Open Access Article
Open Access ArticleThe exogenous delivery of microRNA-449b-5p using spermidine-PLGA nanoparticles efficiently decreases hepatic injury†
Fengli Hu‡
a,
Dongdong Yang‡b,
Bo Qiana,
Shengjie Fana,
Qiankun Zhub,
Haiyang Renb,
Xiaodong Lib and
Bo Zhai *b
*b
aDepartment of Gastroenterology, The Fourth Affiliated Hospital of Harbin Medical University, Nangang District, Harbin, 150001, China
bDepartment of Surgical Oncology and Hepatobiliary Surgery, The Fourth Affiliated Hospital of Harbin Medical University, No. 37, Yiyuan Street, Nangang District, Harbin, 150001, China. E-mail: zhaibo@hrbmu.edu.cn; Fax: +86 451 8257 6777; Tel: +86 451 8257 6777
First published on 30th October 2019
Abstract
A notable liver ischemia/reperfusion (I/R) injury is observed during liver transplantation, shock, trauma and other systemic diseases. The main aim of the present study was to evaluate the fact that HMGB1 acts as an early mediator of inflammation in hepatic injury and the potential of the miR-449b-5p mimic in the restoration of liver disorders. Herein, a miR-449b-5p-loaded spermidine/PLGA nanoparticle system was successfully formulated to improve the systemic delivery and performance of encapsulated miRNA. The major findings of the present study were as follows: (i) the HMGB1 levels were elevated upon the occurrence of I/R in vitro and in vivo; (ii) the inhibition of HMGB1 prevented the spread of inflammation; (iii) miR-449b-5p (PN-miR mimic) increased the cell viability of hepatic cells and decreased cell apoptosis; and (iv) the protective ability of the PN-miR mimic was attributed to the inhibition of the pNF-κB and p-p65 pathways. Compared to the case of the I/R group, the serum AST and ALT levels were significantly reduced in the group treated with miR-449b-5p (PN-miR mimic), indicating the extent of reduction in liver inflammation. The present study highlighted the importance of miR-449b-5p in the treatment of hepatic injury and could serve as a guide to effectively attenuate liver disorders. The application of the proposed nanoparticle system in the systemic delivery of miR-449b-5p further enhances the prospect of this treatment strategy.
Introduction
Liver ischemia reperfusion (I/R) is a pathophysiological event of hypoxic organ damage when blood flow and oxygen delivery return to the organ.1 Episodes of I/R are usually observed during liver transplantation, liver surgery, trauma, and hemorrhagic shock and in medical conditions (such as heart failure or respiratory failure) where blood flow to the liver is low or reduced.2 This leads to the development of a series of biochemical and physiological changes. In the case of ischemia, metabolic reactions damage mitochondrial functions and thereby injure the liver cells; on the other hand, in the case of reperfusion, the generation of reactive oxygen species (ROS) from injured tissues will activate inflammatory cascades and cell apoptosis;3,4 the cellular damage caused by I/R will result in the activation of several inflammatory pathways, which will result in the upregulation of proinflammatory cytokines (TNF-α and IFN-γ), nitric oxide synthase and neutrophil accumulation.5 Moreover, several histopathological changes can be observed including vacuolization, cellular disruption, swelling and hepatocellular necrosis. Therefore, an effective strategy should be developed for the treatment of hepatic injury.6High-mobility group box 1 (HMGB1) has recently been identified as a key link between the hepatic damage to cells and the activation of inflammatory cytokines in the body.7 HMGB1 is a conserved non-histone nuclear chromosomal protein.8 HMGB1 is a proinflammatory mediator that plays an important role in nuclear organization, replication of DNA, DNA repair and the regulation of gene expression. Therefore, it is related to various diseases including cancer (breast and prostate), Alzheimer's disease and arthritis.9,10 HMGB1 consists of 215 amino acids with two box structures (a N-terminal portion as the A-box and a central portion as the B-box). The B-box holds proinflammatory properties, whereas the A-box does not have inflammatory properties; however, it can attenuate the inflammatory cascade.11 HMGB1 is known to be released from damaged tissues and immune cells, such as macrophages or dendritic cells, after stimulation by TNF-α, IL-1 or other oxidative stressors;12 multiple receptors, such as RAGE or Toll-like receptors, would coordinate with HMGB1 and are therefore implicated in multiple diseases. Overall, HMGB1 plays an important role in conditions associated with the hepatic I/R injury; this makes it a potential target in the treatment of hepatic injury.13
MicroRNAs (miRNAs) are small non-coding RNAs comprising 20–24 nucleotides, which play a vital role in controlling the expressions of genes by binding to the 3′-untranslated region of the respective mRNA that suppresses translation.14 miRNAs control various cellular processes such as cell proliferation, differentiation, viability, and apoptosis; therefore, significant changes have been observed in the levels of miRNAs during diseases including autoimmune diseases, cardiovascular diseases and liver diseases.15,16 Studies have reported that several miRNAs, such as miR-203, miR-34a, miR-149, and miR-101, are involved in the inflammatory responses and apoptosis of cells.17,18 Similarly, miR-449b-5p has been reported to be under-expressed in liver cancer and inhibits cell proliferation and cell migration.19 The 3′-UTR of HMGB1 contains the site for miR-449b-5p binding, and therefore, miR-449b-5p has been predicted to be involved in the hepatic I/R injury by controlling HMGB1 and can be a potential target in the effective treatment of liver diseases.20
However, unmodified miRNA is highly unstable in the systemic circulation due to its enzymatic degradation, resulting in its poor therapeutic efficacy. The excessive accumulation of miRNA in vital organs might interfere with normal cell signaling pathways, resulting in little success of the miRNA-based therapy.21 These limitations could be alleviated by the application of a drug delivery system for the systemic delivery of miRNAs.22 In this study, we selected a USFDA-approved PLGA polymer that provided excellent stability to the loaded bioactive therapeutics, biodegradability, sustained release of the loaded therapeutics and surface functionalization of the nanoparticle (NP) system.23,24 Previously, miRNA was complexed with spermidine, and the physical complexes were loaded onto the core of the PLGA NPs. The two-way loading of miRNA onto the nanoparticle system is expected to significantly enhance the stability and systemic delivery of the loaded therapeutics.25
Overall, herein, miRNA-449b-5p was loaded onto a spermidine/PLGA NP system to enhance the therapeutic efficacy of miRNA-449b-5p in hepatic injury; in the present study, we investigated the effect of the correlation between miRNA-449b-5p and HMGB1 on the control of the hepatic I/R injury. Detailed physicochemical studies were performed to characterize the carrier system, followed by biological assays to investigate cell viability and apoptosis. Western blot assays were performed to evaluate the effect of the administered miRNA. In vivo rat models were designed to study the mechanism of action of miRNA-449b-5p and measure systemic parameters.
