Open Access Article
Open Access ArticleIdentification of parameters needed for optimal anaerobic co-digestion of chicken manure and corn stover†
Yilong Yana,
Ziwen Du*a,
Liqiu Zhang *a,
Li Fenga,
Dezhi Suna,
Yan Danga,
Dawn E. Holmesb and
Jessica A. Smithc
*a,
Li Fenga,
Dezhi Suna,
Yan Danga,
Dawn E. Holmesb and
Jessica A. Smithc
aBeijing Key Laboratory for Source Control Technology of Water Pollution, Engineering Research Center for Water Pollution Source Control and Eco-remediation, College of Environmental Science & Engineering, Beijing Forestry University, 35 Tsinghua East Road, Beijing 100083, China. E-mail: ziwendu@bjfu.edu.cn; zhangliqiu@163.com; zhangliqiu@bjfu.edu.cn
bDepartment of Physical and Biological Sciences, Western New England University, 1215 Wilbraham Road, Springfield, Massachusetts 01119, USA
cDepartment of Biomolecular Sciences, Central Connecticut State University, 1615 Stanley Street, New Britain, CT 06050, USA
First published on 19th September 2019
Abstract
While studies have shown that anaerobic co-digestion of chicken manure (CM) and corn stover (CS) is an efficient method to treat these agricultural wastes, the microbial ecology of these systems and optimal parameters for the digestion process are yet to be determined. In this study, the effects of different initial substrate concentrations and CS![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CM mixture ratios on co-digestion and microbial community structure were evaluated. Results demonstrated that both the highest cumulative methane yields and methane production rates were obtained from reactors with a CS
CM mixture ratios on co-digestion and microbial community structure were evaluated. Results demonstrated that both the highest cumulative methane yields and methane production rates were obtained from reactors with a CS![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CM ratio of 1
CM ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 during hemi-solid-state anaerobic digestion (HSS-AD). Cumulative methane yields and methane production rates were 24.8% and 42% lower in solid-state anaerobic digestion (SS-AD) reactors using the same CS
1 during hemi-solid-state anaerobic digestion (HSS-AD). Cumulative methane yields and methane production rates were 24.8% and 42% lower in solid-state anaerobic digestion (SS-AD) reactors using the same CS![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CM ratios. Analysis of microbial community structures revealed that cellulolytic bacteria and a diversity of syntrophic microorganisms capable of direct interspecies electron transfer (DIET) and hydrogen interspecies transfer (HIT) were enriched in the best-performing reactors. Methanosarcina species also dominated during HSS-AD, and their presence was positively correlated with methane production in the reactors.
CM ratios. Analysis of microbial community structures revealed that cellulolytic bacteria and a diversity of syntrophic microorganisms capable of direct interspecies electron transfer (DIET) and hydrogen interspecies transfer (HIT) were enriched in the best-performing reactors. Methanosarcina species also dominated during HSS-AD, and their presence was positively correlated with methane production in the reactors.
Introduction
Agricultural wastes, such as animal manure and crop residue (for example, corn stover), are a major concern to the farming industry and are known to be major contributors to soil pollution and greenhouse gas production.1 Each year, 400 million tons of chicken manure (CM) and 300 million tons of corn stover (CS) are produced in China alone.2 Anaerobic digestion (AD) is a process that can convert these wastes into biogas as a renewable energy source, and any residual solids can be turned into nutrient-rich fertilizers.3 However, mono-digestion of these feedstocks is not desirable because CS contains a high proportion of recalcitrant lignocellulose,4 and CM has a low C/N ratio,5 both factors that lead to poor AD performance.To minimize these inhibitory factors and improve biogas yields, co-digestion of both of these substrates is recommended.6 While studies have already shown that CS![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CM co-digestion strategies improve reactor performance,7 use of large-scale anaerobic digesters has not been frequently implemented due to low energy production and poor economic viability. This limited application is largely because various operating conditions and the effects they have on microbial ecology have not yet been adequately studied.
CM co-digestion strategies improve reactor performance,7 use of large-scale anaerobic digesters has not been frequently implemented due to low energy production and poor economic viability. This limited application is largely because various operating conditions and the effects they have on microbial ecology have not yet been adequately studied.
The type of AD applied often depends on the percentage of total solids (TS) in the waste to be treated. Wet state anaerobic digestion (WS-AD) is used for waste with TS content <10%, hemi-solid-state anaerobic digestion (HSS-AD) is used for waste with TS content between 10–15%, and solid-state anaerobic digestion (SS-AD) is used for waste with TS content >15%. While WS-AD is generally used for treatment of animal manure, HSS-AD and SS-AD are more effective during treatment of complex materials such as lignocellulosic crop residue8 and are likely to be more compatible with co-digestion feedstocks. Other advantages of HSS-AD and SS-AD include higher volumetric methane output, enhanced nutrient balance, sufficient buffer capacity, smaller reactor requirements, and lower energy input and water addition.9
Another major factor that must be taken into consideration when optimizing AD is the microbial ecology of the system. Successful AD relies on a diverse microbial consortium working together to convert complex organic matter into methane using a characteristic four-step process: hydrolysis, acidogenesis, acetogenesis, and methanogenesis.10 Hydrolysis and acidogenesis lead to rapid production of fatty acids and alcohols, and the robust microbes involved in these reactions can survive a wide range of environmental conditions. Further conversion of fatty acids and alcohols relies on interspecies electron transfer (IET) between syntrophic bacteria and methanogens. IET via hydrogen/formate interspecies electron transfer (HFIT) has been recognized as the conventional route of biogas formation for several decades.11–13 A more recently discovered form of IET is direct interspecies electron transfer (DIET), in which syntrophic partners forge biological extracellular protein connections to exchange electrons.14–18 IET often occurs relatively slowly and is extremely sensitive to operating conditions.10,11 Therefore, understanding how to promote these syntrophic interactions, especially in anaerobic digesters with high organic loads, is of great interest to the scientific community.15
Although many studies have shown that supplementation of conductive materials such as granular activated carbon,19 biochar,20 and carbon cloth21 can enhance IET during AD,22–25 studies to determine how to enhance syntrophic metabolism by simply adjusting operational parameters are lacking. Evidence does suggest that altering initial substrate concentrations (ISC) based on total solid content affects IET. For example, extremely high solid content during AD may cause volatile fatty acids (VFAs), such as propionate or butyrate, to accumulate.26,27 Large quantities of VFAs are toxic to methanogens and will lead to high hydrogen partial pressures which inhibit hydrogen interspecies transfer (HIT).28 In addition, data regarding bacterial and methanogenic communities enriched during co-digestion of CS and CM is needed as it will help provide insight into mechanisms of IET that might be occurring in the reactors. This information can then be used to enhance the co-digestion process. For example, studies have shown that co-digestion of CM with microalgae stimulates growth of Methanothrix and Methanosarcina species, both organisms known to participate in DIET.29
The discovery of optimal operational conditions for treatment of agricultural wastes with high TS content, and more importantly, the elucidation of microbial interactions key to this process, are essential for future large-scale application. Therefore, reactors were assembled with a variety of ISC (based on total solid content) and CS![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CM mixture ratios to identify which operational parameters yielded the best results. Bacterial and archaeal communities were also characterized and relationships between community structure, methane production, ISC, and mixture ratios were identified.
