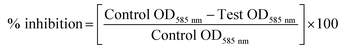Open Access Article
Open Access ArticleHydrocinnamic acid produced by Enterobacter xiangfangensis impairs AHL-based quorum sensing and biofilm formation in Pseudomonas aeruginosa†
Shivangi Sharma,
Venkadesaperumal Gopu‡
 ,
Chandran Sivasankar and
Prathapkumar Halady Shetty*
,
Chandran Sivasankar and
Prathapkumar Halady Shetty*
Department of Food Science and Technology, Pondicherry University, R. V. Nagar, Kalapet, Pondicherry 605014, India. E-mail: pkshalady@yahoo.co.uk; Fax: +91-413-2656743; Tel: +91-413-2656625
First published on 11th September 2019
Abstract
Many of the Gram-negative bacteria regulate their virulence through an AHL-mediated quorum sensing (QS) mechanism. Disruption of this signaling mechanism might be a novel strategy to suppress bacterial virulence. In this report, foodborne bacterial isolates were tested for their QS-inhibitory properties using biosensor strain Chromobacterium violaceum CV026 and the extracted potential active components were evaluated for anti-QS and antibiofilm activity against pathogenic bacteria. The cell-free supernatant of Enterobacter xiangfangensis PUFSTI26 inhibited violacein production in the reporter strain and exhibited a significant reduction in extracellular virulence factors like biofilm formation, pyocyanin production, and motility of Pseudomonas aeruginosa. Characterization of the purified active component by gas chromatography-mass spectrometry (GC-MS) flaunted the resemblance of hydrocinnamic acid (HCA). Treatment of HCA exhibited pronounced attenuation of virulence factors. Further, the biofilm inhibitory activity was evidenced by means of confocal laser microscopy, that evidenced the repression of biofilm biomass. In addition, gene quantification analysis revealed that HCA repressed the expression of major QS-regulated genes. In silico studies showed that HCA competitively interacts with LasR receptor protein. These results clearly indicate the anti-virulence properties of HCA extracted from E. xiangfangensis of food origin. This is also the first report of the QS inhibitor activity of HCA.
1. Introduction
Traditionally, a wide range of antibiotics is in use for the management and treatment of bacterial infection. However, many of these antibiotics have adverse side effects on the subject and more importantly sustained use of these antimicrobial substances results in resistance among pathogens creating drug-resistant strains which lead to difficulties in treating subjects with such infections.1–3 Studies have reported that many pathogenic bacterial strains including P. aeruginosa employ a QS mechanism to govern their virulence factors.4,5 Quorum sensing (QS) is the population-dependent mechanism of bacteria, mediated through signal molecules called auto-inducers that regulates population characteristics such as motility, biofilm formation, pigment production, secretion of virulence factors and antibiotic resistance. Gram-negative bacteria employ acyl-homoserine lactones (AHLs) as their signaling molecules.6 In the LasR–LasI system of P. aeruginosa, AHLs binds to LasR receptor protein to regulate the expression of downstream targets.7Contributing to the importance of QS machinery in bacterial pathogenesis, intervening the signaling mechanism could be a promising approach to control bacterial virulence.8 Quorum sensing inhibitory (QSI) compounds, do not impose selective pressure on bacteria unlike conventional antimicrobials to acquire resistance. Many natural and synthetic QS inhibitors have been investigated for their quorum quenching activity. Researchers have explored QSI compounds from microorganisms with potential ability to inhibit QS system of various pathogenic organism.9–11 The anti-QS activity has been reported in many bacterial strains of Bacillus, Klebsiella, Arthrobacter, and Pseudomonas.12,13
Many natural and synthetic compounds like furanones, cyanidin and aspirin have been reported for their QS inhibitory property including the bacterial metabolite terrein.14–17 Nevertheless, the inadequacy of these molecules for its application has preceded the pursuit of novel quorum sensing inhibitor that can be used in various applications. Previously, we have also evaluated the foodborne isolates with AHL-lactonase activity that attenuated QS mediated virulence factors in C. violaceum and Yersinia enterocolitica.9 Here, we have reported the QS inhibitory potential of foodborne isolate E. xiangfangensis and its metabolite HCA. Hydrocinnamic acid attenuated pyocyanin production, biofilm formation as well as motility of P. aeruginosa. Further, we have demonstrated the molecular mechanism of regulation by gene expression and in silico molecular docking analysis.
