Open Access Article
Open Access ArticleRetracted Article: Knockdown of NEAT1 ameliorated MPP+-induced neuronal damage by sponging miR-221 in SH-SY5Y cells
Lijiao Geng†
a,
Jun Zhao†b,
Wei Liub and
Yong Chen *a
*a
aDepartment of Rehabilitation Medicine, Huaihe Hospital of Henan University, No. 357 Ximen Street, Kaifeng 475000, China. E-mail: vwbkwx@163.com; Tel: +86-371-23906882
bDepartment of Neurology, Huaihe Hospital of Henan University, Kaifeng 475000, China
First published on 13th August 2019
Abstract
Long noncoding RNAs (lncRNAs) have recently attracted increasing attention for their involvement in a wide variety of human neurodegenerative diseases, including Parkinson's disease (PD). The purpose of the present study was to investigate the functional role and underlying mechanism of NEAT1 in PD. qRT-PCR was used to assess the expression of NEAT1 and miR-221, and the expression levels of Bcl-2 and Bax were detected by western blot. Cell viability and apoptosis were determined by CCK-8 assay and flow cytometry, respectively. The changes of oxidative stress and neuroinflammation were evaluated by ELISA assay and qRT-PCR, respectively. The targeted interaction between NEAT1 and miR-221 was verified by dual-luciferase reporter assay and RNA immunoprecipitation assay. Our data supported that MPP+ treatment elevated NEAT1 expression in dose- and time-dependent manners in SH-SY5Y cells, and NEAT1 silencing relieved MPP+-induced suppression of cell viability and enhancement of cell apoptosis in SH-SY5Y cells. Moreover, NEAT1 silencing alleviated MPP+-induced promotion of oxidative stress and neuroinflammation in SH-SY5Y cells. NEAT1 directly targeted miR-221 and negatively regulated miR-221 expression. More importantly, miR-221 mediated the protective effect of NEAT1 knockdown, as evidenced by the restoration of cell viability, cell apoptosis, oxidative stress and neuroinflammation in MPP+-induced SH-SY5Y cells. In conclusion, our study suggested that NEAT1 silencing alleviated MPP+-induced neuronal damage by sponging miR-221 in SH-SY5Y cells, highlighting the role of NEAT1 as a potential molecular target for PD therapy.
1. Introduction
Parkinson's disease (PD), the second most common neurodegenerative disease worldwide with incidence and prevalence on the rise, is characterized by a progressive movement disorder due to the death of dopaminergic neurons in the substantia nigra. The onset of the disease is usually at an age of 65 to 70 years, affecting people mainly in later years of life.1,2 Owing to the complex causes and mechanisms, PD has attracted increasing attention from academic scientists and clinical physicians.3 Chronic neuroinflammation and excessive oxidative stress are the hallmarks of PD pathology and contribute to the cascade leading to dopamine cell degeneration in PD.4,5 A better understanding for PD etiopathogenesis and the underlying mechanisms governing it is still a critical challenge for PD therapy.Long noncoding RNAs (lncRNAs), a diverse class of transcripts commonly defined as being longer than 200 nucleotides, function as crucial regulators in numerous biological processes across every branch of life.6 Emerging evidence has suggested that lncRNAs function as important regulators in a wide variety of human neurodegenerative diseases, including PD.7,8 LncRNAs participate in the development of PD via various mechanisms, particularly focusing on their microRNA (miRNA) binding functions.9–11 Nuclear paraspeckle assembly transcript 1 (NEAT1) is a ubiquitous, highly expressed, nuclear-retained regulatory lncRNA that plays a key role in cellular pathophysiological processes.12,13 Increasing evidence has manifested that dysregulation of NEAT1 is involved in multiple neurodegenerative diseases, such as Huntington's disease and PD.14–16 Previous research has reported that abnormal expression of NEAT1 was found in PD model in vitro and in vivo, and NEAT1 depletion had a protective effect on PD model via regulating α-synuclein expression or neuronal autophagy.16,17 Herein, we aimed to further investigate the functional role and underlying mechanism of NEAT1 on the oxidative stress and neuroinflammation of PD.
MiR-221 has recently been identified as a biomarker for PD evaluation, and its upregulation plays a protective role in PD.18–20 This striking resemblance prompted us to explore miR-221 as a potential molecular mediator of NEAT1 in PD progression. In the present study, our data supported that NEAT1 expression was increased in MPP+-induced SH-SY5Y cells. Furthermore, we firstly demonstrated that NEAT1 silencing alleviated MPP+-induced neuronal damage by sponging miR-221 in SH-SY5Y cells.