Results and discussion
Preparation and characterization of the PN-miR nanosystem
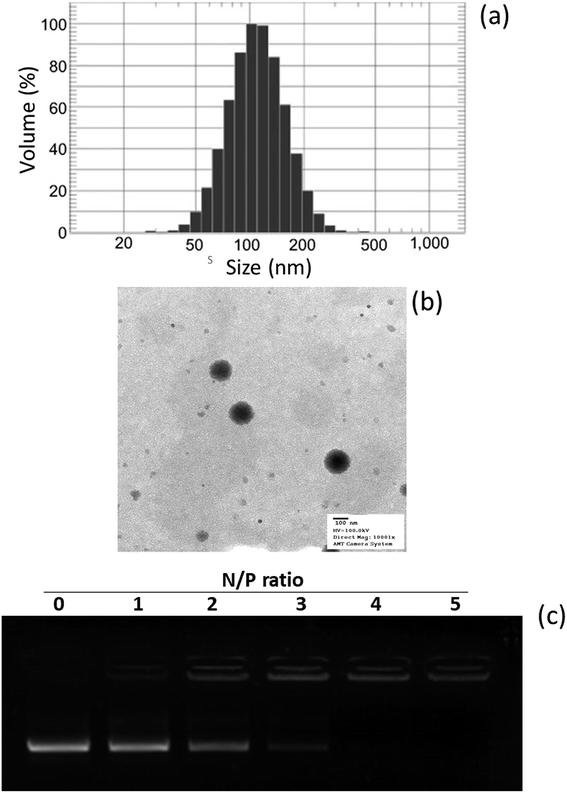
In this study, a novel hybrid nanoparticle system consisting of spermidine and PLGA was prepared to encapsulate miRNA-449b-5p. High-mobility group box 1 (HMGB1) has recently been identified as a key link between hepatic damage to cells and the activation of inflammatory cytokines in the body. The 3′-UTR of HMGB1 contains a site for miR-449b-5p binding; therefore, miR-449b-5p has been predicted to be involved in the hepatic I/R injury by controlling HMGB1 and can be a potential target for the effective treatment of liver injury. miRNA formed a physical complex with spermidine owing to the electrostatic interaction between the negatively charged miRNA and the positively charged spermidine. As the physical complex was unstable in systemic circulation, we loaded the entire physical complex onto the PLGA nanoparticles and made the complex more stable for systemic applications (Fig. 1). The presence of PEG on the surface will lead to enhanced blood circulation properties that would further increase the uptake of the complex by the injured liver cells. The final particle size of PN-miR was observed to be 135.6 ± 1.26 nm (Fig. 2a). The surface charge of the miRNA/spermidine complex was +26.5 ± 1.45 mV, whereas it reversed to −18.4 ± 2.15 mV after the encapsulation of this complex in the PLGA nanoparticles; this indicated the successful entrapment of the miRNA/spermidine complex in the PLGA nanoparticles. The small particle size along with the negatively charged surface enhanced the circulation of particles in the systemic circulation, which had a beneficial effect on the treatment efficacy; the particle morphology was evaluated by transmission electron microscopy (TEM) (Fig. 2b). The particles were perfectly spherical in shape and evenly distributed in the copper grid. The hybrid encapsulation did not change the morphology of the particles. The successful encapsulation of miRNA was confirmed by gel electrophoresis. At the N/P ratio of 5, 100% loading of miRNA was observed, whereas unmodified miRNA (N/P = 0) completely migrated to the opposite electrode (Fig. 2c).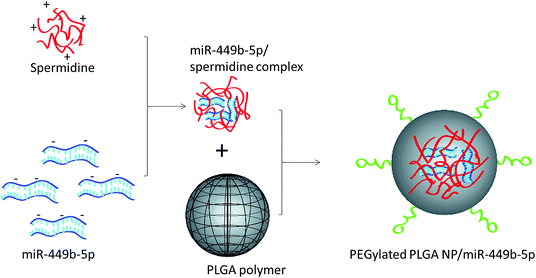 | ||
| Fig. 1 Schematic of the preparation of miRNA-449b-5p-loaded nanoparticles. At first, miRNA-449b-5p was complexed with spermidine, followed by its incorporation in the PEGylated PLGA nanoparticles. | ||
The colloidal stability of PN-miR was studied by the dynamic light scattering (DLS) technique at 4 °C (Fig. S1†). The results clearly indicated the excellent stability of PN-miR for 96 h in PBS buffer. The particle size of PN-miR remained almost the same throughout the study period, whereas the particle size of the spermidine/miR complex significantly increased over time; this indicated the instability of the physical complexes. Similarly, PN-miR exhibited good stability in the culture medium (containing 10% FBS) as compared to the spermidine/miR complex. The results clearly indicate that the encapsulation of the spermidine/miR complex in the PLGA nanoparticles enhances its physical stability, which is one of the important criteria for the successful systemic delivery of miRNAs. The in vitro release of FAM-miRNA was performed under pH 7.4 and pH 5.0 buffer conditions. The results clearly showed a controlled release of miRNA under both the abovementioned pH conditions, with around ∼70% miRNA release at pH 7.4 and ∼80% miRNA release from the nanoparticle system after 48 h. The slight difference in the release pattern of miRNA might be attributed to the diffusion and degradation pattern of the PLGA/spermidine complex under lower pH conditions.