2. Materials and methods
2.1 Bacterial strains
Chromobacterium violaceum CV026 (CECT599) a biosensor strain for short-chain AHLs, wild type C. violaceum (MTCC2656) and P. aeruginosa (MTCC 424) reported for AHL production was used for determining the QS inhibitory potential of thirty-four bacterial isolates from our departmental culture collection. All the strains were cultured in Luria Bertani (LB) broth at 37 °C. CV026 was supplemented with 10 μM of N-hexanoyl-DL-homoserine lactone (Sigma-Aldrich, India) to stimulate violacein pigmentation unless otherwise stated.2.2 Cell-free supernatant
Test bacterial strains seeded into LB broth was incubated at 37 °C for 48 h in a shaker incubator. Bacterial cell pellets were removed by centrifugation at 5000 g for 10 min. Cell-free supernatant was collected and filtered through a 0.22 μm syringe filter which was then stored at −20 °C until further use. All the assays were performed with LB broth as an assay control.2.3 Flask incubation assay
Quantitative analysis of violacein inhibition by the cell-free supernatant of test isolates was determined by flask incubation assay. Inoculated LB broth added with varying concentrations of CFS (0–10%) was incubated overnight at 37 °C. On overnight incubation, violacein pigment was precipitated from 1 ml of culture by spinning at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 g for 5 min. Obtained violacein pellet was resuspended in dimethyl sulfoxide and centrifuged again to remove the cells and quantified at 585 nm using a microplate reader. Cell pellets were re-dissolved in phosphate buffer saline for measuring the cell density at 600 nm. The experiment was repeated for triplicate values and the percentage of inhibition was calculated by the formula as below:
000 g for 5 min. Obtained violacein pellet was resuspended in dimethyl sulfoxide and centrifuged again to remove the cells and quantified at 585 nm using a microplate reader. Cell pellets were re-dissolved in phosphate buffer saline for measuring the cell density at 600 nm. The experiment was repeated for triplicate values and the percentage of inhibition was calculated by the formula as below:2.4 Bacterial identification
The positive isolate was subjected to molecular characterization by amplifying 16S rRNA using universal primers 27F-5′-AGAGTTTGATCCTGGCTCAG-3′ and 1492R-5′-TACGGYTACCTTGTTACGACTT-3. Fifty microliter PCR reaction with initial denaturation of 95 °C for 5 minutes followed by 35 cycles of 94 °C for 1 minute, 50 °C for 45 seconds and 72 °C for 1 minute. The final extension was carried out at 72 °C for 5 min using master cycler (Eppendorf, Germany). Purified PCR products were sequenced and subjected to BLAST analysis for similarity. Similar sequences were subjected to multiple sequence alignment using UPGMA and the phylogenetic tree was constructed using MEGA 7.0.21.2.5 Inhibition of pyocyanin production
Effect of E. xiangfangensis cell-free extract on P. aeruginosa pyocyanin production was quantified as described by Zhang & Chu.18 Briefly, the varying concentration of cell-free extracts (0–10%) was mixed with freshly prepared P. aeruginosa culture and incubated overnight at 37 °C. Chloroform (1.5 ml) was added to the P. aeruginosa culture supernatant and mixed vigorously and centrifuged at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 3 min. The chloroform layer was mixed with 600 μl of 0.2 M HCl and the OD was measured at 520 nm.
000g for 3 min. The chloroform layer was mixed with 600 μl of 0.2 M HCl and the OD was measured at 520 nm.
2.6 Motility assays
Swimming and swarming motility was assessed on 0.3% tryptone agar and 0.5% BM2 glucose plates supplemented with 2.5% of E. xiangfangensis CFS respectively. Overnight culture of P. aeruginosa was inoculated as 1 μl aliquots diluted to 0.6 OD600. Plates were then incubated at for 18 and 24 h respectively for swimming and swarming assays at 37 °C.2.7 Effect of CFL on biofilm formation
Overnight culture of P. aeruginosa was seeded into 12-well plate containing LB broth supplemented with varying concentration of cell-free supernatant (0–10%). Wells without CFS were taken as control and plate were incubated at 37 °C in static condition for 24 h. Following incubation, planktonic cells were removed by rinsing thrice with sterile PBS and dried for 20 min at room temperature. Biofilms formed were tinged with crystal violet (200 μl) for 15 min and again washed gently with PBS to remove the excess dye.19 Bound dye was solubilized in 100 μl decolorizer and the absorbance was recorded at 580 nm using microplate reader (Biotek, USA).2.8 Extraction of active QSI component
The cell-free supernatant of E. xiangfangensis was extracted with an equal amount of ethyl-acetate. The organic phase was collected and concentrated using rotary vacuum evaporator. The crude extract was chromatographed on a silica gel column, eluted with a chloroform/methanol gradient up to five fractions. QS inhibitory activity of extracted fractions was screened as earlier.92.9 Identification of active QSI component by GC-MS
The active fraction was purified using a binary gradient HPLC system equipped with a photodiode array detector. The fraction was separated on a C-18 column using a linear gradient and detected at 272 nm (acetonitrile![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water; flow rate-1 ml min−1; injection volume-25 μl). Purified fractions were collected and screened for its QS inhibitory activity. Further, the active component was subjected to GC-MS (PerklinElmer, Clarus 680/600). The recorded spectra were compared with the GC-MS NIST (2008) library for the identification of bioactive component based on retention indices.
water; flow rate-1 ml min−1; injection volume-25 μl). Purified fractions were collected and screened for its QS inhibitory activity. Further, the active component was subjected to GC-MS (PerklinElmer, Clarus 680/600). The recorded spectra were compared with the GC-MS NIST (2008) library for the identification of bioactive component based on retention indices.
