Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
DNA photo-cross-linking using a pyranocarbazole-modified oligodeoxynucleotide with a D-threoninol linker†
Kenzo
Fujimoto
 *a,
Tsubasa
Yamaguchi
a,
Takahiro
Inatsugi
b,
Masahiko
Takamura
*a,
Tsubasa
Yamaguchi
a,
Takahiro
Inatsugi
b,
Masahiko
Takamura
 b,
Isao
Ishimaru
b,
Ayako
Koto
b and
Shigetaka
Nakamura
b,
Isao
Ishimaru
b,
Ayako
Koto
b and
Shigetaka
Nakamura
 a
a
aDepartment of Advanced Institute Science and Technology, Japan Advanced Institute of Science and Technology, Asahidai 1-1, Nomi, Ishikawa 923-1292, Japan. E-mail: kenzo@jaist.ac.jp
bAdvanced Materials Research Laboratory, Advanced Technology Research Department, In statute of Surface Science and Technology, NICCA CHEMICAL CO., LTD., 23-1, 4-Chome, Bunkyo, Fukui-City, 910-8670, Japan
First published on 27th September 2019
Abstract
An alternative photo-cross-linker having a D-threoninol skeleton instead of the 2′-deoxyribose backbone in 3-cyanovinylcarbazole (CNVK) was investigated to improve the photoreactivity of photo-cross-linkers; the photo-cross-linking rate of 3-cyanovinylcarbazole with D-threoninol (CNVD) was found to be greater than that of CNVK. Therefore, in this study, a novel photo-cross-linker having pyranocarbazole (PCX) and D-threoninol instead of the 2′-deoxyribose backbone in PCX (PCXD) was developed. The PCXD in double-stranded DNA photo-cross-linked to a pyrimidine base at the −1 position of a complementary strand similar to PCX. Furthermore, the photoreactivity of PCXD was significantly higher than that of PCX. The introduction of D-threoninol improved the reactivity of pyranocarbazole to cytosine, the use of PCXD may extend the applicability of the photo-cross-linking reaction for DNA manipulation. In particular, this novel photo-cross-linker can contribute to the photochemical regulation of gene expression or biological events in a living cell.
Introduction
Photo-cross-linking reactions between biomolecules are used for various applications such as screening antigen interactions1 and improving detection sensitivity2 and the stability of biomolecular complexes.3 In particular, the use of DNA photo-cross-linking in the formation of a thymine dimer induced by UV-irradiation4 and interstrand photo-cross-linking with psoralen5 has been reported. In recent years, with the development of nucleic acid medicine and DNA nanotechnology, these have been used for the photochemical regulation of an antisense effect,6 improvement in stability of a DNA nanostructure,7 and other applications. Many photo-cross-linkers such as bromouracil8 and benzophenone9 have also been reported. Among these, DNA photo-cross-linking via [2 + 2] photocycloaddition by psoralens5 and coumarins10 is useful for detection, manipulation, and regulation of nucleic acids because photoreversible manipulation is possible. Compared to the enzymatic method, photo-cross-linking can be used under a wide range of conditions without the addition of reagents. Although the intracellular usage of photo-cross-linking has already been reported, these require UV-irradiation, which pose a limitation for use in living cells because of phototoxicity. We report pyranocarbazole (PCX) as a photo-cross-linker that can photo-cross-link to pyrimidine in complementary DNA or RNA strand under visible light.11 It was anticipated that his photo-cross-linker would