Targeting effect of PN-miR towards HMGB1
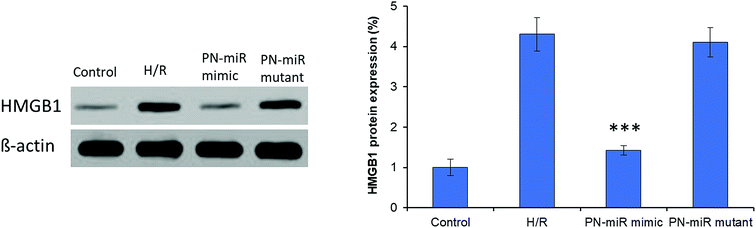
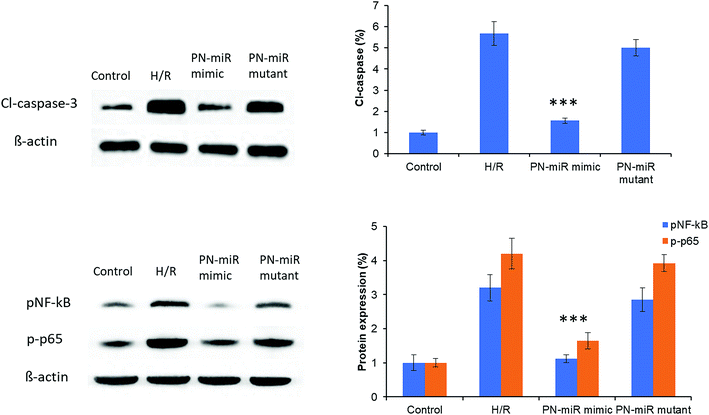
Hypoxia is believed to be the initiating event in the H/R induction, and therefore, its influence on the expression of HMGB1 has been evaluated herein. To evaluate the targeting effect of miR-449b-5p (PN-miR mimic) on HMGB1, LO2 cells were exposed to H/R, the PN-miR mimic and the PN-miR mutant, and western blot assay was performed (Fig. 3). The western blot assay revealed that the transection of the PN-miR mimic significantly decreased the expression of the HMGB1 protein, whereas the PN-miR mutant did not have any effect on the expression of the HMGB1 protein. Note that the level of HMGB1 markedly increases upon the occurrence of H/R as compared to that in the case of the non-treated control; this indicates the potential role of HMGB1 in the hepatic injury; HMGB1 is a typical proinflammatory regulator that controls the NF-κB pathway, which is in turn activated in several medical conditions such as liver transplantation or I/R-related hepatic injuries. The western blot assay showed that the PN-miR mimic effectively down-regulated the expression of HMGB1 in the LO2 cells.Effect of PN-miR on the cellular viability of the LO2 cells
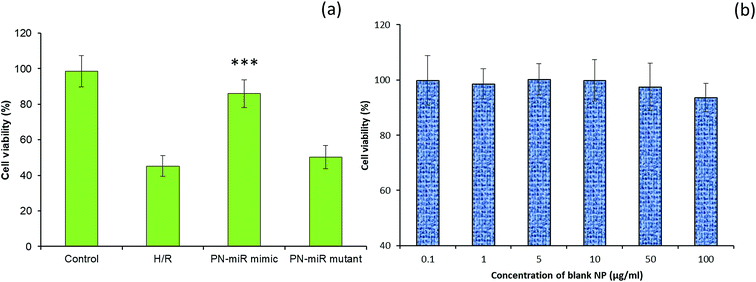
The effect of miR-449b-5p (PN-miR mimic) on the hepatic injury was evaluated via cell viability and apoptosis assay. The cell viability of the LO2 cells was determined via the MTT assay (Fig. 4a). As shown, the induction of H/R significantly decreased the viability of the LO2 cells; this indicated the severe cell death of liver cells. In contrast, the cell viability of the miR-449b-5p (PN-miR mimic)-treated cells was significantly increased as compared to that of the H/R-treated hepatic cells; the cell viability of the miR-449b-5p (PN-miR mimic)-treated cells was almost equal to that of the non-treated control cells, indicating the potential of the proposed system in the treatment of hepatic injury. The importance of the specific sequence of miR-449b-5p was indicated by the fact that the mutant version of miRNA did not have any effect on the cell viability of hepatic cells. To rule out the effect of the nanoparticle system itself, the LO2 cells were treated with blank nanoparticles at different concentrations. As shown, the blank nanoparticles did not exhibit any decrease in the viability of cells at all tested concentrations (Fig. 4b). The lack of any toxicity of the blank nanoparticles suggests the excellent safety and biocompatible properties of the proposed nanosystem, making it suitable for systemic applications.Effect of PN-miR on the apoptosis of LO2 cells
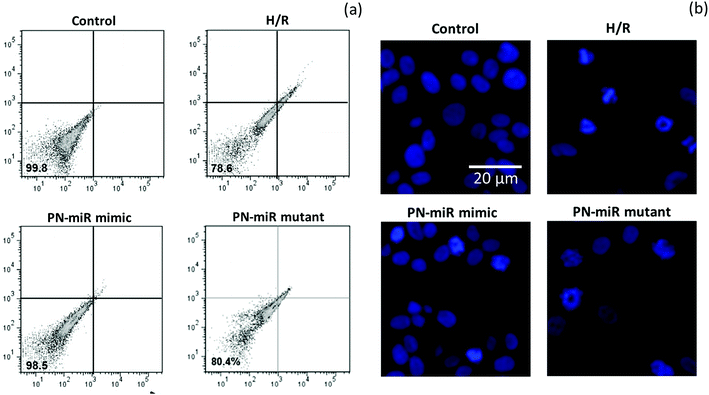