2.10 In situ visualization
Biofilm inhibitory potential of E. xiangfangensis CFS was evaluated as defined.20 Concisely, an overnight culture of P. aeruginosa was inoculated into LB broth with or without E. xiangfangensis CFS (5%). Biofilms on spherical glass slides were formed as described above. Unbound planktonic cells were removed by rinsing with sterile PBS followed by staining with 0.1% acridine orange for 15 min. Images were obtained using a confocal laser scanning microscope (LSM710, Carl Zeiss, Germany).2.11 Regulation of quorum gene expressions
P. aeruginosa supplemented with N-hexanoyl-DL-homoserine lactone (50 μM) was treated with 5 μg ml−1 of HCA overnight. Total RNA was isolated using TRI reagent and cDNA was synthesized using reverse transcription kit (Promega, WI) with random hexamers. All mRNA expression levels were represented against the average expression of the untreated group.2.12 Molecular docking studies
Molecular docking analysis was performed to screen the binding affinity of hydrocinnamic acid and signal molecule to LasR receptor protein. Bond orders and hydrogen molecules were added to the PDB retrieved LasR receptor protein (PDB ID: 2UV0) followed by removal of all crystallized water molecules and co-crystallized ligands.21 Partial atomic charges were assigned according to the OPLS2005 force field to optimize the protein. The grids file for LasR protein were generated around the receptor active site residues (Tyr-56, Trp-60, Asp-73, Thr-75, and Ser-129) using Glide, version 7.8, in Maestro suite version 11.5 (Schrödinger, LLC, New York, NY, 2018), and used for docking studies. Ligands collected from Pubchem database were prepared in LigPrep module in Maestro suite version 11.5 (Schrödinger, LLC, New York, NY, 2018) which generates all possible states at pH of 7.0 and generated all possible states of ionization and tautomerization for all ligands. The prepared proteins and ligands were docked rigidly using molecular docking suit Glide, version 7.8 (Schrodinger, LLC, New York, NY, 2018) with extra precision mode. Resulting 3D images were prepared using chimera version 1.12.3. Results
3.1 Screening and identification of AHL degrading bacteria
To identify quorum sensing inhibitory potential of a bacteria, cell-free supernatants of 34 bacterial isolates from an uncooked batter of a traditional Indian fermented food idli were evaluated for their quorum inhibitory ability by agar overlay and flask incubation assay. Reduction of violacein pigment in the reporter strain (CV026) was regarded as positive for quorum inhibition. Amid 34 isolates screened, PUFSTI26 exhibited quorum inhibitory activity in the agar overlay assay. Further, in quantitative determination, cell-free supernatant of PUFSTI26 inhibited up to 72.45% (p < 0.05) of violacein production in biosensor strain (Fig. 1A). On screening the effect of CFS on bacterial growth (C. violaceum & P. aeruginosa), no significant changes were observed on the growth pattern when treated up to 12% (data not shown). Treatment of cell-free supernatant (CFS) with proteinase K and heating at 95 °C showed no effect on AHL degradation indicates the possibility of non-enzymatic degradation. The positive isolate was tentatively identified as E. xiangfangensis by BLAST assessment of 16S rRNA sequence (Fig. S1†). The sequence was deposited in the National Center for Biotechnology Information (NCBI) database and given with unique identifier number MG371998.3.2 Inhibition of pyocyanin production
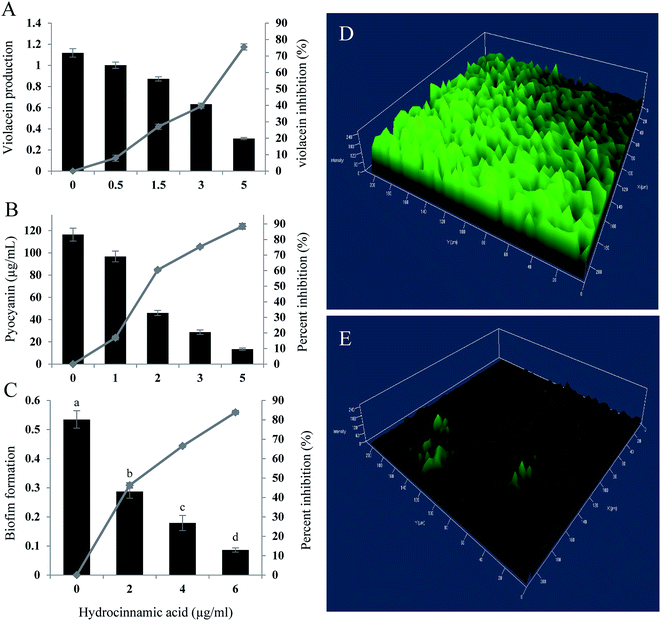
The inhibitory activity of E. xiangfangensis cell-free supernatant against the production of virulence factor, pyocyanin by P. aeruginosa was screened. In the quantitative assay for screening inhibitory activity of cell-free supernatant, results exhibited the concentration-dependent inhibition of pyocyanin production. It was observed that pyocyanin production by test strain in the presence of CFS at different concentration (0.5–5%) was inhibited up to 86.45% (p < 0.05) when compared to untreated control and not showed any influence on cell viability (Fig. 1B).3.3 Inhibition of biofilm formation by P. aeruginosa
