Open Access Article
Open Access ArticleIdentification of the endoplasmic reticulum localization sequence and N-glycosylation of matrix metalloproteinase 26†
Guangji Zhang,
Jinrui Zhang,
Xiang Li,
Xin Meng and
Xuexun Fang *
*
Key Laboratory for Molecular Enzymology and Engineering of Ministry of Education, College of Life Science, Jilin University, 2699 Qianjin Street, Changchun 130012, PR China. E-mail: fangxx@jlu.edu.cn; Tel: +86-431-85155219
First published on 25th July 2019
Abstract
Matrix metalloproteinase 26 (MMP-26), also called endometase and matrilysin-2, belongs to the MMP superfamily. Previous studies have focused on its role in tumor invasion and migration but detailed subcellular localization of MMP-26 remains poorly understood. In this study, sequence deletion mutants of MMP-26 revealed that residues 88–123 function to localize MMP-26 to the endoplasmic reticulum (ER). Moreover, using homologous recombination, we show that exchanging residues 88–123 of secretory MMP-7 with the same region in MMP-26 causes localization of this MMP-7 construct to the ER. Moreover, two (N64, N221) of the three possible N-glycosylation sites in MMP-26 were shown to be N-glycosylated, and N-glycosylation is not required for ER localization. These results demonstrate that the 88–123 region of MMP-26 is a noncanonical ER retention signal and MMP-26 is an N-glycosylated protein, thereby providing novel insights into the properties of MMP-26 within the cell.
1. Introduction
Matrix metalloproteinases (MMPs) are members of the hydrolase superfamily that degrade the extracellular matrix and require Ca2+ and Zn2+ as cofactors. MMPs participate in many important physiological functions in humans, including tissue remodeling, wound repair, bone growth and embryonic development.1 Currently, there are 26 human MMPs that are divided into six groups according to their substrate specificity: collagenase, gelatinase, stromelysin, matrilysin, membrane-type MMPs and furin-activated MMPs.2MMP-26 was first cloned and characterized by four groups in 2000.3–6 In recent years, research on MMP-26 has focused on its role as a tumor marker, identifying its substrates and regulation of MMP-26 gene expression.7,8 These studies have provided considerable information on MMP-26; however, the biological function of MMP-26 remains unclear. Several studies have shown that MMP-26 is largely expressed within the cell and seldom secreted extracellularly.9 In addition, transfected and endogenous MMP-26 is expressed within tumor cells and cannot be detected extracellularly.9 This observation shows clearly that MMP-26 is expressed intracellularly and functions differently to other MMPs. Accordingly, the intracellular distribution of MMP-26 is considered a mislocalization. Our previous research has confirmed that MMP-26 is located in the endoplasmic reticulum (ER). MMP-26 only includes the following minimal characteristic features of the MMP family: a signal peptide (1–17), a pro-peptide domain (18–89) and a catalytic domain (90–261).4 The signal peptide facilitates localization of MMP-26 to the ER; however, it is not the only determinant for localizing MMP-26 to the ER.10–12
MMP-26 is a partially characterized human proteinase. MMP-26 activates pro-MMP-9, which promotes human prostate cancer cell invasion.13 Moreover, the specific expression of MMP-26 in certain tumor cells indicates that MMP-26 may play an important role in tumor progression.10,14 However, MMP-26 also plays protective roles. Savinov et al. reported that in ductal carcinoma in situ expression of MMP-26 was strongly up-regulated. However, the levels of MMP-26 were downregulated in further progression of I–III.15 Moreover, MMP-26 may play a pro-apoptotic role in human prostate cancer cells and tissues. The levels of MMP-26 in androgen-repressed human prostate cancer (ARCaP) cells are strongly correlated with the pro-apoptotic marker Bax.16 Several studies have shown that the expression of MMP-26 is largely within the cell.9 Expression of MMP-26 was observed in MDA-MB-231 cell lysates, whereas MMP-26 was undetected in the cell culture supernatant.17,18 These results clearly show that MMP-26 is expressed intracellularly.
N-Glycosylation is a major post-translational modification of proteins and is essential for protein structure and function. N-Glycans are attached to the asparagine (Asn) side chain of a nascent polypeptide and the consensus sequence of N-glycosylation is Asn-X-Ser/Thr (X! = P), where X represents any amino acid except proline.19 According to this consensus, there are three potential N-glycosylation sites in the MMP-26 sequence: N64 in the pro-peptide domain and N133 and N221 in the catalytic domain.3 However, no experimental data have confirmed this prediction. In addition, based on western blotting analysis of MMP-26-GFP, the molecular mass of MMP-26 is larger than its theoretical mass, indicating that MMP-26 is N-glycosylated. In this report, we identified N-glycosylation of MMP-26 and determined the number of N-glycosylation sites present in the sequence.