2. Materials and methods
2.1. Cell culture and treatment
Human neuroblastoma SH-SY5Y cells obtained from ATCC (Manassas, VA, USA) were maintained in Dulbecco's Modified Eagle's Medium/Nutrient Mixture F-12 (DMEM/F-12, Gibco, Life Technologies Corporation, Grand Island, NY, USA) supplemented with 10% fetal bovine serum (FBS, Gibco), 1% antibiotics (100 U ml−1 penicillin and 100 μg ml−1 streptomycin, Gibco) in a humidified 37 °C, 5% CO2 incubator.MPP+ iodide was purchased from Sigma-Aldrich (St. Louis, MO, USA) and used to establish PD model in vitro as described previously.21 To be brief, SH-SY5Y cells were treated with various concentrations of MPP+ (0, 0.25, 0.5, 1 and 2 mM) for 24 h or different times (0, 12, 24 and 48 h) at a concentration of 1 mM MPP+.
2.2. Cell transfection
To observe the role of NEAT1 in PD, SH-SY5Y cells at 60–70% confluence were transfected with silencer select predesigned siRNA against NEAT1 (si-NEAT1, Ambion, Austin, TX, USA) or negative control siRNA (si-NC, Ambion), NEAT1 overexpression vector (pcDNA-NEAT1, Ambion) or negative control vector (pcDNA-NC, Ambion). For miR-221 overexpression, SH-SY5Y cells were introduced with human modified miR-221 mimic (Ambion) or a scrambled oligonucleotide sequence (miRNA-NC, Ambion) as negative control. MiR-221 depletion was carried on using modified miR-221 inhibitor (anti-miR-221, Ambion) and a scrambled sequence (anti-miR-NC, Ambion) was used as negative control. All transfections were performed using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) following the instructions of manufacturers.2.3. RNA extraction, reverse transcription and quantitative real-time PCR (qRT-PCR)
Total RNA was extracted from SH-SY5Y cells using RNeasy Mini kit (Qiagen, Hilden, Germany) in accordance to the protocols of manufacturers. The quality and quantity of RNA extracts were determined by a NanoDrop ND-2000 spectrophotometer (Nanodrop Technologies, Wilmington, DE, USA). For miR-221 expression detection, RNA was reverse-transcribed into cDNA using the miScript Reverse Transfection kit (Qiagen); qRT-PCR was performed using the miScript SYBR Green PCR kit (Qiagen) with specific primer for miR-221 (Qiagen); and U6 was used as a housekeeping gene for normalization. For gene expression determination, cDNA was synthesized using the M-MLV Reverse Transcriptase (Invitrogen) and then subjected to qRT-PCR using the Power SYBR™ Green PCR Master Mix (Applied Biosystems, Foster City, CA, USA) with GAPDH as the internal reference. The relative expression of each gene and miR-221 was calculated using the 2−ΔΔCt method.2.4. Cell viability assay
Cell viability was determined using a Cell Counting kit-8 (CCK-8, Dojindo Laboratories, Kumamoto, Japan) following the instructions of manufacturers. Briefly, SH-SY5Y cells were seeded into a 96-well plate and then transfected with or without the indicated oligonucleotide for 24 h, followed by the treatment with 1 mM of MPP+. After 24 h treatment, 10 μl of CCK-8 solution was added into each well for 2 h. Cell viability was evaluated by measuring the absorbance at 450 nm using a Biotek Synergy 2 microplate reader (Biotek, Colmar, France).2.5. Flow cytometry
Cell apoptosis was assessed using the Annexin V-FITC/PI Apoptosis Detection kit (EMD Chemicals, Gibbstown, NJ, USA) in accordance with manufacturer's instructions. To be brief, treated SH-SY5Y cells were harvested and washed three times with PBS, and then resuspended in 100 μl of Annexin V binding buffer. Afterward, cells were double stained with Annexin V-FITC and PI in the dark at room temperature. Cell apoptosis was analyzed using a FACS-Calibur flow cytometry (Becton Dickinson, Heidelberg, Germany) with Cell Quest software (Becton Dickinson).2.6. Western blot
Cells were lysed in ice-cold RIPA lysis buffer (50 mM Tris–HCl, pH = 7.4, 1 mM EDTA, 150 mM NaCl, 0.5% NP-40, 1 mM sodium vanadate, 0.5% sodium deoxycholate, 0.05% SDS) containing 1× EDTA-free protease inhibitor cocktail (Roche Diagnostics, Mannheim, Germany). Total protein was extracted from cell lysates by centrifugation (14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 g) at 4 °C for 10 min and quantified using a BCA Protein assay kit (Thermo Fisher Scientific, Waltham, MA, USA) following the protocols of manufacturers. Protein extracts were resolved on a 10% SDS polyacrylamide gel and blotted onto a Hybond-P PVDF membrane (Amersham Bioscience, Piscataway, NJ, USA). The following primary antibodies were used: anti-Bcl-2 (Abcam, Cambridge, UK; dilution 1
000 g) at 4 °C for 10 min and quantified using a BCA Protein assay kit (Thermo Fisher Scientific, Waltham, MA, USA) following the protocols of manufacturers. Protein extracts were resolved on a 10% SDS polyacrylamide gel and blotted onto a Hybond-P PVDF membrane (Amersham Bioscience, Piscataway, NJ, USA). The following primary antibodies were used: anti-Bcl-2 (Abcam, Cambridge, UK; dilution 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000), anti-Bax (Cell Signaling Technology, Danvers, MA, USA; dilution 1
1000), anti-Bax (Cell Signaling Technology, Danvers, MA, USA; dilution 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000) and anti-β-actin (Abcam; dilution 1
1000) and anti-β-actin (Abcam; dilution 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2000). Horseradish peroxidase-conjugated IgG antibody (Abcam; dilution 1
2000). Horseradish peroxidase-conjugated IgG antibody (Abcam; dilution 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5000) was used as a secondary antibody. Protein bands were visualized using the immobilon ECL kit (Millipore, Billerica, MA, USA) and band intensities were quantified by ImageJ software (National Institutes of Health, Bethesda, MD, USA).