The effect of miR-449b-5p (PN-miR mimic) on the apoptosis of the hepatic cells was determined by an apoptosis assay using a flow cytometer and Hoechst 33342 staining. A flow cytometry analysis was performed after staining the cells with Annexin-V and PI (Fig. 5a); the flow cytometry analysis showed that H/R resulted in more than 20% cell apoptosis, whereas miR-449b-5p (PN-miR mimic) significantly reduced cell apoptosis. The cell viability of the control cells and the PN-miR mimic-treated cells remained similar. The mutant miRNA did not have any effect on cell apoptosis and did not decrease the apoptosis of the hepatic cells. The apoptosis effect was further confirmed by Hoechst staining (Fig. 5b). As shown, the control cells were intact with a typical nucleus; however, H/R resulted in the formation of apoptotic bodies and fewer cells were present, indicating the apoptosis of hepatic cells. The PN-miR mimic restored the cell apoptosis, and the cells were intact with a typical nucleus. Both findings clearly indicated that the cells underwent apoptosis upon the induction of H/R, whereas the PN-miR mimic effectively restored the cell apoptosis and cell death.Molecular mechanism of miR-449b-5p in signaling pathways
The molecular mechanism of miR-449b-5p was evaluated by the western blot assay. At first, cleaved-caspase-3 was evaluated. As shown in Fig. 6, H/R significantly increased the protein level of cl-caspase-3 as compared to the case of the control, whereas the PN-miR mimic significantly reduced the expression of cl-caspase-3; this indicated the efficacy of the PN-miR mimic against cell apoptosis. To further determine the underlying mechanisms of the PN-miR mimic, the protein expression levels of pNF-κB and p-p65 were determined. The results showed that H/R significantly increased the expressions of pNF-κB and p-p65 as compared to the case of the control, whereas miR-449b-5p showed significant down-regulation of the levels of pNF-κB and p-p65 in the LO2 cells. The mutant miRNA did not decrease the protein levels and showed an insignificant difference with the H/R-treated group. Note that NF–B activation led to proinflammatory gene expression in leukocytes and endothelial cells. It has been reported that the endocytosis of HMGB1 initiates a cascade of molecular level events that will activate cell apoptosis. Because HMGB1 translocated from the nucleus to the cytoplasm in the immune cells, it played an important role in pyroptosome formation and the activation of caspase-1;26,27 consistent with this finding, our results showed a reduction in the expressions of pNF-κB and p-p65 upon the treatment of the H/R-induced hepatic cells with the miR-449b-5p mimic.Effect of PN-miR on the ALT and AST levels in a rat model
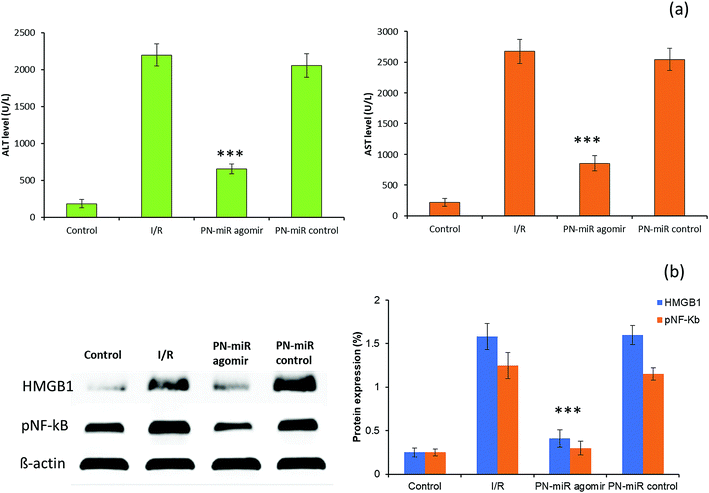
To validate the in vitro findings, studies were performed in a rat model. The serum ALT and AST levels were monitored to evaluate the liver injury (Fig. 7a). It has been reported that ALT is a specific marker for the hepatic I/R injury, whereas AST is a nonspecific marker for hepatic injury. The rat exposed with I/R shows typical pathological symptoms, including a significant increase in the serum ALT and AST levels, which are typical biomarkers of hepatic injury, towards inflammation. Compared to those in the I/R group, the serum AST and ALT levels were significantly reduced in the group treated with miR-449b-5p (PN-miR mimic); this indicated the effective therapeutic potential of the PN-miR mimic. Treatment with the PN-miR mimic attenuated the elevated AST and ALT levels, indicating that the extent of liver