In this study, we generated a series of MMP-26 truncated mutants to gain additional insights into the biochemical properties of MMP-26 within the cell. We found that the 88–123 region of MMP-26 is the key sequence for ER localization. Moreover, exchange of the 88–123 region of MMP-26 with the corresponding region in secretory MMP-7 caused retention of this MMP-7 mutant in the ER. Despite the prediction that the MMP-26 sequence contains three potential N-glycosylation sties,3,20 there are no experimental data to support this prediction and the potential functional role of N-glycosylation is unclear. By inhibiting N-glycosylation and generating site-specific mutants we illustrate that MMP-26 is N-glycosylated and has two N-glycosylation sites (N64 and N221). Furthermore, N-glycosylation is not required for ER localization of MMP-26. These results provide evidence that the 88–123 region of MMP-26 is a noncanonical ER retention signal, and MMP-26 is an N-glycosylated protein, thereby laying the foundation for future research describing the intracellular functions of MMP-26.
2. Materials and methods
2.1 Cell culturing
HeLa cells (ATCC® CCL-2™) and HEK293T cells (ATCC® CRL-3216™) were grown in Dulbecco's modified Eagle's medium (DMEM) (Sigma-Aldrich, USA) with 2 mM L-glutamine, and cultured with 10% fetal bovine serum (BI, Inc.), 100 U mL−1 penicillin (Sigma-Aldrich, USA) and 100 μg mL−1 streptomycin (Sigma-Aldrich, USA).2.2 MMP-26 truncated and N-glycosylation mutant constructs
The DNA of the signal peptide and truncated regions of MMP-26 were obtained by PCR with DNA polymerase Pfu (TransGen Co., Ltd.), and the PCR products were ligated using an In-Fusion cloning kit (Taihe Co., Ltd.). Site-directed mutagenesis (TransGen Co., Ltd.) was used to engineer the desired mutations at sites N64 and N221, and for the N64/N221 double mutation. MMP-26 and the truncated and N-glycosylation mutant constructs were subcloned into pEGFP-N1 (Invitrogen) via the Hind III upstream restriction site and the Kpn I downstream restriction site (TransGen Co., Ltd.). All primers used in this study are listed in the ESI.† The constructs were confirmed by DNA sequencing.2.3 Subcellular localization of MMP-26 constructs
MMP-26 constructs C-terminally tagged with GFP were transiently transfected into HeLa cells. After 24 h transfection, the fluorescence probe (ER-Tracker Red) was added to the culture supernatant (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000) and the culture incubated in the dark for a further 10–20 min. Cells were then stained with Hoechst nuclei stain. Cells were imaged with a Sensicam camera (PCO-Tech Inc.) mounted on an Olympus IX50 microscope (Olympus Corporations) and processed using IPLAB3.6 software (Spectra Services). The pseudo-color of GFP (green), ER-Tracker Red (red) or Hoechst (blue) was accomplished using Adobe Photoshop CC 2015. Lipo6000™ Transfection Reagent, ER-Tracker Red and Hoechst nuclei stain were purchased from Beyotime Biotechnology, Beijing, China. A23187, dithiothreitol (DTT) and thapsigargin (Tg) were purchased from Sigma-Aldrich, USA.
1000) and the culture incubated in the dark for a further 10–20 min. Cells were then stained with Hoechst nuclei stain. Cells were imaged with a Sensicam camera (PCO-Tech Inc.) mounted on an Olympus IX50 microscope (Olympus Corporations) and processed using IPLAB3.6 software (Spectra Services). The pseudo-color of GFP (green), ER-Tracker Red (red) or Hoechst (blue) was accomplished using Adobe Photoshop CC 2015. Lipo6000™ Transfection Reagent, ER-Tracker Red and Hoechst nuclei stain were purchased from Beyotime Biotechnology, Beijing, China. A23187, dithiothreitol (DTT) and thapsigargin (Tg) were purchased from Sigma-Aldrich, USA.