5000) was used as a secondary antibody. Protein bands were visualized using the immobilon ECL kit (Millipore, Billerica, MA, USA) and band intensities were quantified by ImageJ software (National Institutes of Health, Bethesda, MD, USA).
2.7. ELISA assay
SH-SY5Y cells were treated with PBS or 1 mM of MPP+ for 24 h, or transfected with si-NC or si-NEAT1 prior to MPP+ treatment. After 24 h treatment, reactive oxygen species (ROS) generation, lactate dehydrogenase (LDH) activity and superoxide dismutase (SOD) activity were measured using corresponding commercial ELISA assay kits (Beyotime, Shanghai, China) in accordance with the manufacturer's guidance.2.8. Bioinformatics analysis
Analysis for the targeted miRNAs of NEAT1 was performed using the software LncBase Predicted v.2 available at http://carolina.imis.athena-innovation.gr/diana_tools/web/index.php?r=lncbasev2/index-predicted.2.9. Dual-luciferase reporter assay
NEAT1 wild-type luciferase reporter construct (NEAT1-WT) containing miR-221-binding sequence and site-directed mutation of the binding sequence (NEAT1-MUT) were obtained from Ambion. SH-SY5Y cells were transfected with NEAT1-WT or NEAT1-MUT together with miR-221 mimic or miR-NC mimic. After 48 h of transfection, the luciferase activity was determined using a Dual-luciferase Reporter Assay System (Promega, Madison, WI, USA).2.10. RNA immunoprecipitation (RIP) assay
RIP assay was performed using the EZ-Magna RIP kit (Millipore) in accordance with manufacturer's instructions with anti-Argonaute2 (anti-Ago2, Abcam) antibody. Briefly, cell lysates were prepared using ice-cold RIPA lysis buffer and then incubated with anti-Ago2 or IgG antibody for 4 h at 4 °C before adding protein A/G agarose for 2 h. Beads were washed three times and total RNA was extracted using RNeasy Mini kit for the measurement of NEAT1 enrichment by qRT-PCR.2.11. Statistical analysis
The statistical significance of all data was determined by a Student's t-test or one-way ANOVA with Bonferroni post hoc test using GraphPad Prism version 7.0 software (GraphPad Software, La Jolla, CA, USA). All data were expressed as the mean ± standard deviation (SD) of at least three dependent experiments. Statistical significance was defined as *P < 0.05, **P < 0.01 or ***P < 0.001.3. Results
