Open Access Article
Open Access ArticleResponses of seed germination and shoot metabolic profiles of maize (Zea mays L.) to Y2O3 nanoparticle stress†
Chenchen Gong ,
Linghao Wang
,
Linghao Wang ,
Xiaolu Li
,
Xiaolu Li ,
Hongsen Wang
,
Hongsen Wang ,
Yuxin Jiang
,
Yuxin Jiang and
Wenxing Wang
and
Wenxing Wang *
*
College of Life and Health Sciences, Northeastern University, Shenyang 110819, China. E-mail: wangwenxing@126.com; wangwenxing@mail.neu.edu.cn
First published on 3rd September 2019
Abstract
The potential risks of rare-earth nanoparticles (RENPs) to plants in the environment are attracting increasing attention due to their wide-spread application. In this regard, little is known about the effects of Y2O3 NPs as an important member of RENPs on crop plants. Seed germination is vulnerable to environmental stress, which determines the growth and yield of crops. Here, maize seeds were exposed to a Y2O3 NP suspension (0–500 mg L−1) in the dark for 6 days. It was found that the Y2O3 NPs had no significant effect on the germination rates (>93%) in all treatments, but they could reduce seed vitality, delay germination, and inhibit seedling growth in a dose-dependent manner. Further, the inhibition effect of Y2O3 NPs on root elongation was much stronger than that on shoot elongation. Meanwhile, the activities of peroxidase (POD) and catalase (CAT) in shoots were enhanced with the increase in the Y2O3 NP concentration. A high-concentration (≥300 mg L−1) of Y2O3 NPs induced a significant increase in the malondialdehyde (MDA) level in shoots compared to the control, indicating that the membrane lipid peroxidation and permeability were enhanced. 1H NMR-based analysis showed that the polar metabolic profiles were altered significantly after treatment with 0, 10, and 500 mg L−1 of Y2O3 NPs, but there was no marked alteration observed for the non-polar metabolic profiles. The polar metabolites (e.g., sugars, amino acids, and most organic acids) showed a dose-dependent increase to Y2O3 NP stress, indicating that the metabolic pathways of carbohydrate metabolism, the tricarboxylic acid cycle (TCA), and amino acid synthesis were disturbed. There were significantly positive correlations found among the metabolites related with the antioxidant response and osmotic adjustment. The simultaneous accumulation of these metabolites possibly indicated the adaptation of the seedlings to stress at the cost of retarding glycolysis, TCA, and protein synthesis. The retarded effects finally inhibited the apparent growth of the seedlings. These findings reveal the phytotoxicity of Y2O3 NPs and provide physiological and biochemical and molecular-scale perspectives on the response of seedlings to stress.
Introduction
Artificial nanoparticles inevitably enter into the environment in the process of their production, recycling, and waste disposal with their large-scale use, and then may even enter into the food chain through animals and plants, which can cause potential negative impacts on the whole ecosystem.1–4 Therefore, the ecological safety and potential health risks of artificial nanoparticles have attracted increasing attention in recent years.Rare earth nanoparticles (RENPs), as a type of artificial nanoparticles, have excellent physicochemical properties and extensive applications, and are considered as a treasury of opportunities for new light sources, magnetic sources, energy sources, and materials.5–7 With the widespread use of RENPs, the potential risks of their accumulation and migration in the environment are expanding.8 The phytotoxicity of RENPs has been confirmed by some laboratory simulations. For instance, it was found that a 2000 mg L−1 suspension of CeO2 NPs had no effect on root elongation of six higher plants (radish, rape, tomato, wheat, cabbage, and cucumber) except lettuce, but 2000 mg L−1 suspensions of La2O3 NPs, Gd2O3 NPs, and Yb2O3 NPs significantly inhibited the root elongation of all seven plants.9 By comparing the toxicity of Yb2O3 NPs, bulk Yb2O3, and YbCl3·6H2O to cucumbers, it was indicated that the inhibition was dependent on the actual amount of toxic Yb uptake by the cucumber plants.10 A low concentration (0.1–10 mg L−1) of CeO2 NPs slightly promoted the growth and yield of tomato, and high-level Ce concentrations could be detected in roots, stems, and edible tissues.11 Three rice species (high-, moderate-, and low-amylose) were cultured in soils with 0 and 500 mg kg−1 of CeO2 NPs until they completely matured, and it was found that the CeO2 NPs