CM mixture ratios to identify which operational parameters yielded the best results. Bacterial and archaeal communities were also characterized and relationships between community structure, methane production, ISC, and mixture ratios were identified.
Materials and methods
Reactor operating parameters
Reactor operating conditions used in this study were based on settings that have been previously outlined.30 Briefly, CM was collected from the Deqingyuan Ecological Farm in Beijing, China. CS, which included leaves, stalks, and cobs, was obtained from a cornfield near Zhangjiakou City, Hebei, China, and naturally air-dried to a moisture content of less than 10%. Dried CS was crushed with a grinder and passed through a 3 mm sieve.The inoculum used in this study was effluent from the Deqingyuan Ecological Farm full-scale mesophilic liquid anaerobic digester fed with chicken manure (operated by Helee Bio-Energy Co. Ltd, Beijing, China). The effluent was incubated anaerobically at 37 ± 0.2 °C for 14 days to minimize residual biodegradable organic matter before being inoculated into digesters under mesophilic conditions. Detailed characteristics of substrates and inoculum are summarized in the ESI.†
Batch anaerobic digestion experiments were carried out in triplicate using 2 L glass bottles with a working volume of 1 L, and were placed in thermostatic water baths under mesophilic conditions (37 ± 0.2 °C). Both feedstocks were mixed with Milli-Q water (Millipore, USA) and inoculum to obtain a mixture with a total solids content of 10.4–10.7% for HSS-AD, and 17.3–17.8% for SS-AD. The feedstock to inoculum (F/I) ratio of each digester was 3, based on volatile solids (VS). Based on results from a previous report,7 co-digestion of corn stover and chicken manure was applied at ratios of 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 1
1 and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (based on VS) due to higher biodegradability and synergistic effects. The corresponding feedstock carbon to nitrogen (C/N) ratios were 25.7 for CS
1 (based on VS) due to higher biodegradability and synergistic effects. The corresponding feedstock carbon to nitrogen (C/N) ratios were 25.7 for CS![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CM of 3
CM of 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 16.8 for CS
1 and 16.8 for CS![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CM of 1
CM of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, which were both in the preferred C/N range of 15–30.7 Each digester was manually shaken once a day for about 2 min. To monitor the digestion process, 40 mL of digestate from each digester was sampled anaerobically every 5 days (Thermo Scientific, USA). Biogas samples were taken every day during the co-digestion process with 3 L gas collection bags (E-Switch, China).
1, which were both in the preferred C/N range of 15–30.7 Each digester was manually shaken once a day for about 2 min. To monitor the digestion process, 40 mL of digestate from each digester was sampled anaerobically every 5 days (Thermo Scientific, USA). Biogas samples were taken every day during the co-digestion process with 3 L gas collection bags (E-Switch, China).
Analytical methods
TS, VS, total ammonia nitrogen (TAN), and total alkalinity were analyzed according to Standard Methods.31 VFAs (acetate, propionate, and butyrate) were measured with high-performance liquid chromatography (HPLC, Agilent 1260, USA) as previously described.30 Elemental composition (carbon and nitrogen) of freeze-dried CM and CS were measured using an elemental analyzer (Vario EL cube, Germany). A pH-meter was used to measure pH (HACH, USA). Electrical conductivity was measured with a conductivity meter (Mettler Toledo, USA). Free ammonia nitrogen (FAN) was calculated from TAN concentrations according to a previously described formula.32 Gas volume from the gas collection bag was measured with a digital mass flow meter (Omega, USA) every 24 hours. Biogas composition (CO2, CH4) was determined by a 7890B gas chromatograph system (Agilent, USA) equipped with a thermal conductivity detector.High-throughput sequencing
Digestate samples (40 mL) were collected from digesters at the end of operation (day 40). All samples were immediately frozen with liquid nitrogen prior to DNA extraction and stored at −80 °C. For total DNA extraction, frozen samples were ground into a fine powder with a mortar and pestle before processing with the BIO101 FastDNA Soil Kit (MP Biomedical, France), according to the manufacturer's instructions. Extracted DNA was detected and quantified using 1% agarose gel electrophoresis and a micro-volume spectrophotometer (NanoDrop 2000, USA). Archaeal and bacterial 16S rRNA gene fragments were amplified via polymerase chain reaction (PCR) with the following primer sets: (Arch519F/Arch915R) and (515F/806R).33,34 High-throughput sequencing was done on an Illumina Hiseq 2000 platform (Illumina, USA) by Allwegene Biotechnology Co., Ltd. (Beijing, China). Sequences were placed into various operational taxonomic units with Pyrosequencing Pipeline Software (https://pyro.cme.msu.edu). Raw sequence files have been submitted to the NCBI Sequence Read Archive database under accession no. SRP130964.Statistical analysis
Redundancy analysis (RDA) for both community structure and operation parameters were calculated using Canoco (version 5.0).35 Ranked Spearman correlation was applied to determine the correlation between microbial populations and operational conditions or performance.35 All statistical and correlation analyses were performed using IBM SPSS Statistics 23.Results and discussion
Anaerobic digestion performance using varying operating parameters
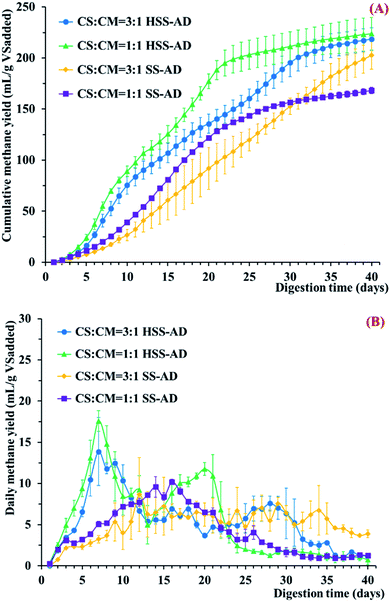
In order to determine which operating parameters promoted the best anaerobic digestion of CS and CM, four different reactor conditions were tested in triplicate; HSS-AD with a CS![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CM ratio of 3
CM ratio of 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (HSS3:1), HSS-AD with a CS
1 (HSS3:1), HSS-AD with a CS![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CM ratio of 1
CM ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (HSS1:1), SS-AD with a CS
1 (HSS1:1), SS-AD with a CS![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CM ratio of 3
CM ratio of 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (SS3:1), and SS-AD with a CS
1 (SS3:1), and SS-AD with a CS![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CM ratio of 1
CM ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (SS1:1). Methane concentrations were monitored during the entire operation period (40 days) (Fig. 1). Cumulative methane production in the HSS-AD reactors was significantly (p < 0.01) higher than the SS-AD reactors (Fig. 1A). The HSS1:1 reactors produced the highest amount of cumulative methane (223.7 ± 16.0 mL per g VSadded), with yields about 33.0% higher than the SS1:1 reactors. Cumulative methane formed by the HSS1:1 reactors was about 15% higher than yields from a similar reactor in a previous study, which may have been due to differences in inoculum.7 The experiments also showed that reactors with 1
1 (SS1:1). Methane concentrations were monitored during the entire operation period (40 days) (Fig. 1). Cumulative methane production in the HSS-AD reactors was significantly (p < 0.01) higher than the SS-AD reactors (Fig. 1A). The HSS1:1 reactors produced the highest amount of cumulative methane (223.7 ± 16.0 mL per g VSadded), with yields about 33.0% higher than the SS1:1 reactors. Cumulative methane formed by the HSS1:1 reactors was about 15% higher than yields from a similar reactor in a previous study, which may have been due to differences in inoculum.7 The experiments also showed that reactors with 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 CS
1 CS![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CM ratios yielded methane production rates that were 35.7 ± 7.7% higher than reactors provided with a 3
CM ratios yielded methane production rates that were 35.7 ± 7.7% higher than reactors provided with a 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 feedstock ratio. These differences can likely be attributed to higher lignocellulose content in the 3
1 feedstock ratio. These differences can likely be attributed to higher lignocellulose content in the 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 feedstock, making it significantly harder to degrade.