accelerate the intracellular application of nucleic acid photo-cross-linking such as photochemical regulation of gene expression12 and detection of RNA strand;13 however, photo-cross-linking using PCX to cytosine requires photoirradiation for 1 min, and it is necessary to speed up this process. Besides, the ribose backbone, D-threoninol backbone,14 and serinol backbone15 have been reported as the backbone of the artificial nucleic acid. It was determined that 3-cyanovinylcarbazole modified D-threoninol (CNVD)16 considerably accelerated the photo-cross-linking reaction with cytosine using the D-threoninol backbone (PCXD) (Fig. 1).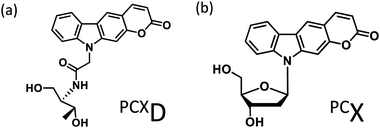 | ||
| Fig. 1 Ultrafast photo-cross-linker (a) pyranocarbazole with D-threoninol (PCXD); (b) pyranocarbazole (PCX). | ||
Results and discussion
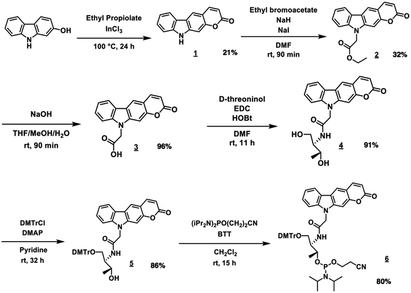
PCXD was successfully synthesized according to the reaction scheme shown in Fig. 2. The PCXD was phosphoramidited using a general method17 after 5′ DMTr protection to synthesize the oligodeoxynucleotide (ODN) containing PCXD. The 9 mer ODN which has A, T, G, C having deoxyribose and PCXD having D-threoninol was synthesized with an automated DNA synthesis machine, and then, it was deprotected using 28% ammonium solution with a general method. After HPLC purification, it was analyzed using Matrix assisted Laser Desorption/Ionization (MALDI) analysis (Table 1).First, we demonstrated the photo-cross-linking of PCXD in double-stranded DNA. The duplex containing PCXD was irradiated with 400 nm and analyzed using ultrahigh-performance liquid chromatography (UPLC). In the case of the pyrimidine base at the −1 position on the complementary ODN (cODN) strand, new peak identical to the photodimer appeared after photoirradiation, although such new peak did not appear in the case of the purine base at the −1 position on the cODN strand. The new peak was identified to photoadduct of PCXD-ODN(C) and its complementary strand (found = 5591.2, calcd [M + H]+ = 5591.0) by MALDI-TOF-MS analysis. This suggests that the photo-cross-linking reaction occurred only in the case of the pyrimidine base at the −1 position on the cODN strand.
| Entry | Sequence (5′–3′) | Calculated [M + H]+ | Found |
|---|---|---|---|
| PCXD-ODN(C) | TGCGPCXDCCGT | 2843.53 | 2843.60 |
| PCX-ODN(C) | TGCGPCXCCGT | 2823.31 | 2823.81 |
| PCXD-ODN(T) | TGCAPCXDCCGT | 2827.84 | 2828.94 |
| PCX-ODN(T) | TGCAPCXCCGT | 2807.52 | 2807.43 |
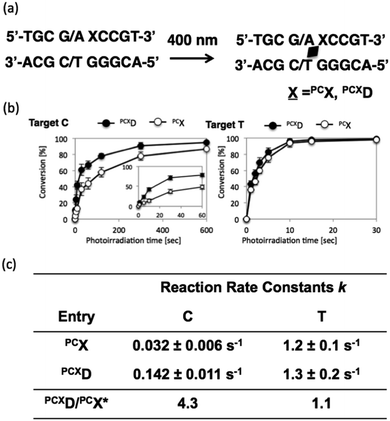
To analyze the photoreactivity of PCXD, the time course of the photoreaction was monitored, and then, the reaction rate constant was analyzed with the assumption of first-order reaction kinetics. As shown in Fig. 3, the reaction rate constant of PCXD with cytosine is 4.3-fold larger than that of PCX, suggesting that the relatively flexible D-threoninol skeleton elevates the accessibility of the reactive double bond to the cytosine base on the complementary strand. The same effect was observed in the case of thymine as the target base of PCXD; the reaction rate constant was 1.1-folds larger than that of PCX. The difference in the reactivity between the thymine and the cytosine bases decreased and the photoreactivity enhanced compared with PCX, which indicates that the use of PCXD extends the applicability of the photo-cross-linking reaction for DNA manipulation.