inflammation was decreased significantly. The liver was obtained and analyzed for the signaling markers. The results obtained from the animal study were concordant with those obtained by the in vitro western blot studies. The western blot analysis results showed that the HMGB1 and p-NF-kB expressions were significantly higher in the I/R group as compared to those in the non-treated control, whereas miR-449b-5p (PN-miR mimic) resulted in a significant down-regulation of the expressions of these proteins (Fig. 7b). Overall, the results indicated that miR-449b-5p manipulated the hepatic injury by manipulating HMGB1 and regulating the NF-kB pathway.Histological analysis of the liver section
We further evaluated the effect of the PN-miR mimic on the liver injury. The histological analysis showed that I/R caused massive necrosis of the liver section as compared to the case of the non-treated rats (Fig. 8). Compared to the control, the I/R-treated rat showed severe hepatic lesions and signs of mononuclear cell infiltration, indicating liver damage or liver injury. In contrast, the miR-449b-5p (PN-miR mimic)-treated rat markedly reduced the hepatic lesions. Histological changes of similar intensity were observed in the mutant miRNA (mock)-treated rat group. The ability of the PN-miR mimic to prevent pathological changes in the liver suggested the liver targeting ability of miR-449b-5p (PN-miR mimic) and the nanocarrier system in general. The results clearly highlighted the potential of the spermidine/PLGA nanoparticle system, which enabled the efficacious delivery of miRNA to the injured liver.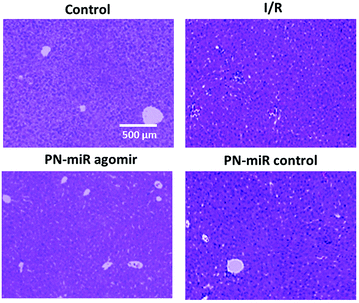 | ||
| Fig. 8 Liver sections were obtained and a H&E staining analysis was performed. The sections were observed for normal liver cell and necrotic cell portions. | ||
HMGB1, originally characterized as a nuclear DNA-binding protein, has also been reported to have an extracellular role in promoting tumor metastasis and inflammation. Wang et al. reported that miR-142-3p could inhibit the HMGB1-mediated NF-kB signaling pathway and reduce inflammation in osteoarthritis.28 Yao et al. showed that miR-26a inhibited the HMGB1 expression and attenuated a cardiac ischemia-reperfusion injury.29 Kim et al. demonstrated that the ascorbic acid inhibition of HMGB1 could improve the survival rate in septic mice by activating the Nrf2/HO-1 signals.30 Sandbothe et al. showed that the microRNA-449 family epigenetically regulated tumor suppressive effects, targeted the specificities of miR-449a, miR-449b, and miR-449c and played crucial roles in HCC progression and metastasis.20 In the current study, we first implied that miR-449b-5p (PN-miR mimic) was a novel miRNA that regulated inflammatory responses by targeting HMGB1 in the LO2 cells. Our findings may broaden and deepen the understanding of the role of miR-449b-5p (PN-miR mimic) in the modulation of biological processes. However, a detailed in vivo experimental analysis is required to further confirm the therapeutic efficacy of the present nanosystem. A toxicity analysis on other vital organs and the fate of the delivery system needs to be conducted in clinical models.