1 feedstock, making it significantly harder to degrade.
In addition to higher overall methane production, the HSS-AD reactors started producing methane early in the digestion process, with the highest daily methane yields being generated around day 7 (Fig. 1B). Conversely, the SS-AD reactors did not show strong start-up performance, and overall daily methane yields never exceeded 10.0 mL per g VSadded. Both daily methane yields and methane content during the start-up period were lowest in the SS3:1 reactors (Fig. 1B and ESI Fig. S1†), and total cumulative methane was only 202.6 ± 13.5 mL per g VSadded in these reactors (Fig. 1A). While the 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 feedstock clearly hindered the start-up period in the SS-AD reactors, it did not appear to impact the start-up period in the HSS-AD reactors (Fig. 1A). These results suggest that hydrolysis/acidification was not a limiting factor in HSS-AD reactors.
1 feedstock clearly hindered the start-up period in the SS-AD reactors, it did not appear to impact the start-up period in the HSS-AD reactors (Fig. 1A). These results suggest that hydrolysis/acidification was not a limiting factor in HSS-AD reactors.
Impact of CS and CM feedstock mixture ratios on volatile fatty acids, nitrogen-containing compounds, and volatile solids
VFAs, FAN, and TAN were also measured throughout the 40 day co-digestion period in order to assess the potential impact of CS and CM feedstock mixture ratios on microbial metabolism (Fig. 2). VFAs accumulated rapidly during the early stages of treatment in all four reactor conditions (Fig. 2A), demonstrating that hydrolytic and acidogenic microorganisms were active.36 However, VFA formation was higher in reactors provided with 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 CS
1 CS![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CM feedstock ratios, indicating that the higher lignocellulose content in the 3
CM feedstock ratios, indicating that the higher lignocellulose content in the 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 reactors may have limited hydrolysis.9,37 In addition, accumulation of VFAs throughout the co-digestion process was significantly greater in SS-AD reactors (Fig. 2A). These results suggest that microorganisms that could cooperatively metabolize VFAs to methane were enriched early in the digestion process in the HSS-AD reactors.
1 reactors may have limited hydrolysis.9,37 In addition, accumulation of VFAs throughout the co-digestion process was significantly greater in SS-AD reactors (Fig. 2A). These results suggest that microorganisms that could cooperatively metabolize VFAs to methane were enriched early in the digestion process in the HSS-AD reactors.
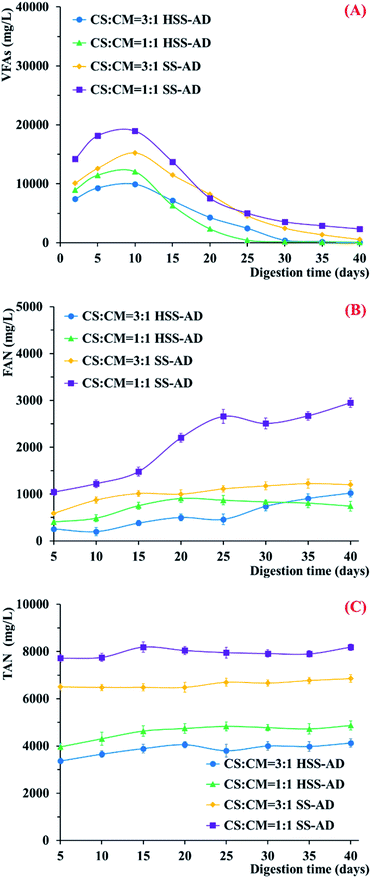 | ||
| Fig. 2 Change in (A) VFAs, (B) FAN, (C) and TAN during anaerobic co-digestion with different mixture ratios and ISCs. Error bars represent standard deviations from triplicate samples. | ||
Two large methane peaks were produced in the HSS-AD reactors over the course of the experiment (Fig. 1B). The first large peak occurred between days 5–10, which was also when VFA concentrations were highest (Fig. 2A), and the second peak occurred between days 19–21 for HSS1:1 reactors and days 27–29 for HSS3:1 reactors when VFAs dropped below 1107 mg L−1 and 2350 mg L−1, respectively. These peaks may have corresponded with shifts in the dominant methanogenic communities over time. Early in the experiment, methanogens were likely to be using hydrogen generated by fermentative bacteria. These hydrogenotrophic methanogens initially kept H2 concentrations below the thermodynamic threshold,38–40 however, in the organically rich HSS-AD reactors, H2 production eventually outpaced H2 consumption and H2 producing pathways are not thermodynamically favorable at high H2 concentrations.41
Therefore, fermentative microorganisms tend to shift their metabolism from H2 producing pathways to VFA (i.e. propionate, butyrate, acetate) producing pathways when H2 concentrations are high.41–44 Homoacetogenesis is also favored over hydrogenotrophic methanogenesis at elevated hydrogen partial pressures.45 Acetoclastic methanogens can then directly utilize acetate as a substrate for methanogenesis,46 and VFAs like propionate and butyrate can serve as electron donors for syntrophic partnerships between bacteria and methanogens.47–49 Evidence supporting the theory that hydrogenotrophic methanogens were dominant when the first peak was formed comes from the finding that VFAs were most abundant during this period as methanogens were not yet able to convert these compounds to methane. During the second peak, on the other hand, VFA concentrations were low because acetoclastic methanogens and methanogens that could participate in DIET became more active. Further investigations into microbial community structure in the HSS-AD reactors during the first and second methane peaks are warranted.
Free ammonia nitrogen concentrations were highest in SS-AD reactors with CS![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CM ratios of 1
CM ratios of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (Fig. 2B), with concentrations remaining well above 1000–2000 mg L−1 throughout the digestion process. The highest TAN was also observed using this operating parameter (Fig. 2C). High FAN and TAN negatively impact methanogenesis and are known to be major inhibitors of anaerobic digestion.50,51 In fact, FAN concentrations >1110 mg L−1 have been shown to completely inhibit anaerobic systems.52 This explains why the SS1:1 reactors produced the lowest cumulative methane yields (Fig. 1A). FAN remained ≤1000 mg L−1 in the other three reactor conditions, indicating that ammonia concentrations did not adversely affect metabolic activity in these digesters (Fig. 2B).