Pyocyanin production has been reported as one of the crucial factors for biofilm production in P. aeruginosa.22 Since, the cell-free supernatant of E. xiangfangensis PUFSTI26 inhibited the production of pyocyanin, its effect on biofilm formation was screened by MTP assay. In the quantitative assay, results showed that P. aeruginosa exhibited strong biofilm formation. However, samples treated with 5% of cell-free supernatant exhibited about 60.11% reduction in biofilm biomass when compared with untreated control (Fig. 1C). It is noted that the cell-free supernatant has not shown any significant influence on planktonic cells and found to retain its biofilm inhibitory activity even after 24 h.3.4 Effects on swimming and swarming motility
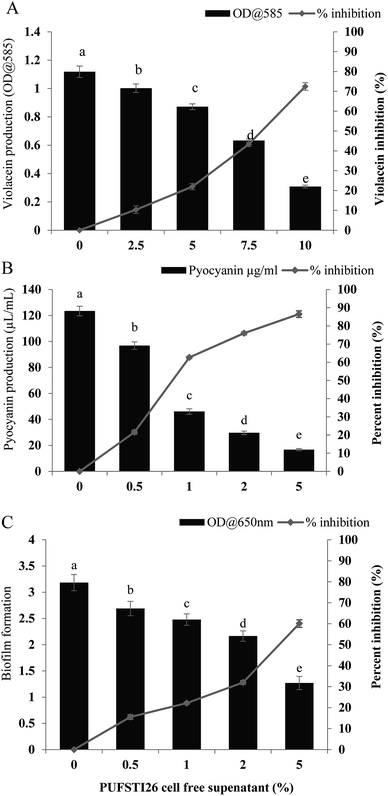
Swimming and swarming motility act as a decisive factor in quorum mediated biofilm formation and development.23 As flagellar-arbitrated motility determines the virulence factor of a pathogen, the effect of test CFS to impede the motility of P. aeruginosa was studied. It was noted that cell-free supernatant of E. xiangfangensis PUFSTI26 significantly inhibited the motility of test strain. Maximum inhibition was recorded at 2.5% substitution which hampered 95.14 ± 0.35% and 88.26 ± 0.25% of swimming and swarming respectively when compared to untreated control. It is apparent from our findings that the bacterial extract substantially hampered the flagellar-mediated motility of test pathogen (Fig. 2).3.5 Extraction and purification of the active component
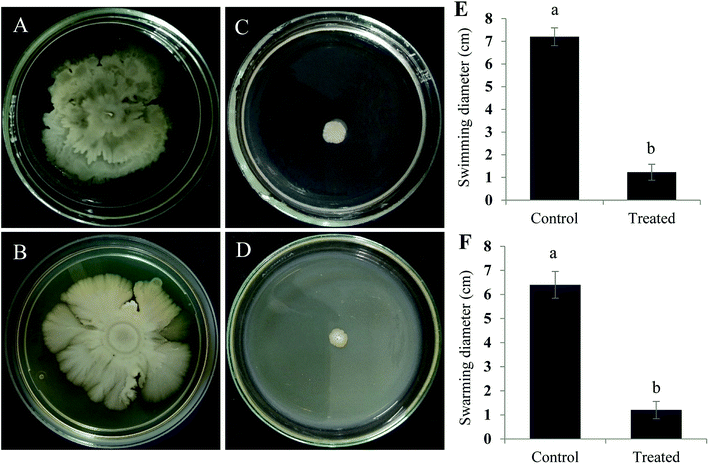
Active component that exhibits quorum quenching potential was purified using silica gel (60–120 mesh) column chromatography eluted with increasing concentration of ethyl acetate and methanol.24 Five different fractions were collected and screened for its activity by violacein inhibition assay. Out of five fractions tested, fraction 2 had exhibited QS inhibitory activity (Fig. 3). The fraction was further purified by TLC by means of chloroform/methanol solvent system (24![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 v/v). On screening the different fractions at Rf 0.84 and 0.52 for QS inhibitory activity, fraction obtained at Rf 0.84 was found to be active (data not shown). Preparative TLC was performed for obtaining a significant quantity of active component from the crude fraction. Fraction of interest on TLC were extracted from silica using small volumes of solvent system until the silica was rendered completely colorless and subjected to further HPLC purification. The chromatogram obtained showed three distinct peaks at 2.476, 2.762 and 3.030 min, were collected, resuspended in DMSO and screened for their anti-QS activity with respective controls. Out of three different fractions, the one collected at the retention time of 3.030 min inhibited violacein production in the reporter strain (Fig. 3).
1 v/v). On screening the different fractions at Rf 0.84 and 0.52 for QS inhibitory activity, fraction obtained at Rf 0.84 was found to be active (data not shown). Preparative TLC was performed for obtaining a significant quantity of active component from the crude fraction. Fraction of interest on TLC were extracted from silica using small volumes of solvent system until the silica was rendered completely colorless and subjected to further HPLC purification. The chromatogram obtained showed three distinct peaks at 2.476, 2.762 and 3.030 min, were collected, resuspended in DMSO and screened for their anti-QS activity with respective controls. Out of three different fractions, the one collected at the retention time of 3.030 min inhibited violacein production in the reporter strain (Fig. 3).