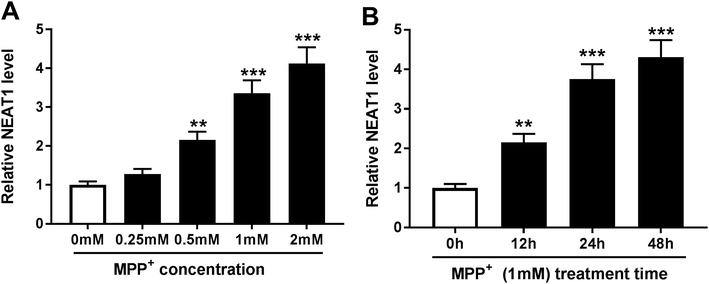
3.1. MPP+ treatment increased NEAT1 expression in SH-SY5Y cells
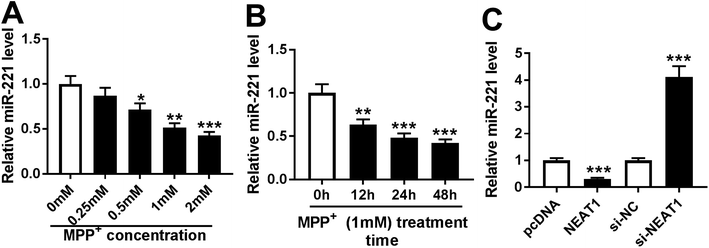
In order to validate the involvement of NEAT1 in PD, we firstly established PD model in vitro. SH-SY5Y cells were treated with various concentrations of MPP+ (0, 0.25, 0.5, 1 and 2 mM) for 24 h or different times (0, 12, 24 and 48 h) at a concentration of 1 mM MPP+. As demonstrated by qRT-PCR, MPP+ treatment induced the expression of NEAT1 in a dose-dependent manner (Fig. 1A). Moreover, MPP+ treatment time-dependently elevated NEAT1 expression in SH-SY5Y cells. These data supported that NEAT1 expression was upregulated by MPP+ in SH-SY5Y cells.3.2. Knockdown of NEAT1 relieved MPP+-induced SH-SY5Y cell injury
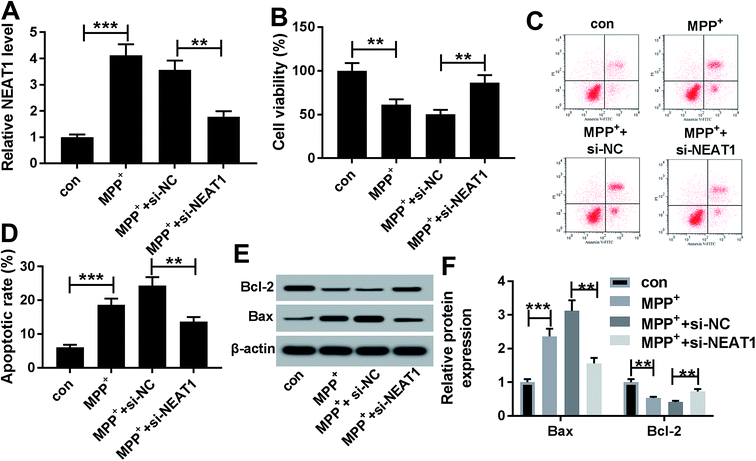
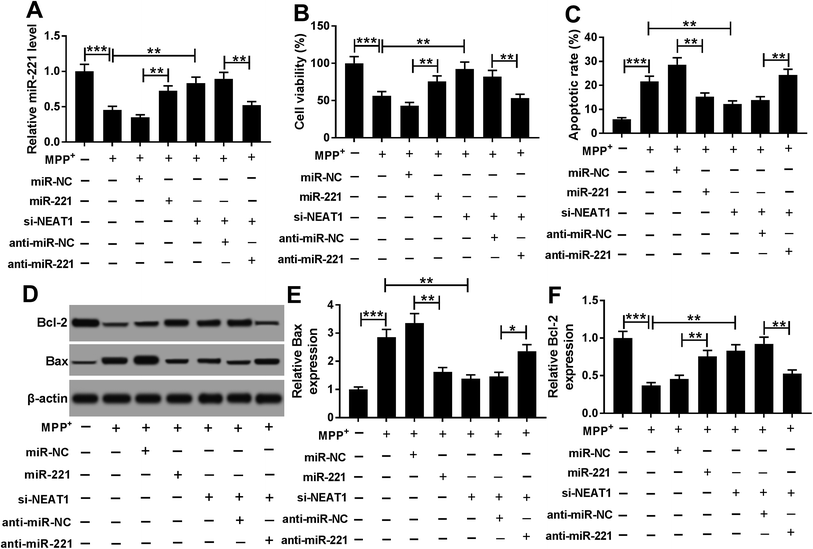
Given our data that NEAT1 expression was elevated by MPP+ in SH-SY5Y cells (Fig. 2A), we further observed whether NEAT1 influenced cell viability and apoptosis. As expected, MPP+ treatment resulted in a suppression of cell viability (Fig. 2B) and a promotion of cell apoptosis (Fig. 2C and D) compared with negative control. Moreover, MPP+ treatment significantly decreased Bcl-2 expression and increased Bax level in SH-SY5Y cells (Fig. 2E and F), supporting the promotional effect of NEAT1 silencing on cell apoptosis. These results together indicated a successful establishment of PD model in vitro.To explore the functional role of NEAT1 in MPP+-induced SH-SY5Y cell injury, loss-of-function experiments were carried out by transfection of siRNA targeting NEAT1 (si-NEAT1). Transient introduction of si-NEAT1, but not a scrambled control sequence, significantly abolished MPP+-induced increased NEAT1 expression in SH-SY5Y cells (Fig. 2A). Subsequently, in contrast to negative group, NEAT1 depletion strikingly antagonized MPP+-induced viability suppression and apoptosis promotion in SH-SY5Y cells (Fig. 2B–F). All these data suggested that NEAT1 knockdown played a protective role on MPP+-induced injury of SH-SY5Y cells.