2.4 Multisequence alignment and homology modeling
Multiple alignment of 11 MMP sequences was carried out by Clustal Omega, and residues are colored according to similarity. For structural modeling, the sequence of MMP-26 (31–256) was submitted to the online homology modeling program (https://swissmodel.expasy.org/). The 3D structure of MMP-7, which shares the highest sequence identity with MMP-26, was selected as the template. The 3D structure of MMP-26 was designed and colored by Discovery Studio.2.5 N-glycosylation inhibition of MMP-26
The MMP-26-GFP plasmid was transfected into HEK293T cells for 24 h. Then, 100 ng mL−1 tunicamycin (Tm; Sigma, Inc.) was added to the cell culture. Cell lysates were harvested after 12 h of administration.2.6 Enzymatic protein deglycosylation
The MMP-26-GFP plasmid was transfected into HEK293T cells. Cell lysates were harvested after 24 h and combined with glycoprotein denaturing buffer and incubated at 100 °C for 10 min. The mixture was incubated with GlycoBuffer, 10% NP-40 and PNGase F or Endo H (New England Biolabs, Inc.) at 37 °C for 2 h.2.7 Western blotting
The expression of MMP-26-GFP and mutants were detected by Western blotting. The cultured cells were harvested and washed with PBS (phosphate buffered saline) buffer. The cells were lysed by RIPA buffer (Beyotime) and boiled for 10 min with the addition of SDS sample buffer. The samples were subjected to SDS-PAGE and proteins were electro-transferred onto 0.45 μm PVDF membranes (Sigma-Aldrich, USA). After blocking in 5% fat-free milk for 0.5 h, the membranes were incubated with the anti-MMP-26 primary antibody (Abcam, Cambridge, UK) at 4 °C overnight. After washing three times (15 min each time), the PVDF membrane was incubated with the secondary antibody (TransGen Biotechnology, China) for protein detection. Immunoreactions were visualized with the Tanon™ High-sig ECL Western Blotting Substrate using a chemiluminescent imaging system (Tanon 4600; Tanon, Shanghai, China).3. Results and discussion
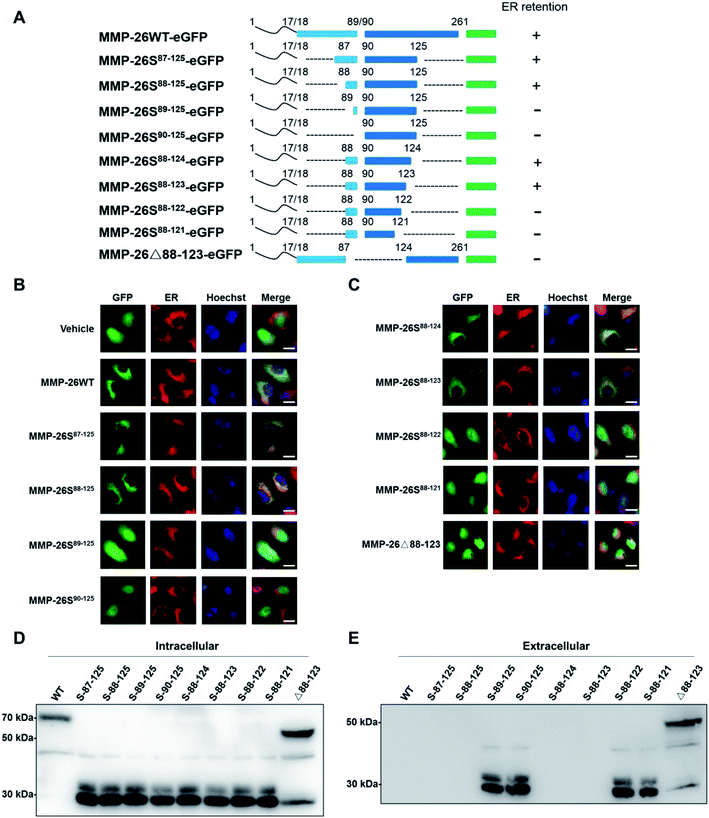
3.1 The 88–123 region of MMP-26 functions as an ER retention signal
In this study we prepared MMP-26-GFP and a series of MMP-26 truncated mutants with a GFP tag at the C-terminus to identify key sequence features required for ER localization of MMP-26 (Fig. 1A). These fusion constructs were transiently expressed in HeLa cells. After transfection for 24 h, ER-Tracker Red (ER-specific fluorescent probe) was added to the culture supernatant. Green fluorescence in cell compartments and the red signal of the ER were observed. The green fluorescence of GFP diffusely expressed in the cytosol and nucleus indicated that there was no disruption of ER retention of MMP-26 (negative control) (Fig. 1B). MMP-26-GFP, MMP-26S87–125-GFP and MMP-26S88–125-GFP were observed to be located in the ER, whereas MMP-26S89–125-GFP and MMP-26S90–125-GFP were expressed diffusely throughout the cytoplasm (Fig. 1B). This observation revealed the importance of Ser88 for MMP-26 ER localization. Residues at the C-terminus were then deleted individually to determine the effect of C-terminal residues to ER localization of MMP-26. Both MMP-26S88–124-GFP and MMP-26S88–123-GFP were found to locate to the ER, whereas mutants MMP-26S88–122-GFP and MMP-26S88–121-GFP