1 (Fig. 2B), with concentrations remaining well above 1000–2000 mg L−1 throughout the digestion process. The highest TAN was also observed using this operating parameter (Fig. 2C). High FAN and TAN negatively impact methanogenesis and are known to be major inhibitors of anaerobic digestion.50,51 In fact, FAN concentrations >1110 mg L−1 have been shown to completely inhibit anaerobic systems.52 This explains why the SS1:1 reactors produced the lowest cumulative methane yields (Fig. 1A). FAN remained ≤1000 mg L−1 in the other three reactor conditions, indicating that ammonia concentrations did not adversely affect metabolic activity in these digesters (Fig. 2B).
Removal of VS and methane conversion rates also varied significantly between the different reactor conditions (Fig. 3). Significantly higher proportions (p < 0.01) of VS were removed in HSS1:1 (60.1 ± 2.4%) and HSS3:1 (59.0 ± 1.4%) reactors than in SS1:1 (51.4 ± 0.8%) and SS3:1 (56.2 ± 2.0%) reactors. Higher methane conversion rates were also observed in HSS1:1 (372.1 ± 27.7 mLmethane per g VSremoved) and HSS3:1 (370.6 ± 19.3 mLmethane per g VSremoved) reactors than in SS1:1 (327.3 ± 6.1 mLmethane per g VSremoved) and SS3:1 (360.2 ± 24.9 mLmethane per g VSremoved) reactors (Fig. 3). Overall, HSS-AD reactors provided with a CS![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CM ratio of 1
CM ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 performed best with the highest cumulative methane yields and methane production rates.
1 performed best with the highest cumulative methane yields and methane production rates.
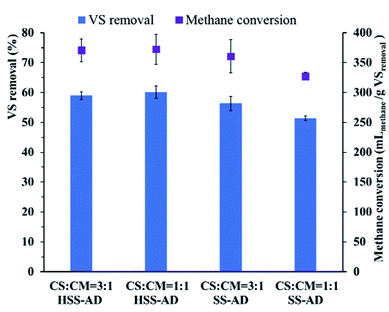 | ||
| Fig. 3 Removal of volatile solids (VS) and methane conversion in anaerobic co-digestion using different mixture ratios and ISCs. Error bars represent standard deviations from triplicate samples. | ||
Microbial community characterization
High-throughput sequencing of bacterial and archaeal 16S rRNA gene fragments was done to determine the impact that reactor operational parameters (ISC and CS![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CM mixture ratios) can have on microbial community structure in reactors treating high-solid agricultural waste (Fig. 4).
CM mixture ratios) can have on microbial community structure in reactors treating high-solid agricultural waste (Fig. 4).
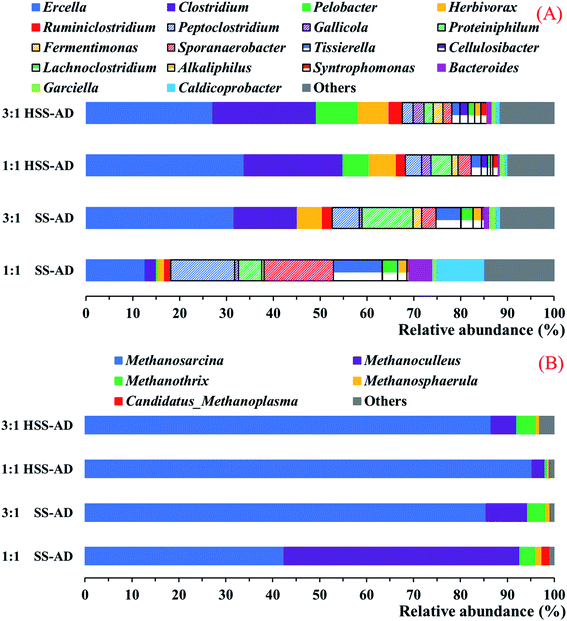 | ||
| Fig. 4 Relative distribution of (A) bacterial and (B) archaeal 16S rRNA gene sequences at the genus level. Sequences that accounted for less than 1.0% of the population were classified as “Others”. | ||
Bacterial community analysis
At the genus taxonomic level, there were several key differences between bacterial communities associated with well-performing HSS-AD reactors and lower methane-yielding SS-AD reactors (Fig. 4A). Both of the HSS-AD communities were abundant in species from the genera Ercella, Clostridium, Pelobacter, and Herbivorax, which accounted for 65.5% ± 0.8% of the population. These same four species accounted for only 16.8% in SS1:1 reactors, which were the poorest performing reactors (Fig. 1).Ercella and Clostridium were the most abundant genera in both of the well-performing HSS-AD reactors and SS3:1 (Fig. 4A). Ercella and Clostridium are members of the Clostridiales order which plays many roles in the anaerobic digestion process, including high-rates of cellulose hydrolysis, protein catabolism, and acidogenesis leading to production of short-chain fatty acids, CO2, and H2.53 Although both of these genera are primarily known for their fermentative metabolisms, some Clostridium species are capable of extracellular electron transfer to such electron acceptors as Fe(III), sulfur, and current-harvesting electrodes54–58 and Ercella can transfer electrons to insoluble sulfur compounds.58
The presence of Clostridium and Syntrophomonas species has been previously reported to be an indicator of AD stability.59,60 This is consistent with the high abundance of Clostridium and Syntrophomonas (p < 0.01) in HSS-AD communities, and the positive correlation between their abundance and cumulative methane production (p < 0.01, R2 = 0.78) (Fig. 1A, and 4A). This correlation between methane production and abundance of these bacteria can be explained by the fact that both of these genera have been shown to form syntrophic partnerships with methanogens.61 In addition to their cooperative growth with methanogens via HFIT,61 it is also possible that these species are capable of DIET to a methanogenic partner.
There are two hallmarks of a bacterium's capacity for DIET, the first is the ability to transfer electrons across the cell membrane to an extracellular electron acceptor (i.e. another microorganism),62,63 and the second is the presence of Geobacter-like type IV pilin proteins. Geobacter pili contain aromatic amino acids that are located at key positions within the protein chain64 and account for ≥9% of the amino acid residues with small gaps between them.65,66 Although the role of pili in extracellular electron transfer is under debate, pili from all of the bacteria known to be capable of DIET thus far have these characteristics.65,66 In fact, recent identification of a Geobacter-like pilin protein in Syntrophus acetitrophicus led to the discovery that this species, once thought to only be capable of HFIT,67,68 also has the capacity for DIET.66 A number of genomes from Clostridiales species known to participate in HFIT,61 including Syntrophomonas, also have genes coding for Geobacter-like pili,66 suggesting that these species might also be capable of DIET. Support for this comes from the finding that Syntrophomonas species were likely to be participating in DIET in anaerobic digesters supplemented with ferroferric oxide.69
In addition, both Ercella and Clostridium species are able to transfer electrons outside of the cell suggesting that it is possible that these genera were not only providing electrons to methanogens through fermentative by-products, but may have also been participating in DIET.