3.6 Identification of active principle by GC-MS
Active component purified through C18-HPLC was subjected to GC-MS analysis. The chromatogram showed a single peak at the retention time 12.657 min. Gas chromatography-mass spectrometry analysis revealed the presence of active component using a direct similarity search. The detected mass spectra showed resemblance to hydrocinnamic acid, in the GS-MS library (NIST, 2008) with a retention time of 12.657 as evident from the GC-MS spectra (Fig. S2†). Retention index and area percentage (concentration) were summarized in Table 1. The calculated (theoretical) or expected molecular mass of compound C9H10O2 is 150.177 (Table 1).| Bioactive metabolite profile of HPLC fraction, identified by GC-MS analysis | |
| Bioactive compound | Hydrocinnamic acid |
| Retention time | 12.657 |
| Molecular formula | C9H10O2 |
| Molecular weight | 150 |
| Peak area (%) | 100 |
3.7 Evaluation of hydrocinnamic acid QSI activity
In quantification assay, HCA inhibited, violacein production in C. violaceum CECT5999 up to 75.49% at the maximum concentration of 5 μg ml−1 (Fig. 4A). Furthermore, at Sub-MIC level tested compound exhibited concentration-dependent regulation of pyocyanin and biofilm production in P. aeruginosa. Hydrocinnamic acid at the concentration of 5 μg ml−1 inhibited 88.47% of pyocyanin production and inhibited 83.86% of biofilm formation at the concentration of 6 μg ml−1 in test strain (Fig. 4B and C). The inhibition of biofilm formation was substantiated with confocal laser scanning microscopy, which again proved the significant reduction in biofilm formation by P. aeruginosa. Three-dimensional images of CLSM (Fig. 4D and E) clearly showed a significant reduction in the thickness of the biofilm in the treated slides when compared with untreated control.3.8 Downregulation of quorum genes by HCA
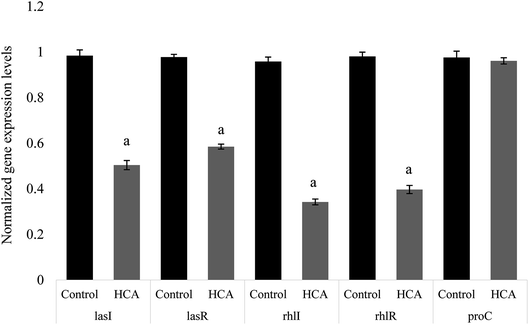
The expression of many virulence factors in P. aeruginosa was found to be dependent on QS regulatory and its controlled genes.25 To further investigate the QS-inhibitory nature of HCA, the relative expression of P. aeruginosa genes like lasI, lasR, rhlI and rhlR were studied to understand the regulatory mechanism of HCA. Real-time PCR revealed that the expression of P. aeruginosa QS-regulatory genes lasI, lasR, rhlI, and rhlR were repressed by 48.76%, 40.16%, 64.24%, and 59.53% respectively on HCA treatment (Fig. 5).3.9 Molecular docking analysis
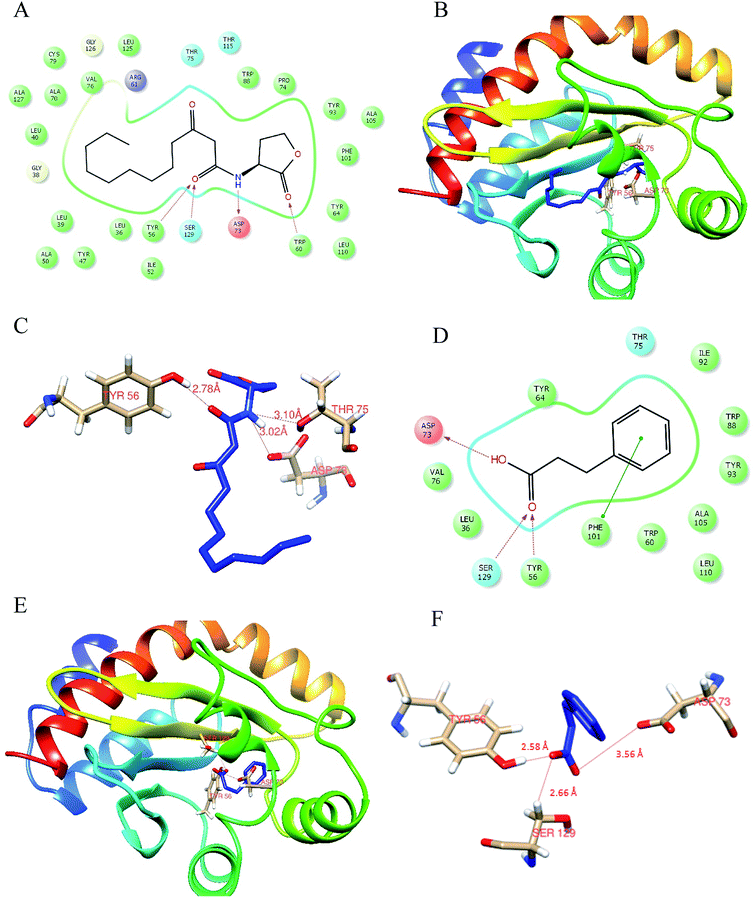
Docking analysis was done to unleash the hotspot residues that interact with signaling molecule and active component for the functional conformation changes. PDB-retrieved three-dimensional structure of the receptor protein, LasR was analyzed for its transcriptional auto-inducer domain. The structural component analysis was done as mentioned in our earlier publication.21 The active site at its second β hairpin turn forms the cavity which was found using PDB database ligand explorer toll.On the first iteration, the docking score of signaling molecule with LasR receptor protein is −6.928 kcal mol−1 and it was submitted to PDBSum database for analysis. The hydrogen bond formed between the residues were visualized with the LigPlot module. The LasR-AHL complex was found to have one hydrogen bond at catalytic residues Tyr 56, Asp 73 and Thr 75 along with 13 hydrophobic interactions. On the second iteration, LasR was docked to hydrocinnamic acid with the same grid constraints used for signaling compound (Fig. 6). LasR-HCA complex that exhibited dock score of −6.356 kcal mol−1 was subjected to further investigation. The complex revealed that one hydrogen bond formed at catalytic residues Tyr 56, Ser 129 and Asp 73 along with 10 hydrophobic interactions. The details of interacting atoms between protein and the active components were listed in Table 2.