were not localized in the ER (Fig. 1C). This result indicates that Ser123 of MMP-26 also plays a key role in the localization of MMP-26 to the ER. Thus, the region 88–123 of MMP-26 is important for subcellular localization because deletion of Ser88 and Ser123 disrupts ER retention of MMP-26. The 88–123 region of MMP-26 was deleted to give the construct MMP-26Δ88–123-GFP, and this MMP-26 construct did not locate to the ER (Fig. 1C), which further supports the notion that the 88–123 region of MMP-26 is a pivotal signal for ER localization. Western blotting to examine the intracellular protein levels of MMP-26-GFP and all constructs were compared using an anti-GFP antibody (Fig. 1D). Examination of the extracellular media showed that MMP-26S89–125-GFP, MMP-26S90–125-GFP, MMP-26S88–122-GFP, MMP-26S88–121-GFP and MMP-26Δ88–123-GFP were secreted from cells because they could not locate to the ER (Fig. 1E). Positively charged residues are vital for directing proteins into cell compartments or secretory pathways,21–23 and altering the subcellular localization of a protein by site-directed insertions and/or deletions of positively charged residues in critical regions should be feasible. A multi-sequence alignment of all secretory human MMPs showed that the 88–123 region of MMP-26 is unique among all secretory MMPs (Fig. S1A†), which contain numerous positive amino acids (including R96, K98, K101, H102, R107, H113, K116 and K121). Eight conserved positively charged residues were mutated to alanine by site-directed mutagenesis and we found that MMP-26S88–123(8A)-GFP (Fig. S1B†), which was diffusely expressed throughout the cytoplasm, cannot locate to the ER (Fig. S1C†). These results indicated that localization of MMP-26 to the ER is dependent on the 88–123 region, and the positively charged residues present in the 88–123 region play a major role in this localization process. Many charged amino acids in this unique unstructured region of MMP-26 are likely to interact with particular non-substrate binding proteins in the ER.3.2 The 88–123 region of MMP-26 is a noncanonical ER retention signal
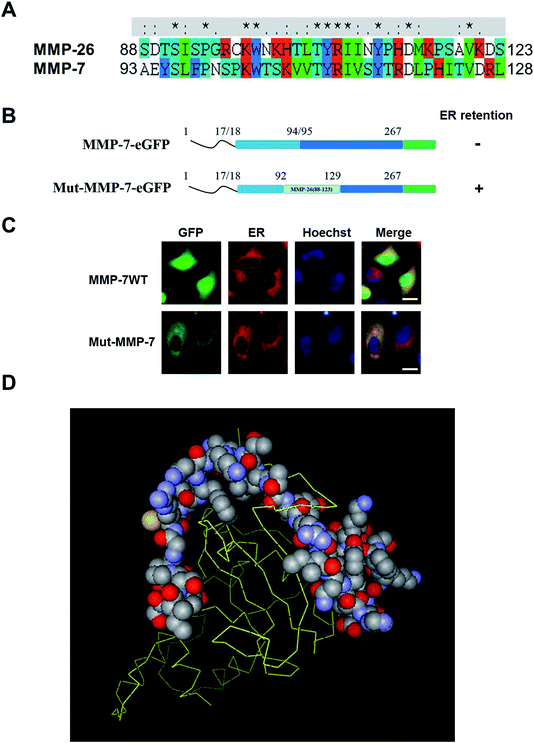
MMP-26 and MMP-7 belong to matrilysin sub-family based on their protein sequences, functional domain architecture and computer modeling. Nevertheless, there are also differences between these two MMPs, including substrate specificity, activation mechanism and functional roles in cancer cells.24,25 For example, MMP-7 is a typical secretory protein that binds to cholesterol sulfate in cell membranes.26,27 MMP-26 has a signal peptide similar to MMP-7, but studies have confirmed that MMP-26 is mainly expressed within human breast and prostate cancer cells, and MMP-26 is not found in culture supernatants.16,25,28,29 This observation indicates that MMP-26 may have a subcellular localization that is different from that of MMP-7. To examine this, the 88–123 region of MMP-26 was exchanged with the same region in MMP-7 through homologous recombination (Fig. 2A) to give the construct Mut-MMP-7. Both MMP-7-GFP and Mut-MMP-7-GFP were prepared (Fig. 2B). Wild-type MMP-7 was not found in the ER as a typical secretory protein, whereas Mut-MMP-7-GFP was retained in the ER (Fig. 2C). A 3D homology model of the MMP-26 structure showed that the 88–123 region of MMP-26 is surface accessible (Fig. 2D), which indicates that this region can readily interact with potential ER-resident proteins. These interactions may mediate the ER retention of MMP-26 and indicate that the 88–123 region of MMP-26 is a noncanonical ER retention signal.3.3 Identification of N-glycosylation in MMP-26