To evaluate the duplex stability of ODN(PCXD), the melting temperature (Tm) of the duplex consisting of ODN(APCXD) and ODN(GT) was evaluated and compared with the duplex including PCX. The sigmoidal melting curve of ODN(PCXD)/ODN(T) duplex was observed, which suggests ODN(PCXD) and cODN(GT) formed a duplex structure. Then, we calculated the thermodynamic parameter from van't Hoff plots.18 The ΔS through ODN(PCXD)/ODN(T) hybridization was smaller than that of ODN(PCX)/ODN(T), suggesting that a relatively flexibility structure of PCXD inhibited the entropic loss via hybridization (Table 2). Therefore, PCXD has a higher flexibility than PCX, and it induces a higher photo-cross-linking ability in cytosine than PCX. In addition, PCX and PCXD containing pyranocarbazole moieties have lower Tm values compared to CNVK and CNVD containing 3-cyanovinylcarbazole moiety. When the polarity of pyranocarbazole (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P = 1.12) and 3-cyanovinylcarbazole (log
P = 1.12) and 3-cyanovinylcarbazole (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P = 1.35) was examined,19 it was found that pyranocarbazole has a smaller log
P = 1.35) was examined,19 it was found that pyranocarbazole has a smaller log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P and is hydrophilic. Therefore, it is considered that the Tm value of the duplex containing 3-cyanovinylcarbazole is high and the entropy loss is large because of the stacking between bases and hydrophobic interactions.
P and is hydrophilic. Therefore, it is considered that the Tm value of the duplex containing 3-cyanovinylcarbazole is high and the entropy loss is large because of the stacking between bases and hydrophobic interactions.
| Entry | T m [°C] | ΔΔG37 (kcal mol−1) | ΔH (kcal mol−1) | ΔS (cal mol−1 K−1) |
|---|---|---|---|---|
| PCXD | 26 | −7.4 | −16.7 | −30.0 |
| PCX | 33 | −0.58 | −21.6 | −67.7 |
| CNVD | 29 | −7.3 | −33.8 | −85.6 |
| CNVK | 34 | −7.5 | −63.6 | −181.1 |
To evaluate the effects of the surrounding bases on the photo-cross-linking of ODNs containing PCXD, double strand with all variations of base pairs at the −1, +1 position of PCXD and four different bases on the counter position to PCXD in cODN were prepared; the conversion after 10 s of 400 nm irradiation was evaluated by UPLC. As shown in Fig. 4, in the case of T at the Z position in cODN, over 90% of the photo-cross-linking was obtained with 400 nm irradiation for 10 s. In the case of C at the Z position, photo-cross-linking was observed. However, in the case of A or G at the Z position, photo-cross-linking was not observed. These results indicate that the photo-cross-linking of PCXD has similar pyrimidine selectivity as those of CNVK, CNVD, and PCX. In addition, some differences in the reactivity were not observed among these duplexes, which indicated that the mechanism of effects such as local duplex stability and local electrostatic environment around the pyranocarbazole moiety remained does not affect the photo-cross-linking.