Pelobacter species were also significantly enriched in HSS-AD reactors (9.1% of total community in HSS3:1 and 5.6% in HSS1:1). However, sequences from this genus were barely detected in SS-AD reactors (0.02% of total community in SS3:1 and 0.5% in SS1:1). Pelobacter has been shown to grow syntrophically with methanogens via HFIT,70–72 but several species from this genus also have traits that are characteristic of bacteria with the capacity for DIET. For example, Pelobacter species are capable of electron transfer to Fe(III) and S0,73–75 and have Geobacter-like type IV pili.76
Species from the genus Sporanaerobacter were also significant members of all four digesters, but had the highest abundance in SS1:1 (14.8%). Sporanaerobacter metabolize proteins and carbohydrates with reduction of elemental sulfur, an extracellular electron acceptor.77 In a previous study, Sporanaerobacter species were enriched in anaerobic digesters supplemented with conductive carbon cloth, and were likely participating in DIET with Methanosarcina species.77,78
Herbivorax and Cellulosibacter, both genera with cellulolytic metabolisms,79,80 were enriched in HSS-AD reactors (Fig. 4). Their abundance was significantly lower in SS-AD reactors suggesting that their scarcity could be linked to impairment of lignocellulose degradation and lower methane production rates.
Overall, results from the bacterial community analysis suggest that a variety of syntrophic bacteria involved in both HFIT and/or DIET were significantly (p < 0.05) enriched in HSS-AD reactors, signifying that syntrophic–methanogenic associations were favored during HSS-AD.
Archaeal community analysis
The predominant methanogenic genera found in all samples included Methanosarcina, Methanoculleus, Methanothrix, and Methanosphaerula, however, the relative abundance of each genus varied between conditions (Fig. 4B). Methanosarcina was dominant in HSS3:1 (86.6% of the overall community), HSS1:1 (95.3%), and SS3:1 (85.5%). However, Methanoculleus (50.2%) were more abundant than Methanosarcina (42.4%) in SS1:1. The relative abundance of Methanosarcina species was positively correlated with cumulative methane production (P < 0.01, R2 = 0.84) (Fig. 1A, and 4B). These results are consistent with previous studies that have shown that Methanosarcina species promote stable digestion, particularly in reactors treating complex organic waste.81,82Methanosarcina species are mixotrophic methanogens that are able to metabolize acetate, hydrogen, and C1 compounds,83 and along with Methanothrix (formerly Methanosaeta), are some of the few methanogens shown to be capable of DIET in culture studies.71 Methanothrix are obligate acetoclastic methanogens, and accounted for <4.2% of the overall archaeal community within all digesters (Fig. 4B). While high acetate concentrations in the reactors should have favored growth of both Methanosarcina and Methanothrix,84–86 Methanothrix are sensitive to high concentrations of VFAs.87,88 Methanosarcina, on the other hand, are typically able to survive exposure to environmental stressors, such as high VFA or FAN concentrations, because they can aggregate to form irregular clumps that protect them from harsh environmental conditions.88,89
Strictly hydrogenotrophic Methanoculleus species predominated SS1:1 reactor archaeal communities (50.2%) (Fig. 4B). Methanoculleus abundance was positively correlated with concentrations of FAN and TAN (P < 0.01, R2 = 0.78) (Fig. 2B, C, and 4B), and negatively correlated with cumulative methane production (P < 0.01, R2 = −0.84) (Fig. 1A, and 4B). This is consistent with reports that Methanoculleus tend to dominate reactors with high concentrations of VFAs and ammonia (Fig. 2).60,90 Sequences from other hydrogenotrophic methanogenic genera, including Methanospaerula and Candidatus-Methanoplasma, were also found in all of the digesters (Fig. 4B). However, their abundance was low, indicating that these hydrogenotrophic methanogens played a lesser role.
It is clear that most of the methanogenic activity detected in efficiently operating reactors (HSS3:1, HSS1:1, SS3:1) could be attributed to Methanosarcina. Not only is this genus able to survive harsh conditions found in anaerobic digesters with high organic loads, it is also capable of syntrophic growth with syntrophic bacteria through both HFIT and DIET72,91 and can use a variety of fermentative products as substrates for methanogenesis (acetate, H2/CO2, formate, C1 compounds).83
Correlation between microbial community composition and reactor performance
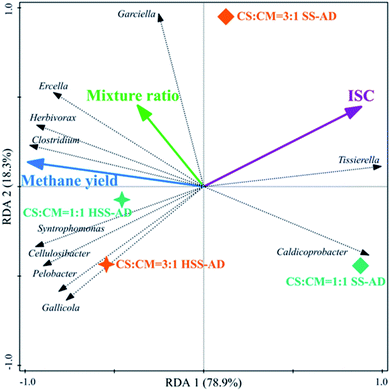
Redundancy analysis (RDA) was conducted to elucidate relationships between microbial community structure, operational conditions (ISC and mixture ratios), and reactor performance (cumulative methane production) (Fig. 5 and 6). RDA showed that both ISC and mixture ratio had a significant impact on bacterial community composition (Fig. 5). The relative abundance of bacterial sequences from Herbivorax, Cellulosibacter, Ercella, Syntrophomonas, and Pelobacter positively corresponded with methane yields (Fig. 5). As previously discussed, these genera are associated with degradation of cellulose and syntrophic metabolism of alcohols or short-chain fatty acids, both of which enhance reactor performance by preventing the accumulation of inhibitory intermediates. On the other hand, these genera were negatively correlated with ISC and thus primarily clustered with the HSS-AD samples. This indicates that a decrease in ISC promotes the enrichment of these beneficial genera which aid in the production of methane (Fig. 1). Abundance of Ercella, Herbivorax, and Clostridium species were partially associated with an increase of CS in feedstocks, which makes sense because these are cellulolytic bacteria that would be enriched when lignocellulose content is high.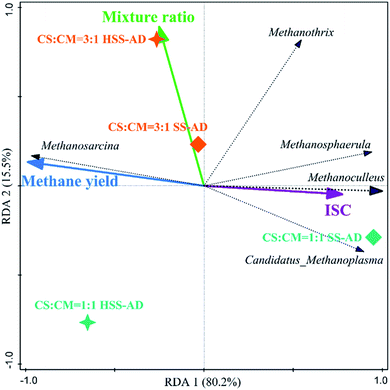 | ||
| Fig. 6 Redundancy analysis triplot ordination diagram using the archaeal community at the genus level (archaeal genera fitting greater than 90% were displayed). | ||
The presence of such fermentative genera as Clostridium, Gallicola, and Garciella, were also significantly influenced by operational parameters and their presence appeared to promote methanogenesis (Fig. 5). In contrast, the relative abundance of sequences from Caldicoprobacter and Tissierella were positively correlated with ISC and negatively correlated with mixture ratio (Fig. 5), suggesting that these bacteria are likely favored in digesters with high ammonia concentrations (SS1:1). This is consistent with previous reports that showed that Caldicoprobacter and Tissierella were dominant in anaerobic digesters with excess ammonia or alkaline conditions.92,93
ISC and mixture ratio also directly influenced archaeal community structure (Fig. 6). For example, Methanoculleus and Methanosphaerula showed a significant (P < 0.01) positive correlation with ISC, and were clustered closely with the SS1:1 sample, which were parameters that yielded the lowest methane productions. The high prevalence of obligate hydrogenotrophic methanogens in SS1:1 reactors suggests that hydrogen was the main electron carrier between bacteria and methanogens.84 In this syntrophic scenario, the hydrogen partial pressure must typically be kept lower than 10−5 atm by hydrogen consumers, and the value of Gibbs free energy discharge of the entire methanogenic process is thereafter negative, making syntrophic methanogenesis thermodynamically feasible.94 However, in SS1:1 reactors, excess FAN likely inhibited hydrogenotrophic methanogenesis, and diffusion limitations may have also resulted in local hydrogen accumulation, all of which may cause a significant increase of hydrogen partial pressure in these reactors, further inhibiting the syntrophic metabolism.51,84,95
Methanosarcina showed a significant (P < 0.01) positive correlation with cumulative methane production, and was negatively correlated with ISC (Fig. 6). The metabolic plasticity and morphological characteristics of Methanosarcina are likely to give them a competitive advantage in these systems. They can utilize DIET for methane production when conditions (i.e. high ammonia concentrations) are inhibitory for hydrogenotrophic methanogenesis.95
Conclusions
The HSS-AD reactor provided with feedstock at a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 CS
1 CS![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CM ratio performed better than the other three operational parameters (HSS-AD3:1, SS-AD1:1, SS-AD3:1). These optimal parameters promoted methane production while suppressing the accumulation of VFAs and FAN, both compounds known to inhibit the biomethanogenic process. Analysis of bacterial and archaeal communities demonstrated conditions associated with the highest performing reactors enriched for bacteria that could form syntrophic partnerships with methanogens (i.e. Methanosarcina). Results from this study should be taken into consideration in future attempts to optimize full-scale digester performance.