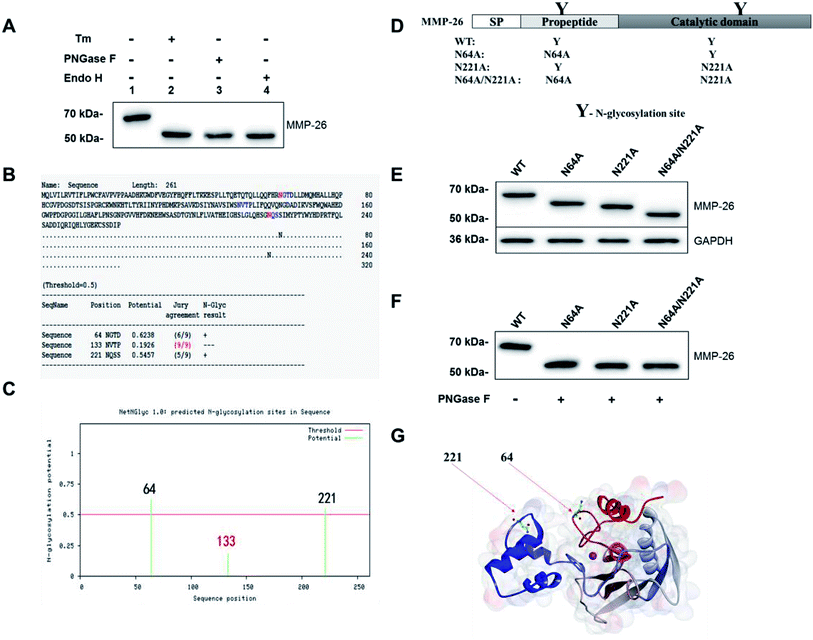
Sang et al. reported that MMP-26 has three potential N-glycosylation sites; residue N64 in the pro-peptide domain and N133 and N221 located within the catalytic domain.3 Mobashery et al. generated a preliminary model of MMP-26 to investigate the structural features of N-glycosylation and showed that N64 and N133 may be N-glycosylation sites.20 In the Western blotting analysis of MMP-26-GFP we observed that the molecular mass is larger than the theoretical mass (Fig. 1D). Thus, N-glycosylation inhibition of MMP-26 was performed to determine whether MMP-26 is N-glycosylated. The culture supernatant of MMP-26-GFP-expressing HEK293T cells was incubated with tunicamycin (Tm). Proteins were harvested from the MMP-26-GFP-expressing HEK293T cells, and PNGase F and Endo H were used to cleave the link between MMP-26-GFP and N-glycans directly. The Western blotting results showed that the molecular mass of MMP-26-GFP was reduced by approximately 57 kDa when treated with Tm, PNGase F and Endo H, indicating that MMP-26 is N-glycosylated (Fig. 2A). We then attempted to identify the N-glycosylation sites in MMP-26. There are three potential N-glycosylation sites based on the sequence motif Asn-X-Ser/Thr (X! = P). According to the NetNGlyc 1.0 server (http://www.cbs.dtu.dk/services/NetNGlyc/) prediction, N133 is probably not a N-glycosylation site (Fig. 2B) because the potential of the site should be higher than 0.5 to be considered an N-glycosylation site (Fig. 2C). The “potential” score is the average output of nine neural networks and the jury agreement column revealed that all nine networks do not support Asn133 N-glycosylation (Fig. 3B). Thus, two single point mutants (N64A, N221A) and a double point mutant (N64A/N221A) were generated (Fig. 2D). These constructs were subcloned into pEGFP-N1 and transfected into HEK293T cells and the total lysates were harvested, and Western blotting was carried out using the anti-MMP-26 antibody. The bands of MMP-26 constructs illustrated the expected molecular mass and the result is consistent with the loss of one (N64A or N221A) or two (N64A/N221A) N-glycosylation sites (Fig. 2E). To further examine whether N133 is N-glycosylated, the MMP-26 N-glycosylation-deficient constructs were treated with PNGase F. If N133 is N-glycosylated, then the molecular mass should be greater in N64A and N221A than in N64A/N221A. After treatment with PNGase F, the molecular masses of these three molecules were equal (Fig. 2F). Thus, N133 can be excluded as a potential N-glycosylation site. Fig. 2G illustrates the 3D homology modeling of MMP-26. Collectively, these results showed that MMP-26 is N-glycosylated and has two N-glycosylation sites, and these data are consistent with the prediction work by Sang et al. and the NetNGlyc 1.0 server.3.4 Effect of N-glycosylation on MMP-26 ER localization
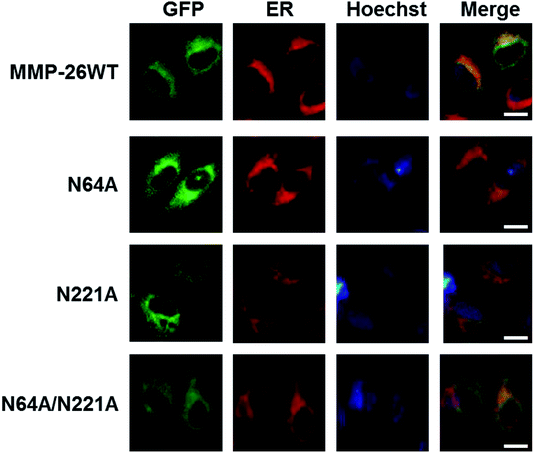