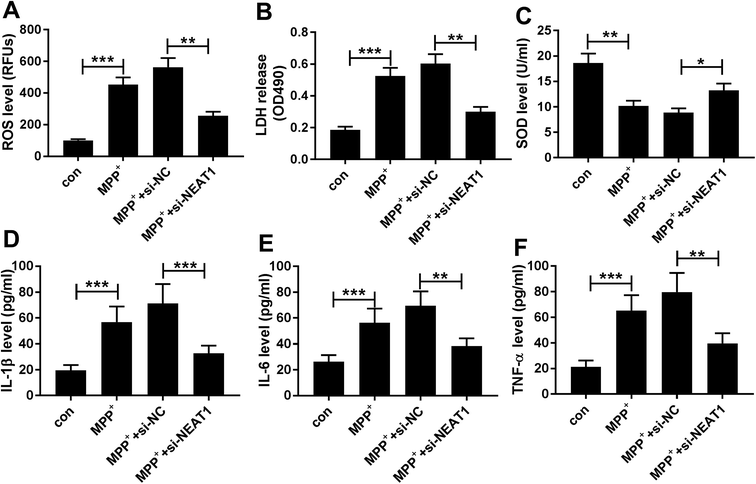
3.3. Silencing of NEAT1 alleviated MPP+-induced promotion of oxidative stress and neuroinflammation in SH-SY5Y cells
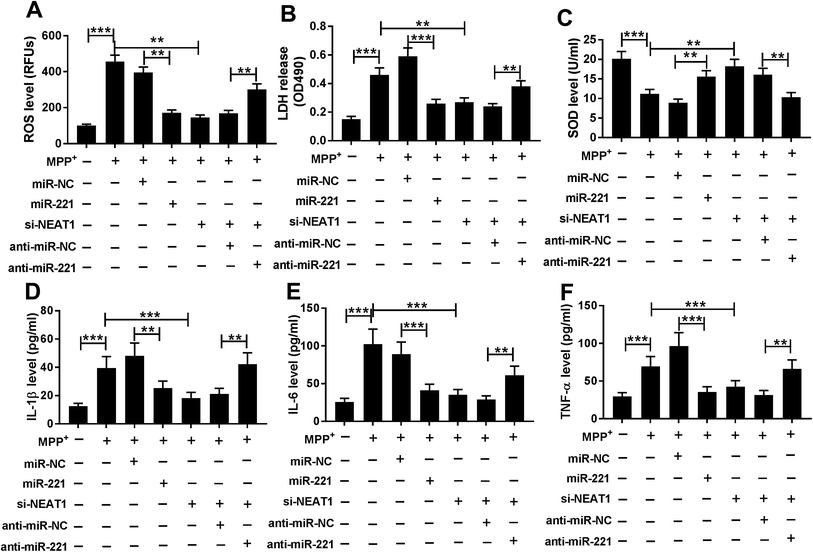
Then, we further investigated the effect of NEAT1 on MPP+-induced SH-SY5Y cells cytotoxicity by transfection of si-NEAT1 prior to MPP+ treatment, followed by the measurement of ROS generation, LDH activity, SOD activity, and the mRNA levels of interleukin-1β (IL-1β), tumor necrosis factor α (TNF-α) and IL-6. In comparison to negative control, MPP+ treatment resulted in a significant enhancement of ROS generation (Fig. 3A), LDH release (Fig. 3B) and a distinct suppression of SOD level (Fig. 3C), as well as a high increase of IL-1β (Fig. 3D), IL-6 (Fig. 3E) and TNF-α (Fig. 3F) mRNA levels. However, MPP+-triggered these effects were dramatically alleviated by NEAT1 knockdown in SH-SY5Y cells (Fig. 3A–F). Together, these results implied that NEAT1 depletion alleviated MPP+-induced cytotoxicity in SH-SY5Y cells.3.4. NEAT1 directly targeted miR-221
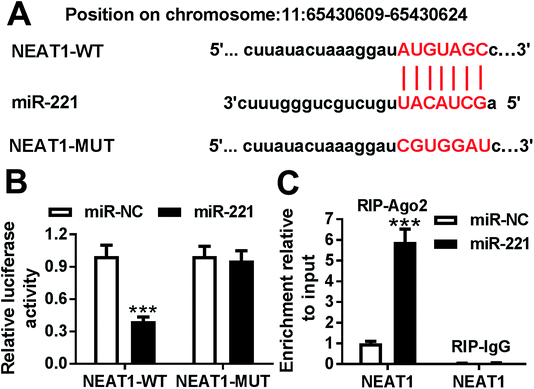
To further explore the mechanism by which NEAT1 regulated MPP+-induced SH-SY5Y cell injury, we carried out a detailed analysis for the targeted miRNAs of NEAT1. Using the software LncBase Predicted v.2, these data showed a putative miR-221-binding sequence in NEAT1 (Fig. 4A). To validate this, we performed a dual-luciferase reporter assay using NEAT1 luciferase reporter (NEAT1-WT) containing miR-221-binding sequence. With the NEAT1-WT construct and miR-221 overexpression caused a significant reduction in luciferase activity (Fig. 4B), suggesting that ectopically expressed miR-221 specifically binded into NEAT1. To verify whether NEAT1 binding sites were required for this effect, a mutant reporter (NEAT1-MUT), in which all predicted miR-221 interaction sites were mutated, was tested. Notably, the mutant completely abolished the effect of miR-221 on luciferase activity under the same conditions (Fig. 4B). Then, RIP assays were performed to verify the endogenous interrelation between NEAT1 and miR-221 using anti-Ago2 antibody. In comparison to a scrambled control sequence, NEAT1 was preferentially enriched by miR-221 overexpression in SH-SY5Y cells (Fig. 4C). These data together pointed to a role of NEAT1 as a molecular sponge for miR-221 in SH-SY5Y cells.3.5. MiR-221 expression was inhibited by MPP+ treatment and negatively modulated by NEAT1 in SH-SY5Y cells
Then, we determined the expression of miR-221 in MPP+-induced SH-SY5Y cells. As demonstrated by qRT-PCR, MPP+ treatment resulted in a significant decrease of miR-221 expression in dose- and time-dependent manners (Fig. 5A and B). Given the data that NEAT1 directly targeted miR-221, we further explored whether, if so, how NEAT1 modulated miR-221 expression in SH-SY5Y cells. As expected, in contrast to their counterparts, miR-221 expression was strikingly reduced by transfection of pcDNA-NEAT1 and evidently elevated following NEAT1 silencing (Fig. 5C). These results indicated that NEAT1 negatively modulated miR-221 expression in SH-SY5Y cells.3.6. MiR-221 was involved in the protective effect of NEAT1 knockdown in MPP+-induced SH-SY5Y cells