| Compound name | Docking score kcal mol−1 | Hydrogen bond | Hydrophobic interactions | Glide-Emodel score kcal mol−1 |
|---|---|---|---|---|
| 3-Oxo-C12 HSL | −6.928 | Tyr 56, Asp 73, Thr 75 | Leu 36, Leu 40, Try 47, Arg 61, Tyr 64, Val 70, Val 76, Trp 88, Tyr 93, Ala 105, Phe 101, Leu 110, Ser 129 | −73.830 |
| Hydrocinnamic acid | −6.356 | Tyr 56, Ser 129, Asp 73 | Leu 36, Val 76, Trp 60, Tyr 64, Leu 110, Thr 75, Ala 105, Tyr 93, Trp 88, Ile 92 | −37.393 |
4. Discussion
Amidst the desperate need for novel anti-microbials, anti-infective therapies targeting virulence mechanism may be a promising strategy that is more sustainable. The crucial role of quorum sensing in the bacterial pathogenesis has made this machinery an attractive target for bacterial control in the recent years.26–29 This study explored the interference of various bacterial extracts with AHL-mediated quorum sensing and demonstrated that hydrocinnamic acid purified from the cell-free supernatant of E. xiangfangensis PUFSTI26 suppresses the regulation of QS mediated virulence factors like pyocyanin production, biofilm formation and motility in P. aeruginosa. This compound was also found to repress the expression of QS regulatory and its controlled genes. In order to provide insight into the specific mode of action, we have used molecular and in silico tools to support our conclusions.Out of the 34 bacterial isolates evaluated for QS-inhibitory potential against C. violaceum CV026, cell-free supernatant of E. xiangfangensis PUFSTI26 alone exhibited a significant reduction in violacein pigmentation at different tested concentrations. In flask incubation assay, test isolate inhibited about 72.45% of violacein production at 10% supplementation. Our results are in accordance with,30 who reported that cell-free lysates of Bacillus firmus PT18 and Enterobacter asburiae PT39 inhibited about 80% violacein production in biosensor strain. Pyocyanin production and biofilm formation are known to be regulated through quorum sensing machinery in many pathogens including P. aeruginosa.22,31 It is reported that bacterial lysate inhibits biofilm formation in Y. enterocolitica9 and pyocyanin production.32 Here, we have demonstrated that crude extract of E. xiangfangensis, significantly inhibited the pyocyanin production and biofilm formation in test strain without restricting the growth of bacterial cells. Pulmonary infections have shown that P. aeruginosa forms organized communities that requites swarming motility for its dispersion.33 Our results showed that the cell-free extract of E. xiangfangensis inhibited swimming and swarming motility suggesting impaired flagellar biogenesis and biofilm dispersion.
Having ascertained the potential of crude extract to repress the QS-regulated phenotypes, the cell-free supernatant was purified using thin-layer chromatography. Two fractions obtained at Rfs 0.84 and 0.52 were screened for its quorum inhibitory activity by flask incubation method. The potential fraction was further subjected to preparative HPLC and the fraction with QS inhibitory property was purified to obtain a single distinctive peak. Pre-HPLC purified fraction was subjected to GC-MS analysis. The mass spectra obtained was compared for its resemblance using NIST library and the active principle component was identified as hydrocinnamic acid. The probability of HCA to inhibit QS machinery was evidenced by flask incubation assay in which AHL-mediated violacein production was inhibited up to 75.49% at the concentration of 5 μg ml−1. The first evidence that QS regulates biofilm formation, a key player in the pathogenesis of several bacteria was reported in P. aeruginosa.34 Hence, intervening QS mechanism could be a promising approach to inhibit biofilm formation in P. aeruginosa. Obtained results showed that HCA not only inhibited biofilm formation in PAO1 but also lowered the swimming and swarming motility. Flagella-mediated swimming motility is known to confer enhanced biofilm formation by instigating the cell-to-surface attachment.35 Therefore, inhibition of swimming motility could be one of the reasons for decreased biofilm formation. Venkadesaperumal and colleagues also examined a similar effect wherein quercetin inhibited the biofilm formation of P. aeruginosa by interfering with its swimming motility.36 Also, HCA repressed the production of extracellular virulence factor pyocyanin. Our results are in agreement with Li and colleagues who reported pyocyanin inhibition by homogentisic acid γ lactone.5
LasR receptor protein interacts with signaling molecule to form a transcriptional activator complex at high cell population to regulate the expressions of genes including lasI, lasR, rhlI, and rhlR.27 Our results indicate that HCA impedes the quorum-sensing circuit of P. aeruginosa by downregulating the QS regulon genes. Repression of these genes could reflect on inhibition of several virulence factors. Obtained results are in line with that of Hossain et al.37 wherein methyl gallate has shown anti-QS activity in PAO1 by inhibiting the expression of QS regulated genes. Collectively, these data suggest that HCA interfere directly with the QS machinery in P. aeruginosa.