We then examined whether N-glycosylation deficiency disrupts the ER localization of MMP-26. The three N-glycosylation deficient mutants of MMP-26 were found to locate to the ER (Fig. 4), indicating that N-glycosylation of MMP-26 is not a retention factor for ER localization, presumably because N64 and N221 do not reside in the 88–123 region. Moreover, N-glycosylation is a post-translational modification of nascent proteins, and this modification occurs in the ER, which is consistent with MMP-26-GFP residing in the ER. The unfolded protein response (UPR) is activated when misfolded or unfolded proteins accumulate in the ER, or intracellular Ca2+ homeostasis is lost. The UPR is a cellular stress response related to ER stress.30 We aimed to determine whether ER stress would influence MMP-26 localization to the ER, and the functional role of N-glycosylation of MMP-26 under ER stress conditions. A23187, DTT, Tg and Tm were added to the cultured supernatant to mimic ER stress. After treatment with A23187, DTT and Tg, MMP-26 and its N-glycosylation-deficient mutants transferred to the perinuclear region, whereas Tm treatment did not change the ER localization of MMP-26 (Fig. S2†).4. Conclusions
In this report, we showed that localization of MMP-26-GFP in the ER of HeLa cells is through the 88–123 region and positively charged residues present in this region play a major role in ER localization. In addition, the 88–123 region of MMP-26, which is surface exposed in an MMP-26 model, potentially mediates interactions between MMP-26 and ER-resident proteins. Moreover, an MMP-7 mutant that included the 88–123 region of MMP-26 was retained in the ER. Thus, the 88–123 region of MMP-26 appears to be a novel and noncanonical ER retention signal.Based on prediction and analysis of the MMP-26 protein sequence, we experimentally identified N-glycosylated sites of MMP-26. We showed that N64 and N221 are N-glycosylation sites of MMP-26. The localization of MMP-26-GFP to the ER is also consistent with the observation that MMP-26 is N-glycosylated because N-glycosylation of proteins occurs in the ER. However, inhibiting N-glycosylation of MMP-26 did not disrupt ER localization. This result is presumably because N64 and N221 do not reside in the 88–123 region of MMP-26. Thus, N-glycosylation of MMP-26 is not an essential prerequisite for ER localization.
The biological significance of MMP-26 residing in the ER remains unknown. We have confirmed that the 88–123 region of MMP-26 is important in ensuring MMP-26 locates to the ER, but we have not determined whether this localization is through interaction with ER-resident proteins. Future efforts aim to identify MMP-26-interacting proteins in the ER. This approach will help explain the specific functions of MMP-26 in the ER. In addition, we have shown that MMP-26 is an N-glycosylated protein. This is the first study to present the post-translational modification of MMP-26. Future research focusing on the functional roles of N-glycosylation of MMP-26 is required.
Conflicts of interest
The authors have no conflicts of interest with the content of this article.Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 31370742, No. 31070669 and No. 81603459).References
- I. Stamenkovic, Matrix metalloproteinases in tumor invasion and metastasis, J. Pathol., 1999, 189, 300 CrossRef.