Next, gain-of-function experiments were performed to explore the functional role of miR-221 in MPP+-induced SH-SY5Y cells. In contrast to negative control, transfection of miR-221 mimic significantly increased miR-221 expression in MPP+-induced SH-SY5Y cells (Fig. 6A). Subsequent experiment data showed that miR-221 overexpression resulted in a significant enhancement of cell viability and a distinct suppression of cell apoptosis in MPP+-induced SH-SY5Y cells (Fig. 6B–F). All these data implied that miR-221 overexpression relieved MPP+-induced SH-SY5Y cell injury.To provide further mechanistic insight into the link between NEAT1 and miR-221 on MPP+-induced SH-SY5Y cell injury, cells were cotransfected with si-NEAT1 and anti-miR-221 prior to MPP+ treatment. Transient transfection of anti-miR-221, but not a negative control sequence, significantly abolished si-NEAT1-mediated increased effect on miR-221 expression in MPP+-induced SH-SY5Y cells (Fig. 6A). Moreover, the restoration of miR-221 expression remarkably antagonized si-NEAT1-mediated pro-viability and anti-apoptosis effect compared with negative control (Fig. 6B–F). Together, these results strongly established that miR-221 mediated the protective effect of NEAT1 silencing on MPP+-induced SH-SY5Y cell injury.
3.7. MiR-221 was involved in the alleviative effect of NEAT1 silencing on MPP+-induced oxidative stress and neuroinflammation in SH-SY5Y cells
In comparison to a negative control sequence, miR-221 overexpression triggered a significant repression of ROS generation (Fig. 7A), LDH release (Fig. 7B) and a distinct enhancement of SOD level (Fig. 7C), as well as an obvious suppression of IL-1β (Fig. 7D), IL-6 (Fig. 7E) and TNF-α (Fig. 7F) mRNA levels in MPP+-induced SH-SY5Y cells, suggesting the protective role of miR-221 overexpression on MPP+-induced SH-SY5Y cell injury. Subsequent experiment results revealed that compared with negative control, miR-221 restoration significantly reversed the alleviative effect of NEAT1 silencing on MPP+-induced promotion of oxidative stress and neuroinflammation in SH-SY5Y cells (Fig. 7A–F). All these data together suggested that NEAT1 silencing exerted its protective role on MPP+-induced oxidative stress and neuroinflammation by miR-221 in SH-SY5Y cells.4. Discussion
In the present study, our results verified a successful establishment for PD model by MPP+ in SH-SY5Y cells, as evidenced by the suppression of cell viability and the promotion of cell apoptosis, oxidative stress and neuroinflammation, in accordance with previous researches.22,23 LncRNAs have attracted increasing attention for their functions involved in the mechanisms of pathogenesis in neurodegenerative diseases, including PD.24,25 Our study firstly illuminated that NEAT1 silencing ameliorated MPP+-induced neuronal damage by sponging miR-221 in SH-SY5Y cells.NEAT1 is an abundant, ubiquitously expressed lncRNA, which forms a scaffold for a specific RNA granule in the nucleus, or nuclear body of the paraspeckle.26 Accumulating evidence has demonstrated that NEAT1 fine-tunes the function of various neurodegeneration-associated pathways, such as neuronal apoptosis and neuroinflammation.27,28 A recent literature reported that NEAT1 was upregulated in Huntington's disease, and NEAT1 overexpression enhanced cell viability under oxidative stress.15 Moreover, aberrant NEAT1 expression was identified to modulate MPP+-induced SH-SY5Y cell autophagy by stabilization of PINK1 protein.29 In the present study, our data indicated that MPP+ elevated the expression of NEAT1 and NEAT1 knockdown relieved MPP+-induced cytotoxicity in SH-SY5Y cells, consistent with former works.29,30 More interestingly, we firstly manifested that NEAT1 knockdown alleviated MPP+-induced promotion of oxidative stress and neuroinflammation in SH-SY5Y cells. Previous research had reported that NEAT1 depletion repressed oxidized low-density lipoprotein (ox-LDL)-induced inflammation and oxidative stress in macrophages.31