Conclusion
The main aim of the present study was to evaluate the fact that HMGB1 acts as an early mediator of inflammation in hepatic injury and the potential of the miR-449b-5p mimic in the restoration of liver disorders. The miR-449b-5p-loaded spermidine/PLGA nanoparticle system was successfully formulated to improve the systemic delivery and performance of the encapsulated miRNA. The major findings of the present study were as follows: (i) the HMGB1 levels were elevated upon the occurrence of I/R in vitro and in vivo; (ii) the inhibition of HMGB1 prevented the spread of inflammation; (iii) miR-449b-5p (PN-miR mimic) increased the cell viability of hepatic cells and decreased cell apoptosis; and (iv) the protective ability of the PN-miR mimic was attributed to the inhibition of the pNF-κB and p-p65 pathways. Compared to the case of the I/R group, the serum AST and ALT levels were significantly reduced in the group treated with miR-449b-5p (PN-miR mimic); this indicated the extent of reduction of liver inflammation. The present study highlighted the importance of miR-449b-5p in the treatment of hepatic injury and could serve as a guide to effectively attenuate liver disorders. The application of the proposed nanoparticle system in the systemic delivery of miR-449b-5p further enhances the prospect of this treatment strategy.Materials and methods
Preparation of miRNA-449b-5p-loaded spermidine/PLGA complex nanoparticles
Poly(D,L-lactide-co-glycolide)/poly(L-lactide) (PLGA, 75![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 25) and spermidine were purchased from Sigma-Aldrich Chemicals, China. At first, miRNA-449b-5p was reconstituted in RNAse-free water and incubated with spermidine at the N/P ratio of 5
25) and spermidine were purchased from Sigma-Aldrich Chemicals, China. At first, miRNA-449b-5p was reconstituted in RNAse-free water and incubated with spermidine at the N/P ratio of 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 for 1 h with gentle shaking at room temperature. The spermidine/miRNA complex was centrifuged, and the supernatant was discarded. The miRNA complex was reconstituted in 1× PBS. Next, 100 mg of PLGA was dissolved in 2 ml of dichloromethane (DCM) and stirred well. To this organic solution, the spermidine/miRNA complex was added in a drop-wise manner and immediately sonicated at a 50% amplitude for 2 min in an ice-cold bath. The primary emulsion was then poured into a 5% polyvinyl alcohol (PVA) solution and stirred for 2 min. The mixture was again sonicated at a 50% amplitude for 2 min in an ice-cold bath. The secondary emulsion was stirred for 6 h followed by rotary evaporation for 1 h to remove all organic solvents. The optimized nanoparticles were stored at 4 °C until further use.
1 for 1 h with gentle shaking at room temperature. The spermidine/miRNA complex was centrifuged, and the supernatant was discarded. The miRNA complex was reconstituted in 1× PBS. Next, 100 mg of PLGA was dissolved in 2 ml of dichloromethane (DCM) and stirred well. To this organic solution, the spermidine/miRNA complex was added in a drop-wise manner and immediately sonicated at a 50% amplitude for 2 min in an ice-cold bath. The primary emulsion was then poured into a 5% polyvinyl alcohol (PVA) solution and stirred for 2 min. The mixture was again sonicated at a 50% amplitude for 2 min in an ice-cold bath. The secondary emulsion was stirred for 6 h followed by rotary evaporation for 1 h to remove all organic solvents. The optimized nanoparticles were stored at 4 °C until further use.
Characterization of the miRNA-449b-5p-loaded spermidine/PLGA complex nanoparticles
The particle size distribution and surface charge of the optimized nanoparticle were determined by the size and zeta potential of the particles using Nicomp 380 ZLS. The particles were appropriately diluted before the experiment was performed. The experiment was performed at room temperature. The particle morphology was investigated by a transmission electron microscope (TEM) (EM-200CX, JEOL Ltd., Tokyo, Japan). The particles were counterstained with 2% phosphotungstic acid (PTA) and observed using an electron microscope.Gel electrophoresis
The binding and loading efficiency of miRNA to spermidine was evaluated by gel electrophoresis. The spermidine/miRNA complex was prepared as mentioned above and purified. The different N/P ratios of 0, 1, 2, 3, 4, and 5 were used, where 0 is the unmodified miRNA. The spermidine/miRNA complexes with different N/P ratios were loaded onto the 2% agarose gel, run at the current voltage of 80 V for 30 min in a buffer solution and visualized by gel documentation. The images were observed using an ultraviolet transilluminator and a digital imaging system (GL 200; Kodak, Windsor, CO, USA).Cell culture
The hepatic LO2 cells were cultured in RPMI growth media and supplemented with 10% fetal bovine serum and a 1% streptomycin and penicillin antibiotic mixture. The cells were grown under an ambient atmosphere of 5% CO2 and 95% air circulation at 37 °C. For hypoxia/reoxygenation (H/R), the LO2 cells were kept in a hypoxia chamber with 5% CO2 and 95% N2 for 4 h. Then, the cells were immediately reoxygenated under 95% air and 5% CO2 for the remaining 10 h at 37 °C.Cell transfection and cell viability analysis
The cell viability analysis was performed by the MTT assay. The cells were grown in a T75 flask. After 80% confluence, the cells were harvested and plated in a 96-well plate at a density of 5 × 103 cells per well and incubated for 24 h. The cells were washed and treated with different concentrations of the PN-miR mimic and mutant and incubated for 24 h. The cells exposed to H/R were considered a control and related to the non-treated control cells. The cells were washed two times with PBS and treated with 100 μL of a 5 mg ml−1 stock MTT solution and incubated for 4 h. The cells were added with DMSO to extract the formazan crystal, and the absorbance was read at 570 nm using a microplate reader (Versamax, Molecular Devices, Inc.).Apoptosis assay by flow cytometer & Hoechst 33342 staining
The cells were grown in a T75 flask, after 80% confluence, the cells were harvested and plated in a 96-well plate at a density of 5 × 103 cells per well and incubated for 24 h. The cells were washed and treated with the PN-miR mimic and mutant and incubated for 24 h. The cells exposed to H/R were considered a control and were related to the non-treated control cells. Apoptosis was first detected by the flow cytometer after staining with Annexin-V and propidium iodide (PI) (Sigma-Aldrich, China). After treatment of the cells with respective formulations, the cells were washed and extracted in a 2 mL centrifuge tube and centrifuged at 1200 rpm for 5 min. The cells were reconstituted with 100 μL of PBS and stained with 2.5 μL of each, Annexin-V and PI, and incubated for 15 min under dark conditions. The cells were then studied for 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 events using the flow cytometer (BD FACS, CA, USA). Apoptosis was also observed after staining with 10 μg ml−1 of Hoechst 33342 for 15 min. The cells were washed and fixed with 4% paraformaldehyde and observed under a confocal laser scanning microscope (Nikon A1, Japan).