- H. Nagase, Substrate Specificity of MMPs, in Matrix Metalloproteinase Inhibitors in Cancer Therapy, ed. N. J. Clendeninn and K. Appelt, 2001 Search PubMed.
- H. I. Park, J. Ni, F. E. Gerkema, D. Liu, V. E. Belozerov and Q. X. Sang, Identification and Characterization of Human Endometase (Matrix Metalloproteinase-26) from Endometrial Tumor, J. Biol. Chem., 2000, 275, 20540 CrossRef CAS PubMed.
- A. B. D. Coignac, G. Elson, Y. Delneste, G. Magistrelli, P. Jeannin, J. P. Aubry, O. Berthier, D. Schmitt, J. Y. Bonnefoy and J. F. Gauchat, Cloning of MMP-26, FEBS J., 2000, 267, 3323–3329 Search PubMed.
- G. N. Marchenko, B. I. Ratnikov, D. V. Rozanov, A. Godzik, E. I. Deryugina and A. Y. Strongin, Characterization of matrix metalloproteinase-26, a novel metalloproteinase widely expressed in cancer cells of epithelial origin, Biochem. J., 2001, 356, 705–718 CrossRef CAS PubMed.
- J. A. Uría and C. López-Otín, Matrilysin-2, a new matrix metalloproteinase expressed in human tumors and showing the minimal domain organization required for secretion, latency, and activity, Cancer Res., 2000, 60, 4745 Search PubMed.
- Q. L. Hu, C. J. Yan, C. F. Xu, H. Yan, L. Qing, Y. J. Pu, Z. Y. He and X. J. Li, Matrilysin-2 expression in colorectal cancer is associated with overall survival of patients, Tumor Biol., 2014, 35, 3569–3574 CrossRef CAS PubMed.
- T. Mu, W. Liang, Y. Ju, Z. Wang, Z. Wang, M. D. Roycik, Q.-X. A. Sang, D. Yu, H. Xiang and X. Fang, Efficient soluble expression of secreted matrix metalloproteinase 26 in Brevibacillus choshinensis, Protein Expression Purif., 2013, 91, 125–133 CrossRef CAS PubMed.
- A. Y. Strongin, Mislocalization and unconventional functions of cellular MMPs in cancer, Cancer Metastasis Rev., 2006, 25, 87–98 CrossRef CAS PubMed.
- Q. X. A. Sang, J. Ni, F. E. Gerkema, D. Liu, V. E. Belozerov and H. I. Park, Biochemical properties of human endometase/MMP-26, a unique new metalloproteinase specifically expressed in uterus, placenta, and cancer cells, FASEB J., 2001, 15, A189 Search PubMed.
- N. D. Marchenko, G. N. Marchenko and A. Y. Strongin, Unconventional activation mechanisms of MMP-26, a human matrix metalloproteinase with a unique PHCGXXD cysteine-switch motif, J. Biol. Chem., 2002, 277, 18967–18972 CrossRef CAS PubMed.
- Y. G. Zhao, A. Z. Xiao, H. I. Park, R. G. Newcomer, M. Yan, Y. G. Man, S. C. Heffelfinger and Q. X. A. Sang, Endometase/matrilysin-2 in human breast ductal carcinoma in situ and its inhibition by tissue inhibitors of metalloproteinases-2 and -4: a putative role in the initiation of breast cancer invasion, Cancer Res., 2004, 64, 590–598 CrossRef CAS PubMed.
- H. Yamamoto, A. Vinitketkumnuen, Y. Adachi, H. Taniguchi, T. Hirata, N. Miyamoto, K. Nosho, A. Imsumran, M. Fujita and M. Hosokawa, Association of matrilysin-2 (MMP-26) expression with tumor progression and activation of MMP-9 in esophageal squamous cell carcinoma, Carcinogenesis, 2012, 5, 420–426 Search PubMed.
- L. Jing, B. Cao, Y. X. Li, X. Q. Wu and Y. L. Wang, GnRH I and II up-regulate MMP-26 expression through the JNK pathway in human cytotrophoblasts, Reprod. Biol. Endocrinol., 2010, 8, 1–10 CrossRef PubMed.
- A. Y. Savinov, A. G. Remacle, V. S. Golubkov, M. Krajewska, S. Kennedy, M. J. Duffy, D. V. Rozanov, S. Krajewski and A. Y. Strongin, Matrix metalloproteinase 26 proteolysis of the NH2-terminal domain of the estrogen receptor beta correlates with the survival of breast cancer patients, Cancer Res., 2006, 66, 2716–2724 CrossRef CAS PubMed.