LncRNAs involve in the pathogenesis of PD via various mechanisms, including their miRNA binding functions. For example, lncRNA MALAT1 regulated MPP+-induced MN9D cell apoptosis through sponging miR-205-5p.32 LncRNA HOTAIR contributed to the progression of PD via targeting miR-216-5p. Therefore, we analyzed the potential targeted miRNAs of NEAT1 using LncBase Predicted v.2 software. Among the predicted candidates, miR-221 was of interest in the present study, considering its crucial role as a biomarker for PD evaluation.18,19 Additionally, downregulated miR-221 was found in PD model in vitro, and miR-221 overexpression played a protective role in PD through regulation of PC12 cell viability and apoptosis by targeting PTEN.20 A recent document demonstrated that miR-221 overexpression protected SH-SY5Y cell against MPP+-induced neurite retraction and cell death through targeting transcripts of apoptotic proteins, highlighting the protective role of miR-221 in PD.20 Subsequently, we validated that NEAT1 directly targeted miR-221 in SH-SY5Y cells. Our data also indicated that MPP+ treatment reduced miR-221 expression in dose- and time-dependent manners in SH-SY5Y cells. Moreover, miR-221 expression was negatively regulated by NEAT1 in SH-SY5Y cells, providing an evidence for the role of NEAT1 as a molecular sponge for miR-221. Based on the above, we made a hypothesis that NEAT1 silencing alleviated MPP+-induced SH-SY5Y cell injury by targeting miR-221. Subsequently, we firstly manifested that miR-221 mediated the protective effect of NEAT1 silencing in MPP+-induced SH-SY5Y cells. Similar with our findings, Chen et al. manifested that NEAT1 enhanced ox-LDL-induced oxidative stress and inflammation via sponging miR-128.31 The present research is limited to in vitro study, and more researches in vivo using PD model will be performed to confirm this novel mechanism in the further.
5. Conclusion
In conclusion, our study suggested that NEAT1 silencing alleviated MPP+-induced SH-SY5Y cell injury by regulation of cell viability, cell apoptosis, oxidative stress and neuroinflammation through targeting miR-221. Targeting NEAT1 might be a potential therapeutic target for PD treatment.Conflicts of interest
The authors declare no conflicts of interest.Acknowledgements
Not applicable.References
- O.-B. Tysnes and A. Storstein, J. Neural Transm., 2017, 124, 901–905 CrossRef PubMed.
- Z. Goodarzi, K. J. Mrklas, D. J. Roberts, N. Jette, T. Pringsheim and J. Holroyd-Leduc, Neurology, 2016, 87, 426–437 CrossRef PubMed.
- A. De Virgilio, A. Greco, G. Fabbrini, M. Inghilleri, M. I. Rizzo, A. Gallo, M. Conte, C. Rosato, M. C. Appiani and M. de Vincentiis, Autoimmun. Rev., 2016, 15, 1005–1011 CrossRef PubMed.
- Q. Wang, Y. Liu and J. Zhou, Transl. Neurodegener., 2015, 4, 19, DOI:10.1186/s40035-015-0042-0.
- A. Sarrafchi, M. Bahmani, H. Shirzad and M. Rafieian-Kopaei, Curr. Pharm. Des., 2016, 22, 238–246 CrossRef PubMed.
- J. J. Quinn and H. Y. Chang, Nat. Rev. Genet., 2016, 17, 47–62 CrossRef CAS PubMed.
- T. F. J. Kraus, M. Haider, J. Spanner, M. Steinmaurer, V. Dietinger and H. A. Kretzschmar, Mol. Neurobiol., 2017, 54, 2869–2877 CrossRef CAS PubMed.
- M. Elkouris, G. Kouroupi, A. Vourvoukelis, N. Papagiannakis, V. Kaltezioti, R. Matsas, L. Stefanis, M. Xilouri and P. K. Politis, Front. Cell. Neurosci., 2019, 13, 58 CrossRef PubMed.