CM ratio performed better than the other three operational parameters (HSS-AD3:1, SS-AD1:1, SS-AD3:1). These optimal parameters promoted methane production while suppressing the accumulation of VFAs and FAN, both compounds known to inhibit the biomethanogenic process. Analysis of bacterial and archaeal communities demonstrated conditions associated with the highest performing reactors enriched for bacteria that could form syntrophic partnerships with methanogens (i.e. Methanosarcina). Results from this study should be taken into consideration in future attempts to optimize full-scale digester performance.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This research was financially supported by the “Major Science and Technology Program for Water Pollution Control and Treatment” (No. 2015ZX07203005), the Fundamental Research Funds for the Central Universities (No. BLX201725), the National Natural Science Foundation of China (No. 51578066 and 51608036), and the Natural Science Foundation of Beijing Municipality (No. 8182037 and 8184079).References
- J. J. Owen and W. L. Silver, Glob. Chang. Biol., 2015, 21, 550–565 CrossRef PubMed.
- P. Zhang, Y. Yang, Y. Tian, X. Yang, Y. Zhang, Y. Zheng and L. Wang, Renewable Sustainable Energy Rev., 2009, 13, 2571–2579 CrossRef.
- L. Wu, Y. Yang, S. Chen, M. Zhao, Z. Zhu, S. Yang, Y. Qu, Q. Ma, Z. He, J. Zhou and Q. He, Water Res., 2016, 104, 1–10 CrossRef CAS PubMed.
- R. A. Labatut, L. T. Angenent and N. R. Scott, Bioresour. Technol., 2011, 102, 2255–2264 CrossRef CAS PubMed.
- H. M. El-Mashad and R. Zhang, Bioresour. Technol., 2010, 101, 4021–4028 CrossRef CAS PubMed.
- J. Pagés-Díaz, I. Pereda-Reyes, M. J. Taherzadeh, I. Sárvári-Horváth and M. Lundin, Chem. Eng. J., 2014, 245, 89–98 CrossRef.
- Y. Li, R. Zhang, C. Chen, G. Liu, Y. He and X. Liu, Bioresour. Technol., 2013, 149, 406–412 CrossRef CAS PubMed.
- Y. Li, R. Zhang, C. Chen, G. Liu, Y. He and X. Liu, Bioresour. Technol., 2013, 149, 406–412 CrossRef CAS PubMed.
- X. Ge, F. Xu and Y. Li, Bioresour. Technol., 2016, 205, 239–249 CrossRef CAS PubMed.
- A. J. Stams and C. M. Plugge, Nat. Rev. Microbiol., 2009, 7, 568–577 CrossRef CAS PubMed.
- M. Bryant, E. Wolin, M. Wolin and R. Wolfe, Arch. Mikrobiol., 1967, 59, 20–31 CAS.
- M. J. McInerney, J. R. Sieber and R. P. Gunsalus, Curr. Opin. Biotechnol., 2009, 20, 623–632 CrossRef CAS PubMed.
- A. J. Stams and C. M. Plugge, Nat. Rev. Microbiol., 2009, 7, 568 CrossRef CAS PubMed.
- Z. M. Summers, H. E. Fogarty, C. Leang, A. E. Franks, N. S. Malvankar and D. R. Lovley, Science, 2010, 330, 1413–1415 CrossRef CAS PubMed.
- D. R. Lovley, Energy Environ. Sci., 2011, 4, 4896–4906 RSC.
- P. M. Shrestha, A. E. Rotaru, Z. M. Summers, M. Shrestha, F. Liu and D. R. Lovley, Appl. Environ. Microbiol., 2013, 79, 2397–2404 CrossRef CAS PubMed.
- A. E. Rotaru, P. M. Shrestha, F. Liu, M. Shrestha, D. Shrestha, M. Embree, K. Zengler, C. Wardman, K. P. Nevin and D. R. Lovley, Energy Environ. Sci., 2014, 7, 408–415 RSC.
- Q. Lin, J. De Vrieze, J. Li and X. Li, Bioresour. Technol., 2016, 209, 228–236 CrossRef CAS PubMed.
- F. Liu, A. E. Rotaru, P. M. Shrestha, N. S. Malvankar, K. P. Nevin and D. R. Lovley, Energy Environ. Sci., 2012, 5, 8982–8989 RSC.
- S. Chen, A. E. Rotaru, P. M. Shrestha, N. S. Malvankar, F. Liu, W. Fan, K. P. Nevin and D. R. Lovley, Sci. Rep., 2014, 4, 5019 CrossRef CAS PubMed.
- S. Chen, A. E. Rotaru, F. Liu, J. Philips, T. L. Woodard, K. P. Nevin and D. R. Lovley, Bioresour. Technol., 2014, 173, 82–86 CrossRef CAS PubMed.
- Y. Dang, D. E. Holmes, Z. Zhao, T. L. Woodard, Y. Zhang, D. Sun, L.-Y. Wang, K. P. Nevin and D. R. Lovley, Bioresour. Technol., 2016, 220, 516–522 CrossRef CAS PubMed.