000 events using the flow cytometer (BD FACS, CA, USA). Apoptosis was also observed after staining with 10 μg ml−1 of Hoechst 33342 for 15 min. The cells were washed and fixed with 4% paraformaldehyde and observed under a confocal laser scanning microscope (Nikon A1, Japan).
Western blotting analysis
The cells were grown in a T75 flask. After 80% confluence, the cells were harvested and plated in a 96-well plate at a density of 5 × 103 cells per well and incubated for 24 h. The cells were washed and treated with the PN-miR mimic and mutant and incubated for 24 h. The cells exposed to H/R were considered a control and were related to the non-treated control cells. The cells were washed and extracted with a scrapper. The cells were centrifuged and lysed using a lysis buffer under an ice-cold bath for 30 min. The cells were centrifuged at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 15 min and the cell supernatant was collected for further analysis. The protein levels in the individual samples were calculated from bicinchoninic acid assays (Pierce, Rockford, IL). Approximately, 25 μg of protein from each sample was loaded on a sodium dodecyl sulfate polyacrylamide gel for electrophoresis (SDS-PAGE) and run at 120 V for 1 h (approximately). The proteins were then transferred to a PVDF membrane for 1 h. The membranes were blocked with 5% skim milk for 1 h at room temperature and then incubated with primary antibodies overnight at 4 °C. The primary antibodies included HMGB1, cleaved caspase-3, NF-kB and p-p65 (Abcam, China). The membranes were treated with the respective mouse or rabbit secondary antibodies the next day for 1 h. The proteins were visualized using a LAS-4000 mini imager (Fujifilm, Japan).
000 rpm for 15 min and the cell supernatant was collected for further analysis. The protein levels in the individual samples were calculated from bicinchoninic acid assays (Pierce, Rockford, IL). Approximately, 25 μg of protein from each sample was loaded on a sodium dodecyl sulfate polyacrylamide gel for electrophoresis (SDS-PAGE) and run at 120 V for 1 h (approximately). The proteins were then transferred to a PVDF membrane for 1 h. The membranes were blocked with 5% skim milk for 1 h at room temperature and then incubated with primary antibodies overnight at 4 °C. The primary antibodies included HMGB1, cleaved caspase-3, NF-kB and p-p65 (Abcam, China). The membranes were treated with the respective mouse or rabbit secondary antibodies the next day for 1 h. The proteins were visualized using a LAS-4000 mini imager (Fujifilm, Japan).
Animal model experiment
All animal procedures were performed in accordance with the guidelines framed by the Institutional Animal Care and Use Committee of the Fourth Affiliated Hospital of Harbin Medical University, China and duly approved by the Animal Ethics Committee of the Fourth Affiliated Hospital of Harbin Medical University, China. Male SD rats (200–220 g) were procured from the animal facility of the Fourth Affiliated Hospital of Harbin Medical University, China and caged under a pathogen-free guided atmosphere. The animals were given free access to food and water throughout the study period with a standard light/dark cycle. At first, to induce the hepatic I/R, the rats were administered 40 mg kg−1 pentobarbital for anesthesia. Then, a 3 cm incision was made to expose the liver and produce a 70% hepatic ischemia. After this, reperfusion was performed for 1 h, and the abdomen was closed. The untreated rat was not treated with any therapeutics and not subjected to any procedure. The treatment group of rats was administered the miR-449b-5p agomir (PN-miR agomir) and its respective mutant miRNA. The formulations were administered by tail vein injection at a dose of 80 mg per kg per day and surgery was performed after 3 days. Blood was obtained to evaluate the enzyme levels, and the liver was harvested to perform the histological analysis of each group. The liver was homogenized using an automated homogenizer, and the lysate was centrifuged to obtain the protein lysate, which was then subjected to western blot analysis.Statistical analysis
The data are presented as mean ± standard deviation, and a statistical difference of p < 0.05 was considered significant. The differences between groups were analyzed by ANOVA, followed by the Newman–Keuls post hoc test for multiple groups.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This study was supported by the Heilongjiang Provincial Scientific Foundation (Grant No: H2018028), the Heilongjiang Provincial Scientific Foundation (Grant No: LH2019H018), the Spark Research Fund from the Fourth Affiliated Hospital of Harbin Medical University (Grant No: HYDSYXH201904), the General Research Project from the Fourth Affiliated Hospital of Harbin Medical University (Grant No: HYDSYJQ201605), the Heilongjiang Province Health Bureau (Grant No: 2014-399), the Heilongjiang Academy of Medical Sciences (Grant No: 201815), and the Heilongjiang Province Education Department (Grant No: 12541459).Notes and references
- Y. Zhai, H. Petrowsky, J. C. Hong, R. W. Busuttil and J. Kupiec-Weglinski, Nat. Rev. Gastroenterol. Hepatol., 2013, 10, 79–89 CrossRef CAS.
- J. J. Lemasters and R. G. Thurman, Annu. Rev. Pharmacol. Toxicol., 1997, 37, 327–338 CrossRef CAS.
- R. F. van Golen, T. M. van Gulik and M. Heger, Cytokine Growth Factor Rev., 2012, 23, 69–84 CrossRef CAS.