To further test this hypothesis at the atomic level, the potential interactions of HCA with LasR was studied using docking tools. HCA and the autoinducer molecules were docked into potential binding sites of LasR protein. Docking analysis showed that HCA interact with LasR receptor, thus averting autoinducer biding which could modulate the expression of quorum sensing mediated phenotypes. Nevertheless, these results strengthen our speculation that HCA acts as a competitive inhibitor for autoinducer molecule. The findings fully agree with the results of this study.
5. Conclusion
This part of the study revealed that Enterobacter xiangfangensis PUFSTI26 and its active component hydrocinnamic acid efficiently intervened the QS machinery of C. violaceum and P. aeruginosa and could attenuate the virulence factors that are crucial for bacterial pathogenicity. Quorum inhibitory potential of HCA has not been reported earlier and hence is the first report. HCA showed the repression of autoinducer synthase and its cognate receptor genes of P. aeruginosa. In silico analysis showed the competitive binding nature of HCA against natural autoinducer molecule suggesting this natural molecule could have far-reaching application in the management of pathogenesis in humans, domestic animals as well as in aquaculture industry.Conflicts of interest
Authors declare that they do not have any conflict of interest.Acknowledgements
Author CS acknowledge SERB for providing financial assistance in the form of Post-Doctoral Fellowship and Central Instrumentation facility (CIF) of Pondicherry University for providing necessary facilities.References
- G. Brackman, P. Cos, L. Maes, H. J. Nelis and T. Coenye, Quorum sensing inhibitors increase the susceptibility of bacterial biofilms to antibiotics in vitro and in vivo, Antimicrob. Agents Chemother., 2011, 55, 2655–2661 CrossRef CAS.
- E. Sánchez, et al., Antibacterial and Antibiofilm Activity of Methanolic Plant Extracts against Nosocomial Microorganisms, J. Evidence-Based Complementary Altern. Med., 2016, 2016, 1572697 Search PubMed.
- P. Jiang, et al., Antibiofilm activity of an exopolysaccharide from marine bacterium Vibrio sp. QY101, PLoS One, 2011, 6, e18514 CrossRef CAS.
- J. Lee and L. Zhang, The hierarchy quorum sensing network in Pseudomonas aeruginosa, Protein Cell, 2015, 6, 26 CrossRef CAS.
- T. Li, et al., Inhibition of quorum sensing-controlled virulence factors and biofilm formation in Pseudomonas fluorescens by cinnamaldehyde, Int. J. Food Microbiol., 2018, 269, 98–106 CrossRef CAS.
- C. M. Waters and B. L. Bassler, Quorum sensing: cell-to-cell communication in bacteria, Annu. Rev. Cell Dev. Biol., 2005, 21, 319–346 CrossRef CAS.
- K. Papenfort and B. L. Bassler, Quorum sensing signal-response systems in Gram-negative bacteria, Nat. Rev. Microbiol., 2016, 14, 576–588 CrossRef CAS.
- A. L. Adonizio, K. Downum, B. C. Bennett and K. Mathee, Anti-quorum sensing activity of medicinal plants in southern Florida, J. Ethnopharmacol., 2006, 105, 427–435 CrossRef PubMed.
- V. Gopu, C. Kumar Meena, A. Murali and P. Halady Shetty, Quorum quenching activity in the cell-free lysate of Enterobacter ludwigii isolated from beef and its effect on quorum sensing regulation in Yersinia enterocolitica, RSC Adv., 2016, 6, 21277–21284 RSC.
- M. Manefield, et al., Halogenated furanones inhibit quorum sensing through accelerated LuxR turnover, Microbiology, 2002, 148, 1119–1127 CrossRef CAS.
- T. B. Rasmussen, et al., Identity and effects of quorum-sensing inhibitors produced by Penicillium species, Microbiology, 2005, 151, 1325–1340 CrossRef CAS.
- Y. H. Dong, et al., Quenching quorum-sensing-dependent bacterial infection by an N-acyl homoserine lactonase, Nature, 2001, 411, 813–817 CrossRef CAS.
- V. Gopu and P. H. Shetty, Cyanidin inhibits quorum signalling pathway of a food borne opportunistic pathogen, J. Food Sci. Technol., 2016, 53, 968–976 CrossRef CAS.
- B. Kim, J.-S. Park, H.-Y. Choi, S. S. Yoon and W.-G. Kim, Terrein is an inhibitor of quorum sensing and c-di-GMP in Pseudomonas aeruginosa: a connection between quorum sensing and c-di-GMP, Sci. Rep., 2018, 8, 8617 CrossRef.
- K. Tateda, et al., Azithromycin inhibits quorum sensing in Pseudomonas aeruginosa, Antimicrob. Agents Chemother., 2001, 45, 1930–1933 CrossRef CAS.
- S. Sarabhai, P. Sharma and N. Capalash, Ellagic acid derivatives from Terminalia chebula Retz. downregulate the expression of quorum sensing genes to attenuate Pseudomonas aeruginosa PAO1 virulence, PLoS One, 2013, 8, e53441 CrossRef CAS.
- T. H. Jakobsen, et al., Ajoene, a sulfur-rich molecule from garlic, inhibits genes controlled by quorum sensing, Antimicrob. Agents Chemother., 2012, 56, 2314–2325 CrossRef CAS.
- A. Zhang and W.-H. Chu, Anti-Quorum Sensing Activity of Forsythia suspense on Chromobacterium violaceum and Pseudomonas aeruginosa, Pharmacogn. Mag., 2017, 13, 321–325 Search PubMed.