- Y. Chen, Y. J. Lian, Y. Q. Ma, C. J. Wu, Y. K. Zheng and N. C. Xie, NeuroToxicology, 2018, 68, 212–221 CrossRef CAS PubMed.
- B. Cao, T. Wang, Q. Qu, T. Kang and Q. Yang, Neuroscience, 2018, 388, 118–127 CrossRef CAS PubMed.
- Q. Lin, S. Hou, Y. Dai, N. Jiang and Y. Lin, Biol. Chem., 2019, 400, 1217–1228 Search PubMed.
- C. M. Clemson, J. N. Hutchinson, S. A. Sara, A. W. Ensminger, A. H. Fox, A. Chess and J. B. Lawrence, Mol. Cell, 2009, 33, 717–726 CrossRef CAS PubMed.
- X. Yu, Z. Li, H. Zheng, M. T. Chan and W. K. Wu, Cell Prolif., 2017, 50 DOI:10.1111/cpr.12329.
- H. An, N. G. Williams and T. A. Shelkovnikova, Non-coding RNA Res., 2018, 3, 243–252 CrossRef CAS PubMed.
- J. S. Sunwoo, S. T. Lee, W. Im, M. Lee, J. I. Byun, K. H. Jung, K. I. Park, K. Y. Jung, S. K. Lee and K. Chu, Mol. Neurodegener., 2017, 54, 1577–1586 CAS.
- W. Yan, Z. Y. Chen, J. Q. Chen and H. M. Chen, Biochem. Biophys. Res. Commun., 2018, 496, 1019–1024 CrossRef CAS PubMed.
- Y. Liu and Z. Lu, Clin. Exp. Pharmacol. Physiol., 2018, 45, 841–848 CrossRef CAS PubMed.
- W. Ma, Y. Li, C. Wang, F. Xu, M. Wang and Y. Liu, Cell Biochem. Funct., 2016, 34, 511–515 CrossRef CAS PubMed.
- H. Ding, Z. Huang, M. Chen, C. Wang, X. Chen, J. Chen and J. Zhang, Park. Relat. Disord., 2016, 22, 68–73 CrossRef PubMed.
- L. Li, J. Xu, M. Wu and J. Hu, Bratisl. Lek. Listy, 2018, 119, 22–27 CAS.
- H. Xicoy, B. Wieringa and G. J. Martens, Mol. Neurodegener., 2017, 12, 10 CrossRef PubMed.
- Z. Wang, L. Sun, K. Jia, H. Wang and X. Wang, Neurosci. Lett., 2019, 701, 226–233 CrossRef CAS PubMed.
- N. Xie, J. Qi, S. Li, J. Deng, Y. Chen and Y. Lian, J. Cell. Biochem., 2019, 120, 5790–5801 CrossRef CAS.
- P. Riva, A. Ratti and M. Venturin, Curr. Alzheimer Res., 2016, 13, 1219–1231 CrossRef CAS PubMed.
- T. F. Kraus, M. Haider, J. Spanner, M. Steinmaurer, V. Dietinger and H. A. Kretzschmar, Mol. Neurobiol., 2017, 54, 2869–2877 CrossRef CAS PubMed.
- C. M. Clemson, J. N. Hutchinson, S. A. Sara, A. W. Ensminger, A. H. Fox, A. Chess and J. B. Lawrence, Mol. Cell, 2009, 33, 717–726 CrossRef CAS PubMed.
- C. Adriaens, L. Standaert, J. Barra, M. Latil, A. Verfaillie, P. Kalev, B. Boeckx, P. W. Wijnhoven, E. Radaelli and W. Vermi, Nat. Med., 2016, 22, 861–868 CrossRef CAS.
- F. Zhang, L. Wu, J. Qian, B. Qu, S. Xia, T. La, Y. Wu, J. Ma, J. Zeng and Q. Guo, J. Autoimmun., 2016, 75, 96–104 CrossRef CAS.
- W. Yan, Z.-Y. Chen, J.-Q. Chen and H.-M. Chen, Biochem. Biophys. Res. Commun., 2018, 496, 1019–1024 CrossRef CAS PubMed.
- Y. Liu and Z. Lu, Clin. Exp. Pharmacol. Physiol., 2018, 45, 841–848 CrossRef CAS PubMed.
- D. D. Chen, L. L. Hui, X. C. Zhang and Q. Chang, J. Cell. Biochem., 2019, 120, 2493–2501 CrossRef CAS PubMed.
- Q. Chen, X. Huang and R. Li, Am. J. Transl. Res., 2018, 10, 563–572 CAS.
Footnote |
| † These authors contributed equally to this work equally as co-first authors. |
| This journal is © The Royal Society of Chemistry 2019 |