- Y. Dang, D. Sun, T. L. Woodard, L. Y. Wang, K. P. Nevin and D. E. Holmes, Bioresour. Technol., 2017, 238, 30–38 CrossRef CAS PubMed.
- J. Y. Lee, S. H. Lee and H. D. Park, Bioresour. Technol., 2016, 205, 205–212 CrossRef CAS PubMed.
- G. Baek, J. Kim and C. Lee, Bioresour. Technol., 2016, 222, 344–354 CrossRef CAS PubMed.
- N. Duan, B. Dong, B. Wu and X. Dai, Bioresour. Technol., 2012, 104, 150–156 CrossRef CAS PubMed.
- C. Luo, F. Lü, L. Shao and P. He, Water Res., 2015, 68, 710–718 CrossRef CAS PubMed.
- I. Siegert and C. Banks, Process Biochem., 2005, 40, 3412–3418 CrossRef CAS.
- R. Li, N. Duan, Y. Zhang, Z. Liu, B. Li, D. Zhang and T. Dong, Waste Manag., 2017, 68, 120–127 CrossRef CAS PubMed.
- Y. Yan, L. Zhang, L. Feng, D. Sun and Y. Dang, Bioresour. Technol., 2018, 247, 660–668 CrossRef CAS PubMed.
- A. Apha, WPCF 2005, American Public Health Association, Washington, DC, USA., 2012 Search PubMed.
- K. H. Hansen, I. Angelidaki and B. K. Ahring, Water Res., 1998, 32, 5–12 CrossRef CAS.
- Y. Lei, D. Sun, Y. Dang, H. Chen, Z. Zhao, Y. Zhang and D. E. Holmes, Bioresour. Technol., 2016, 222, 270–276 CrossRef CAS PubMed.
- Y. Lei, L. Wei, T. Liu, Y. Xiao, Y. Dang, D. Sun and D. E. Holmes, Chem. Eng. J., 2018, 348, 992–999 CrossRef CAS.
- J. Lee, G. Han, S. G. Shin, T. Koo, K. Cho, W. Kim and S. Hwang, Chem. Eng. J., 2016, 300, 291–299 CrossRef CAS.
- K. Venkiteshwaran, B. Bocher, J. Maki and D. Zitomer, Microbiol. Insights, 2015, 8, 37–44 Search PubMed.
- R. Kinet, J. Destain, S. Hiligsmann, P. Thonart, L. Delhalle, B. Taminiau, G. Daube and F. Delvigne, Bioresour. Technol., 2015, 189, 138–144 CrossRef CAS PubMed.
- S. Tanisho, N. Kamiya and N. Wakao, Biochim. Biophys. Acta, Bioenerg., 1989, 973, 1–6 CrossRef CAS.
- B. Schink, Microbiol. Mol. Biol. Rev., 1997, 61, 262–280 CAS.
- L. T. Angenent, K. Karim, M. H. Al-Dahhan, B. A. Wrenn and R. Domíguez-Espinosa, Trends Biotechnol., 2004, 22, 477–485 CrossRef CAS PubMed.
- P. H. Janssen, Anim. Feed Sci. Technol., 2010, 160, 1–22 CrossRef CAS.
- D. Arslan, K. Steinbusch, L. Diels, H. De Wever, C. Buisman and H. Hamelers, Bioresour. Technol., 2012, 118, 227–234 CrossRef CAS PubMed.
- C. M. Spirito, H. Richter, K. Rabaey, A. J. Stams and L. T. Angenent, Curr. Opin. Biotechnol., 2014, 27, 115–122 CrossRef CAS PubMed.
- K. J. Steinbusch, H. V. Hamelers, C. M. Plugge and C. J. Buisman, Energy Environ. Sci., 2011, 4, 216–224 RSC.
- O. R. Kotsyurbenko, M. V. Glagolev, A. N. Nozhevnikova and R. Conrad, FEMS Microbiol. Ecol., 2001, 38, 153–159 CrossRef CAS.
- D. Holmes and J. Smith, Adv. Appl. Microbiol., 2016, 97, 1–61 CAS.
- Z. Zhao, Y. Zhang, D. E. Holmes, Y. Dang, T. L. Woodard, K. P. Nevin and D. R. Lovley, Bioresour. Technol., 2016, 209, 148–156 CrossRef CAS PubMed.
- F. De Bok, C. Plugge and A. Stams, Water Res., 2004, 38, 1368–1375 CrossRef CAS PubMed.
- H. Li, J. Chang, P. Liu, L. Fu, D. Ding and Y. Lu, Environ. Microbiol., 2015, 17, 1533–1547 CrossRef PubMed.
- Q. Niu, Y. Takemura, K. Kubota and Y. Y. Li, Waste Manag., 2015, 43, 114–122 CrossRef CAS PubMed.
- R. Rajagopal, D. I. Masse and G. Singh, Bioresour. Technol., 2013, 143, 632–641 CrossRef CAS PubMed.
- Q. Lu, J. Yi and D. Yang, J. Microbiol. Biotechnol., 2016, 26, 110 CrossRef CAS PubMed.
- L. T. Angenent, H. Richter, W. Buckel, C. M. Spirito, K. J. Steinbusch, C. M. Plugge, D. P. Strik, T. I. Grootscholten, C. J. Buisman and H. V. Hamelers, Environ. Sci. Technol., 2016, 50, 2796–2810 CrossRef CAS PubMed.
- S. Ahmed and S. Alexander, Int. J. Syst. Evol. Microbiol., 2009, 59, 1661–1665 CrossRef PubMed.
- P. S. Dobbin, J. P. Carter, G. S. S. Juan, M. V. Hobe, A. K. Powell and D. J. Richardson, FEMS Microbiol. Lett., 1999, 176, 131–138 CrossRef CAS PubMed.
- H. E. Guadalupe, F. Marie-Laure, C. Jean-Luc, B. K. C. Patel, T. Pierre, M. Hervé, G. Jean-Louis and O. Bernard, Int. J. Syst. Evol. Microbiol., 2002, 52, 1461–1468 Search PubMed.
- C. List, Z. Hosseini, K. Lederballe Meibom, V. Hatzimanikatis and R. Bernier-Latmani, Environ. Microbiol., 2019 DOI:10.1111/1462-2920.14640.
- A. H. Van Gelder, D. Z. Sousa, W. I. Rijpstra, J. S. Damste, A. J. Stams and I. Sanchez-Andrea, Int. J. Syst. Evol. Microbiol., 2014, 64, 2449–2454 CrossRef CAS PubMed.
- Y. M. Amha, P. Sinha, J. Lagman, M. Gregori and A. L. Smith, Water Res., 2017, 123, 277–289 CrossRef CAS PubMed.
- J. De Vrieze and W. Verstraete, Environ. Microbiol., 2016, 18, 2797–2809 CrossRef CAS PubMed.
- B. Schink, Microbiol. Mol. Biol. Rev., 1997, 61, 262–280 CAS.
- D. R. Lovley, Annu. Rev. Microbiol., 2017, 71, 643–664 CrossRef CAS PubMed.
