- K. Karimi, M. Tabatabaei, I. S. Horváth and R. Kumar, Biofuel Res. J., 2015, 8, 301–308 CrossRef.
- E. I. Lan and J. C. Liao, Metab. Eng., 2011, 13, 353–363 CrossRef CAS PubMed.
- S. Atsumi, A. F. Cann, M. R. Connor, C. R. Shen, K. M. Smith, M. P. Brynildsen, K. J. Y. Chou, T. Hanai and J. C. L. Ã, Metab. Eng., 2008, 10, 305–311 CrossRef CAS PubMed.
- L. H. Reyes, M. P. Almario, J. Winkler, M. M. Orozco and K. C. Kao, Metab. Eng., 2012, 14, 579–590 CrossRef CAS PubMed.
- K. A. Zingaro and E. Terry, Metab. Eng., 2013, 15, 196–205 CrossRef CAS PubMed.
- J. K. Guterl, D. Garbe, J. Carsten, F. Steffler, B. Sommer, S. Reiße, A. Philipp, M. Haack, B. Rühmann, A. Koltermann, U. Kettling, T. Brück and V. Sieber, ChemSusChem, 2012, 5, 2165–2172 CrossRef CAS PubMed.
- S. Torres, M. A. Martínez, A. Pandey and G. R. Castro, Bioresour. Technol., 2009, 100, 896–902 CrossRef CAS PubMed.
- T. Koudelakova, R. Chaloupkova, J. Brezovsky, Z. Prokop, E. Sebestova, M. Hesseler, M. Khabiri, M. Plevaka, D. Kulik, I. K. Smatanova, P. Rezacova, R. Ettrich, U. T. Bornscheuer and J. Damborsky, Angew. Chem., Int. Ed., 2013, 52, 1959–1963 CrossRef CAS PubMed.
- D. Gocke, T. Graf, H. Brosi, I. Frindi-Wosch, L. Walter, M. Müller and M. Pohl, J. Mol. Catal. B: Enzym., 2009, 61, 30–35 CrossRef CAS.
- A. S. Bommarius and M. F. Paye, Chem. Soc. Rev., 2013, 42, 6534–6565 RSC.
- S. Sutiono, J. Carsten and V. Sieber, ChemSusChem, 2018, 11, 3335–3344 CrossRef CAS PubMed.
- E. Vazquez-Figueroa, V. Yeh, J. M. Broering, J. F. Chaparro-Riggers and A. S. Bommarius, Protein Eng., Des. Sel., 2008, 21, 673–680 CrossRef CAS PubMed.
- M. T. Reetz, P. Soni and L. Ferna, Chem. Commun., 2010, 46, 8657–8658 RSC.
- H. Zhao, L. Giver, Z. Shao, J. A. Affholter and F. H. Arnold, Nat. Biotechnol., 1998, 16, 258–261 CrossRef CAS PubMed.
- B. P. L. Wintrode and F. H. Arnold, in Advances in Protein Chemistry, ed. F. H. Arnold, Academic Press, 1st edn, 2001, vol. 55, pp. 161–225 Search PubMed.
- M. Kanno, T. Katayama, H. Tamaki, Y. Mitani, X. Y. Meng, T. Hori, T. Narihiro, N. Morita, T. Hoshino, I. Yumoto, N. Kimura, S. Hanada and Y. Kamagata, Appl. Environ. Microbiol., 2013, 79, 6998–7005 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9ra06251c |
| ‡ Current address: Pharma Stulln GmbH, Werksstraße 3, 92551 Stulln. |
| This journal is © The Royal Society of Chemistry 2019 |