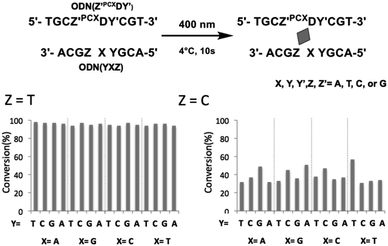 | ||
| Fig. 4 Effect of surrounding base of PCXD on its photoreactivity in dsDNA. 10 μM DNA in 50 mM cacodylate buffer (pH 7.4) containing 100 mM NaCl was 400 nm-irradiated for 10 s at 4 °C. | ||
Experimental
General
1H NMR spectra were measured on a Bruker AVANCE III 400 system. Mass spectra were recorded on a Voyager PRO-SF, Applied Biosystems and HRMS were measured on a Solarix-JA (Bruker) HPLC was performed on a Chemcosorb 5-ODS-H column with JASCO PU-980, HG-980-31, DG-980-50 system equipped with a JASCO UV 970 detector at 260 nm. Reagents for synthesis for compound 1 to 5 were purchase from Aldrich, Wako, and TCI. Phosphoramidites of PCXD were synthesized by the procedure as follows. Other phosphoramidite reagents for the DNA sysnthesizer such as A, G, C, T-β-cyanoethyl phosphoramidite, and CPG support were purchased form Glen research. Other oligonucleotides were purchased from Fasmac (Japan) and used without farther purification. The synthesized ODN having deoxyribose backbone except PCXD.Synthetic procedures
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 18 mL) and stirred for 90 min at room temperature. After the addition of 1 N HCl (150 mL), reaction mixture was extracted with EtOAc (150 mL) and washed with 1 N HCl. Organic layer was dried over MgSO4 and evaporated to afford 3 (yellow solid, 1.2 g, 4.4 mmol, 96%). 1H NMR (400 MHz, DMSO-d6) 13.17 (s, 1H), 8.50 (s, 1H), 8.19 (d, 2H, J = 9.2 Hz), 7.65 (s, 1H), 7.59 (d, 1H, J = 8.4 Hz), 7.50 (m, 1H), 7.30 (t, 1H, J = 6.8 Hz), 6.36 (m, 1H), 5.30 (d, 2H). 13C NMR (100 MHz, DMSO-d6) 169.8, 160.6, 152.8, 145.4, 143.0, 141.6, 126.5, 121.9, 120.4, 120.3, 120.1, 120.0, 112.0, 111.9, 109.7, 96.4, 44.2. ESI-FT-ICR MS(HRMS) calcd for [M + H]+: 294.0760, found: 294.0762.
1, 18 mL) and stirred for 90 min at room temperature. After the addition of 1 N HCl (150 mL), reaction mixture was extracted with EtOAc (150 mL) and washed with 1 N HCl. Organic layer was dried over MgSO4 and evaporated to afford 3 (yellow solid, 1.2 g, 4.4 mmol, 96%). 1H NMR (400 MHz, DMSO-d6) 13.17 (s, 1H), 8.50 (s, 1H), 8.19 (d, 2H, J = 9.2 Hz), 7.65 (s, 1H), 7.59 (d, 1H, J = 8.4 Hz), 7.50 (m, 1H), 7.30 (t, 1H, J = 6.8 Hz), 6.36 (m, 1H), 5.30 (d, 2H). 13C NMR (100 MHz, DMSO-d6) 169.8, 160.6, 152.8, 145.4, 143.0, 141.6, 126.5, 121.9, 120.4, 120.3, 120.1, 120.0, 112.0, 111.9, 109.7, 96.4, 44.2. ESI-FT-ICR MS(HRMS) calcd for [M + H]+: 294.0760, found: 294.0762.
Synthesis of oligodeoxynucleotide
The oligonucleotides having PCXD were synthesized by a 3400 DNA synthesizer (Applied Biosystems) and purified by a HPLC system (JASCO PU-980, HG-980-31, DG-980-50, UV-970) with an InertSustainTM C18 column (GL Science, 5 μm, 10 × 150 mm). Synthesized oligonucleotides was identified by MALDI-TOF-MS.Photoirradiation and UPLC analysis
Photoirradiation was performed with an LED lamp (OminiCure, 400 nm, 8600 mW cm−2) on thermal cycler. The photoirradiated samples were analyzed with a UPLC system (Aquity, waters) equipped with BEH Shield RP18 column (1.7 μm, 2.1 × 50 mm, elution was with 0.05 M ammonium formate containing 1–10% CH3CN, linear gradient (10 min) at a flow rate of 0.4 mL min−1, 60 °C).Thermodynamic analysis of the hybridization
Thermodynamic parameters were obtained according to a method in the literature: where TM is the melting temperature of duplex, ΔS° is entropy and ΔH is the entropy of duplex formation, respectively. R is the gas constant and CT is the total strand concentration. TM was measured at various concentrations of duplex in 50 mM Na-cacodylate buffer (pH 7.4) containing 100 mM NaCl by a spectrophotometer (V-630bio, Jasco) equipped with a temperature controller.Conclusions
The PCXD in double-stranded DNA photo-cross-linked to the pyrimidine base at the −1 position of the complementary strand was similar to that for PCX, and the photoreactivity of PCXD was significantly higher than that of PCX. The photo-cross-linking rate of PCXD having D-threoninol and cytosine was faster than that of PCX and cytosine, the applicability of the photo-cross-linking reaction for DNA manipulation may be expanded by the use of PCXD. We have already reported the importance of photo-cross-linking rate in photochemical antisense method in living cell and RNA FISH to E. coli 16S rRNA with a secondary structure. Therefore, this novel photo-cross-linker would contribute to the photoregulation of gene expression or biological events in living cells.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This study was supported by Grant-in-Aid for Scientific Research (B) (No. 17H03085) of Japan Society of Promotion of Science.Notes and references
- D. S. Ziemianowicz, D. Ng, A. B. Schryvers and D. C. Schriemer, J. Proteome Res., 2019, 18, 934–946 CrossRef CAS.