- A. E. Rotaru, T. L. Woodard, K. P. Nevin and D. R. Lovley, Front. Microbiol., 2015, 6, 744 Search PubMed.
- M. Vargas, N. S. Malvankar, P.-L. Tremblay, C. Leang, J. A. Smith, P. Patel, O. Synoeyenbos-West, K. P. Nevin and D. R. Lovley, mBio, 2013, 4, e00105–00113 CrossRef PubMed.
- D. J. Walker, R. Y. Adhikari, D. E. Holmes, J. E. Ward, T. L. Woodard, K. P. Nevin and D. R. Lovley, ISME J., 2018, 12, 48–58 CrossRef CAS PubMed.
- D. J. F. Walker, K. P. Nevin, D. E. Holmes, A.-E. Rotaru, J. E. Ward, T. L. Woodard, J. Zhu, T. Ueki, S. S. Nonnenmann, M. J. McInerney and D. R. Lovley, bioRxiv, 2018, 479683, DOI:10.1101/479683.
- B. E. Jackson, V. K. Bhupathiraju, R. S. Tanner, C. R. Woese and M. J. Mcinerney, Arch. Microbiol., 1999, 171, 107–114 CrossRef CAS PubMed.
- M. J. Mcinerney, R. Lars, M. Housna, K. Unmi, R. S. Krupp, R. H. Luis, S. Jessica, C. G. Struchtemeyer, B. Anamitra and J. W. Campbell, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 7600–7605 CrossRef PubMed.
- Z. Zhao, Y. Li, Q. Yu and Y. Zhang, Bioresour. Technol., 2017, 250, 79–85 CrossRef PubMed.
- P. Parameswaran, H. Zhang, C. I. Torres, B. E. Rittmann and R. Krajmalnik-Brown, Biotechnol. Bioeng., 2010, 105, 69–78 CrossRef CAS PubMed.
- A. E. Rotaru, P. M. Shrestha, F. Liu, B. Markovaite, S. Chen, K. P. Nevin and D. R. Lovley, Appl. Environ. Microbiol., 2014, 80, 4599–4605 CrossRef PubMed.
- D. E. Holmes, A. E. Rotaru, T. Ueki, P. M. Shrestha, J. G. Ferry and D. R. Lovley, Front. Microbiol., 2018, 9, 3109 CrossRef PubMed.
- S. Haveman, D. Holmes, Y. Ding, J. Ward, R. Didonato and D. Lovley, Appl. Environ. Microbiol., 2006, 72, 6980–6985 CrossRef CAS PubMed.
- D. J. Lonergan, H. L. Jenter, J. D. Coates, E. J. Phillips, T. M. Schmidt and D. R. Lovley, J. Bacteriol., 1996, 178, 2402–2408 CrossRef CAS PubMed.
- D. R. Lovley, E. J. Phillips, D. J. Lonergan and P. K. Widman, Appl. Environ. Microbiol., 1995, 61, 2132–2138 CAS.
- R. Mei, M. K. Nobu, T. Narihiro, J. Yu, A. Sathyagal, E. Willman and W. T. Liu, Water Res., 2018, 147, 403–412 CrossRef CAS PubMed.
- G. Hernandez-Eugenio, M. L. Fardeau, J. L. Cayol, B. K. Patel, P. Thomas, H. Macarie, J. L. Garcia and B. Ollivier, Int. J. Syst. Evol. Microbiol., 2002, 52, 1217–1223 CAS.
- Y. Dang, D. E. Holmes, Z. Zhao, T. L. Woodard, Y. Zhang, D. Sun, L. Y. Wang, K. P. Nevin and D. R. Lovley, Bioresour. Technol., 2016, 220, 516–522 CrossRef CAS PubMed.
- D. E. Koeck, M. Mechelke, V. V. Zverlov, W. Liebl and W. H. Schwarz, Int. J. Syst. Evol. Microbiol., 2016, 66, 4458–4463 CrossRef CAS PubMed.
- A. Watthanalamloet, C. Tachaapaikoon, Y. S. Lee, A. Kosugi, Y. Mori, S. Tanasupawat, K. L. Kyu and K. Ratanakhanokchai, Int. J. Syst. Evol. Microbiol., 2012, 62, 2330–2335 CrossRef CAS PubMed.
- C. Anne, S. H David and F. John, Water Environ. Res., 2006, 78, 486–496 CrossRef.
- G. Capson-Tojo, E. Trably, M. Rouez, M. Crest, N. Bernet, J. P. Steyer, J. P. Delgenes and R. Escudie, Waste Manag., 2018, 76, 423–430 CrossRef CAS PubMed.
- T. Kern, M. A. Fischer, U. Deppenmeier, R. A. Schmitz and M. Rother, Int. J. Syst. Evol. Microbiol., 2016, 66, 1533–1538 CrossRef CAS PubMed.
- X. Guo, C. Wang, F. Sun, W. Zhu and W. Wu, Bioresour. Technol., 2014, 152, 420–428 CrossRef CAS PubMed.
- K. S. Smith and C. Ingram-Smith, Trends Microbiol., 2007, 15, 150–155 CrossRef CAS PubMed.
- D. Zheng and L. Raskin, Microb. Ecol., 2000, 39, 246–262 CAS.
- Y. Dang, D. Sun, T. L. Woodard, L. Y. Wang, K. P. Nevin and D. E. Holmes, Bioresour. Technol., 2017, 238, 30–38 CrossRef CAS PubMed.
- B. Demirel and P. Scherer, Rev. Environ. Sci. Bio/Technol., 2008, 7, 173–190 CrossRef CAS.
- B. Calli, B. Mertoglu, B. Inanc and O. Yenigun, Environ. Technol., 2005, 26, 85–91 CrossRef CAS PubMed.
- I. H. Franke-Whittle, A. Walter, C. Ebner and H. Insam, Waste Manag., 2014, 34, 2080–2089 CrossRef CAS PubMed.
- R. Amelia-Elena, S. Pravin Malla, L. Fanghua, M. Beatrice, C. Shanshan, K. P. Nevin and D. R. Lovley, Appl. Environ. Microbiol., 2014, 80, 4599 CrossRef PubMed.
- X. Huang, W. Dong, H. Wang and Y. Feng, Bioresour. Technol., 2018, 249, 943–952 CrossRef CAS PubMed.
- S. Poirier, E. Desmond-Le Quemener, C. Madigou, T. Bouchez and O. Chapleur, Bioresour. Technol., 2016, 207, 92–101 CrossRef CAS PubMed.
- L. Shen, Q. Zhao, X. Wu, X. Li, Q. Li and Y. Wang, Renewable Sustainable Energy Rev., 2016, 54, 1358–1367 CrossRef CAS.
- Z. Zhao, Y. Zhang, Y. Li, Y. Dang, T. Zhu and X. Quan, Chem. Eng. J., 2017, 313, 10–18 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available: Characterization of substrates and inoculum (Table S1); daily methane content of biogas in digesters containing different mixture ratios and ISCs. Error bars represent standard deviations from triplicate samples (Fig. S1). See DOI: 10.1039/c9ra05556h |
| This journal is © The Royal Society of Chemistry 2019 |