- Z. I. Khamis, K. A. Iczkowski, Y. G. Man, M. J. Bou-Dargham and Q. X. A. Sang, Evidence for a Proapoptotic Role of Matrix Metalloproteinase-26 in Human Prostate Cancer Cells and Tissues, J. Cancer, 2016, 7, 80–87 CrossRef CAS PubMed.
- L. Hegedus, H. Cho, X. Xie and G. L. Eliceiri, Additional MDA-MB-231 breast cancer cell matrix metalloproteinases promote invasiveness, J. Cell. Physiol., 2008, 216, 480–485 CrossRef CAS PubMed.
- S. Lee, D. Terry, D. R. Hurst, D. R. Welch and Q.-X. A. Sang, Protein Signatures in Human MDA-MB-231 Breast Cancer Cells Indicating a More Invasive Phenotype Following Knockdown of Human Endometase/Matrilysin-2 by siRNA, J. Cancer, 2011, 2, 165–176 CrossRef CAS PubMed.
- B. Imperiali and S. E. O'Connor, Effect of N-linked glycosylation on glycopeptide and glycoprotein structure, Curr. Opin. Chem. Biol., 1999, 3, 643–649 CrossRef CAS PubMed.
- L. P. Kotra, L. Zhang, R. Fridman, R. Orlando and S. Mobashery, N-Glycosylation pattern of the zymogenic form of human matrix metalloproteinase-9, Bioorg. Chem., 2002, 30, 356–370 CrossRef CAS PubMed.
- M.-Y. Li, C.-Q. Xiao, Z.-Q. Xu, M.-M. Yin, Q.-Q. Yang, Y.-L. Yin and Y. Liu, Role of surface charge on the interaction between carbon nanodots and human serum albumin, Spectrochim. Acta, Part A, 2018, 204, 484–494 CrossRef CAS PubMed.
- M. Zhang, J. Qiao and L. Qi, Dual-functional polymer-modified magnetic nanoparticles for isolation of lysozyme, Anal. Chim. Acta, 2018, 1035, 70–76 CrossRef CAS PubMed.
- J. Perrin, A. Bary, A. Vernay and P. Cosson, Role of the HIV-1 envelope transmembrane domain in intracellular sorting, BMC Cell Biol., 2018, 19, 3 CrossRef PubMed.
- P. Pittayapruek, J. Meephansan, O. Prapapan, M. Komine and M. Ohtsuki, Role of Matrix Metalloproteinases in Photoaging and Photocarcinogenesis, Int. J. Mol. Sci., 2016, 17, 8 CrossRef PubMed.
- N. D. Marchenko, G. N. Marchenko, R. N. Weinreb, J. D. Lindsey, A. Kyshtoobayeva, H. C. Crawford and A. Y. Strongin, β-Catenin regulates the gene of MMP-26, a novel matrix metalloproteinase expressed both in carcinomas and normal epithelial cells, Int. J. Biochem. Cell Biol., 2004, 36, 942–956 CrossRef CAS PubMed.
- H. Shouichi, O. Miwa, Y. Kazuhiro and M. Kaoru, Identification of amino acid residues of matrix metalloproteinase-7 essential for binding to cholesterol sulfate, J. Biol. Chem., 2008, 283, 35735–35744 CrossRef PubMed.
- S. Vimbai, Y. Kiyoshi and I. Kuniyo, Effects of heparin and cholesterol sulfate on the activity and stability of human matrix metalloproteinase 7, Biosci., Biotechnol., Biochem., 2014, 78, 41–48 CrossRef PubMed.
- H. Yang, Y. Wang, Y. Li, L. Zhang, Y. Deng, D. Qi, Y. Li and W. Li, Roles of matrix metalloproteinase-26 in the growth, invasion and angiogenesis of breast cancer, Oncol. Lett., 2012, 4, 832–836 CrossRef CAS PubMed.
- L. Seakwoo, D. Kevin K, I. Kenneth A, N. Robert G, W. Kevin J, Z. Yun-Ge, T. Winston W, R. Mark D and S. Qing-Xiang Amy, Coordinated peak expression of MMP-26 and TIMP-4 in preinvasive human prostate tumor, Cell Res., 2006, 16, 750–758 CrossRef PubMed.
- P. Walter and D. Ron, The Unfolded Protein Response: From Stress Pathway to Homeostatic Regulation, Science, 2011, 334, 1081–1086 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9ra05222d |
| This journal is © The Royal Society of Chemistry 2019 |