- W. G. Van Riel, R. F. van Golen, M. J. Reiniers, M. Heger and T. M. van Gulik, Hepatobiliary Surgery and Nutrition, 2016, 1, 58–71 Search PubMed.
- R. F. van Golen, T. M. van Gulik and M. Heger, Free Radical Biol. Med., 2012, 52, 1382–1402 CrossRef CAS.
- Y. Zhang, X. Lei, W. Li, X. Ding, J. Bai, J. Wang and G. Wu, Biochem. Biophys. Res. Commun., 2018, 501, 186–192 CrossRef CAS.
- D. Messmer, H. Yang, G. Telusma, F. Knoll, J. Li and B. Messmer, et al., J. Immunol., 2004, 173, 307–313 CrossRef CAS.
- H. Yang, M. Ochani, J. Li, X. Qiang, M. Tanovic and H. E. Harris, et al., Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 296–330 CrossRef CAS.
- X. Wu, W. Wang, Y. Chen, X. Liu, J. Wang, X. Qin and D. Pei, BioMed Res. Int., 2018, 3453706–3453707 Search PubMed.
- E. Yagmur, L. Buendgens, U. Herbers, A. Beeretz, R. Weiskirchen, G. H. Koek and A. Koch, J. Clin. Lab. Anal., 2018, 32, e22584 CrossRef.
- T. Sung, N. Zheng, G. Jeyabalan, K. Izuishi, J. R. Klune and D. A. Geller, et al., J. Leukocyte Biol., 2007, 81, 119–128 CrossRef.
- R. Pullerits, I. M. Jonsson, M. Verdrengh, M. Bokarewa, U. Andersson and H. Erlandsson-Harris, et al., Arthritis Rheum., 2003, 48, 1693–1700 CrossRef CAS PubMed.
- U. Andersson and K. J. Traeey, Scand. J. Infect. Dis., 2003, 35, 577–584 CrossRef CAS.
- Z. Bai, J. Wei, C. Yu, X. Han, X. Qin, C. Zhang, W. Liao and L. Li, J. Mater. Chem. B, 2019, 7, 1209–1225 RSC.
- W. Yang, C. Yu, C. Wu, S. Q. Yao and S. Wu, Polym. Chem., 2017, 8, 4043–4051 RSC.
- M. J. Wang, Y. Y. Xu, R. Y. Huang, X. M. Chen, H. M. Chen, L. Han and C. J. Lu, Oncotarget, 2017, 8, 5498–5507 Search PubMed.
- C. Yu, L. Qian, M. Uttamchandani, L. Li and S. Q. Yao, Angew. Chem., Int. Ed., 2015, 54, 10574–10578 CrossRef CAS.
- A. M. Ardekani and M. M. Naeini, Journal of Medical Biotechnology, 2010, 2, 161–179 CAS.
- E. Indersie, S. Lesjean, K. B. Hooks, F. Sagliocco, T. Ernault, S. Cairo and C. F. Grosset, Hepatol. Commun., 2017, 1, 168–183 CrossRef CAS PubMed.
- M. Sandbothe, R. Buurman, N. Reich, L. Greiwe, B. Vajen, E. Gürlevik and B. Skawran, J. Hepatol., 2017, 66, 1012–1021 CrossRef CAS PubMed.
- T. Ramasamy, H. B. Ruttala, K. Kaliraj, K. Poudel, S. G. Jin, H. G. Choi, S. K. Ku, C. S. Yong and J. O. Kim, ACS Biomater. Sci. Eng., 2019, 5, 5159–5168 CrossRef CAS.
- E. Van Rooij and E. N. Olson, Nat. Rev. Drug Discovery, 2012, 11, 860–872 CrossRef CAS.
- T. Ramasamy, H. B. Ruttala, B. Gupta, B. K. Poudel, H. G. Choi, C. S. Yong and J. O. Kim, J. Controlled Release, 2017, 258, 226–253 CrossRef CAS.
- Y. Chen, D. Y. Gao and L. Huang, Adv. Drug Delivery Rev., 2015, 81, 128–141 CrossRef CAS.
- R. Devulapally, K. Foygel, T. V. Sekar, J. K. Willmann and R. Paulmurugan, ACS Appl. Mater. Interfaces, 2016, 8, 33412–33422 CrossRef CAS.
- D. Liu, X. Liu, T. Zhou, W. Yao, J. Zhao, Z. Zheng and Y. Chen, J. Mol. Cell Biol., 2016, 8, 144–156 CrossRef CAS.
- J. Zhu, F. Zhu, W. Song, B. Zhang, X. Zhang, X. Jin and H. Li, Gene, 2017, 607, 23–30 CrossRef CAS.
- X. Wang, Y. Guo, C. Wang, H. Yu, X. Yu and H. Yu, Inflammation, 2016, 39, 1718–1728 CrossRef CAS.
- L. Yao, X. Lv and X. Wang, J. Pharmacol. Sci., 2016, 131, 6–12 CrossRef CAS.
- S. R. Kim, Y. M. Ha, Y. M. Kim, E. J. Park, J. W. Kim, S. W. Park, H. J. Kim, H. T. Chung and K. C. Chang, Biochem. Pharmacol., 2015, 95, 279–289 CrossRef CAS.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9ra06129k |
| ‡ These two authors contributed to this work equally. |
| This journal is © The Royal Society of Chemistry 2019 |