- G. Fu and Z. Dai, Talanta, 2012, 97, 438–444 CrossRef CAS.
- M. M. Felczak, J. M. Sage, K. Huper-Kocurek, S. Aykui and J. M. Kaguni, J. Biol. Chem., 2016, 291, 4803–4812 CrossRef CAS.
- D. S. Goodsell, Oncologist, 2001, 6, 298–299 CrossRef CAS.
- F. P. Gasparro, P. A. Havre, G. A. Olack, E. J. Gunther and P. M. Glazer, Nucleic Acids Res., 1994, 22, 2845–2852 CrossRef CAS.
- M. Higuchi, A. Kobori, A. Yamayoshi and A. Murakami, Bioorg. Med. Chem., 2009, 17, 475–483 CrossRef CAS.
- A. Rajendran, M. Endo, Y. Katsuda, K. Hidaka and H. Sugiyama, J. Am. Chem. Soc., 2011, 133(37), 14488–14491 CrossRef CAS PubMed.
- C. D. Churchil, L. A. Eriksson and S. D. Wetmore, J. Phys. Chem. B, 2016, 120, 1195 CrossRef.
- K. Nakatani, T. Yoshida and I. Saito, J. Am. Chem. Soc., 2002, 124, 2118 CrossRef CAS.
- H. Sun, H. Fan, H. Eom and X. Peng, ChemBioChem, 2016, 17, 2046–2053 CrossRef CAS.
- K. Fujimoto, S. Sasago, J. Mihara and S. Nakamura, Org. Lett., 2018, 20(10), 2802–2805 CrossRef CAS.
- K. Fujimoto, H. Yung and S. Nakamura, Chem.–Asian J., 2019, 14, 1912–1916 CrossRef CAS.
- K. Fujimoto, M. Hashimoto, N. Watanabe and S. Nakamura, Bioorg. Med. Chem. Lett., 2019, 29, 2173–2177 CrossRef CAS.
- H. Asanuma, T. Takarada, T. Yoshida, D. Tamura, X. Liang and M. Komiyama, Angew. Chem., Int. Ed., 2001, 40, 2671–2673 CrossRef CAS.
- K. Murayama, Y. Tanaka, T. Toda, H. Kashida and H. Asanuma, Chemistry, 2013, 19, 14151–14518 CrossRef CAS.
- T. Sakamoto, Y. Tanaka and K. Fujimoto, Org. Lett., 2015, 17, 936–939 CrossRef CAS.
- S. L. Beaucage and M. H. Caruthers, Tetrahedron Lett., 1981, 22, 1859–1862 CrossRef CAS.
- J. L. Mergny and L. Lacroix, Oligonucleotides, 2003, 13, 515–537 CrossRef CAS.
- OECD Guideline for testing of chemicals 117, http://www.oecd.org/chemicalsafety/risk-assessment/1948177.pdf Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9ra06145b |
| This journal is © The Royal Society of Chemistry 2019 |