- G. G. Anderson and G. A. O'Toole, Innate and induced resistance mechanisms of bacterial biofilms, Curr. Top. Microbiol. Immunol., 2008, 322, 85–105 CAS.
- L. D. R. Melo, et al., Development of a Phage Cocktail to Control Proteus mirabilis Catheter-associated Urinary Tract Infections, Front. Microbiol., 2016, 7, 1024 Search PubMed.
- V. Gopu, C. Kumar Meena, A. Murali and P. Halady Shetty, Petunidin as a competitive inhibitor of acylated homoserine lactones in Klebsiella pneumoniae, RSC Adv., 2016, 6, 2592–2601 RSC.
- D. N. Naik, S. Wahidullah and R. M. Meena, Attenuation of Pseudomonas aeruginosa virulence by marine invertebrate-derived Streptomyces sp, Lett. Appl. Microbiol., 2013, 56, 197–207 CrossRef CAS.
- D. Damte, Evaluation of Anti-Quorum Sensing Activity of 97 Indigenous Plant Extracts From Korea through Bioreporter Bacterial Strains Chromobacterium violaceum and Pseudomonas aeruginosa, J. Microb. Biochem. Technol., 2013, 05, 042–046 Search PubMed.
- H. S. Vasavi, A. B. Arun and P. D. Rekha, Anti-quorum sensing activity of flavonoid-rich fraction from Centella asiatica L. against Pseudomonas aeruginosa PAO1, J. Microbiol., Immunol. Infect., 2016, 49, 8–15 CrossRef CAS.
- A. Latifi, M. Foglino, K. Tanaka, P. Williams and A. Lazdunski, A hierarchical quorum-sensing cascade in Pseudomonas aeruginosa links the transcriptional activators LasR and RhIR (VsmR) to expression of the stationary-phase sigma factor RpoS, Mol. Microbiol., 1996, 21, 1137–1146 CrossRef CAS.
- H.-S. Kim, S.-H. Lee, Y. Byun and H.-D. Park, 6-Gingerol reduces Pseudomonas aeruginosa biofilm formation and virulence via quorum sensing inhibition, Sci. Rep., 2015, 5, 8656 CrossRef CAS.
- B. N. Singh, et al., Lagerstroemia speciosa fruit extract modulates quorum sensing-controlled virulence factor production and biofilm formation in Pseudomonas aeruginosa, Microbiology, 2012, 158, 529–538 CrossRef CAS.
- C. Arevalo-Ferro, et al., Identification of quorum-sensing regulated proteins in the opportunistic pathogen Pseudomonas aeruginosa by proteomics, Environ. Microbiol., 2003, 5, 1350–1369 CrossRef CAS.
- M. Hentzer, et al., Inhibition of quorum sensing in Pseudomonas aeruginosa biofilm bacteria by a halogenated furanone compound, Microbiology, 2002, 148, 87–102 CrossRef CAS.
- P. S. Rajesh and V. Ravishankar Rai, Quorum quenching activity in cell-free lysate of endophytic bacteria isolated from Pterocarpus santalinus Linn., and its effect on quorum sensing regulated biofilm in Pseudomonas aeruginosa PAO1, Microbiol. Res., 2014, 169, 561–569 CrossRef CAS.
- K.-F. Kong, et al., Pseudomonas aeruginosa AmpR is a global transcriptional factor that regulates expression of AmpC and PoxB beta-lactamases, proteases, quorum sensing, and other virulence factors, Antimicrob. Agents Chemother., 2005, 49, 4567–4575 CrossRef CAS.
- Z.-P. Ma, et al., Anti-quorum Sensing Activities of Selected Coral Symbiotic Bacterial Extracts From the South China Sea, Front. Cell. Infect. Microbiol., 2018, 8, 144 CrossRef.
- D. J. Hassett, L. Charniga, K. Bean, D. E. Ohman and M. S. Cohen, Response of Pseudomonas aeruginosa to pyocyanin: mechanisms of resistance, antioxidant defenses, and demonstration of a manganese-cofactored superoxide dismutase, Infect. Immun., 1992, 60, 328–336 CAS.
- D. G. Davies, et al., The involvement of cell-to-cell signals in the development of a bacterial biofilm, Science, 1998, 280, 295–298 CrossRef CAS.
- M. H. Rashid and A. Kornberg, Inorganic polyphosphate is needed for swimming, swarming, and twitching motilities of Pseudomonas aeruginosa, Proc. Natl. Acad. Sci. U.S.A., 2000, 97, 4885–4890 CrossRef CAS.
- V. Gopu, C. K. Meena and P. H. Shetty, Quercetin Influences Quorum Sensing in Food Borne Bacteria: In-Vitro and In-Silico Evidence, PLoS One, 2015, 10, e0134684 CrossRef PubMed.
- M. A. Hossain, et al., Impact of phenolic compounds in the acyl homoserine lactone-mediated quorum sensing regulatory pathways, Sci. Rep., 2017, 7, 10618 CrossRef PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9ra05725k |
| ‡ Current affiliation: Department of Microbiology and Molecular Genetics, Institute for Medical Research Israel-Canada, The Hebrew University-Hadassah Medical School, POB 12272, The Hebrew University of Jerusalem, 91120, Jerusalem, Israel. |
| This journal is © The Royal Society of Chemistry 2019 |