could lower the rice quality.12 The toxicity of three types of CeO2 NPs (lab-synthesized 7 and 25 nm CeO2 NPs, and commercial CeO2 NPs) to three kinds of Lactuca genus plants showed that small parts of CeO2 NPs were transformed from Ce(IV) to Ce(III) in the roots of the plants.13,14 Different degrees of biotransformation among the three types of CeO2 NPs accounted for the discrepancy in their toxicity to Lactuca plants due to the differences in their sizes and zeta potentials.14 Besides, the translocation of Ce resulting from NP-root-exposure was found to be species dependent.15 Based on the luminescence characteristics of upconversion NaYF4:Yb, Er NPs, it was visible that the NPs quickly reached all the organs of pumpkin seedlings in 3 h, and their migration rate was negatively correlated with their size.16 These reports show that the phytotoxicity of RENPs is influenced by the type, size, charge, and concentration of NPs as well as the plant species. This means that RENPs may have complex and diverse toxic effects on plants in the environment.
Current studies mainly focus on the phytotoxicity of RENPs as well as their internalization, transportation, distribution, and biotransformation in plants, indicating an abiotic stress of RENPs to plants. Under abiotic stress, plants have a quick response in gene expression level first, and then make different transcriptional regulations. The post-transcription RNA controls the synthesis of the corresponding proteins. Finally, the metabolic balance in plants is adjusted by controlling the metabolite synthesis.17 Metabolites are the final products of this genetic transcription and protein modification, and belong to specific metabolic pathways.18 Thus, the plant metabolism may be perturbed under such stress.19 However, the metabolic responses of plants to RENP stress still remains unknown. Internal regulation and pathways of plants in response to the stress can be understood comprehensively by analyzing the composition and level of the metabolites in plants. Metabonomics can provide a more sensitive and mechanistic understanding of the biological response to a particular stressor.20–22 Nuclear magnetic resonance (NMR), one of major tools for metabonomics, causes neither damage to the structures and properties of samples nor radiation damage, and is highly suited for investigating molecular interactions under approximately physiological conditions.20,23 Moreover, it provides unbiased detection, whereby the response coefficients of different metabolites in the mixture are consistent.
Y2O3 NPs are an important kind of RENPs that are widely applied in ceramic stabilizers, fluorescent powder in color TVs, metal surface coatings, lubricants, petroleum cracking catalysts, and solid laser materials due to their good thermostability, and mechanical and chemical durance.24,25 Such important industrial raw materials inevitably spread to the environment in the form of dust or migrate to plants and animals through water and soil in the process of production and waste treatment, thus causing potential impacts on the ecological environment. According to an existing report, Y2O3 nanotubes could be accumulated in the lateral roots of hydroponic cabbage plants, but not into stems through the principal roots.26 The blockage of Y2O3 nanotubes in a root system was considered to be a primary and potentially fatal factor. However, the effects of Y2O3 NPs on plants are not yet clear.
Maize (Zea mays L.) is of the most important agricultural crops, and is often used as a model organism for basic and applied research in plant biology.27 Seed germination is the most important and vulnerable stage in a plant life cycle.28 During this period, seeds are subjected to environmental stress, which will determine the time the plants will enter the natural and agricultural ecosystems, and directly affects the yield and quality of crops.29 Especially for annual plants, such as maize, wheat, rice, and so on, the success of seed germination is more important. Besides, maize seeds mainly transform nutrients stored in endosperm into the substances and energy needed for the germination and growth of seedlings through respiration, which is different from the metabolism of seedlings under photosynthesis.
In the present study, maize seeds were exposed to Y2O3 NPs suspensions in the range of 0 to 500 mg L−1, and germinated in the dark for 6 days. We assessed the toxicity of Y2O3 NPs to seed germination, and the phenotypic, physiological, and metabolic responses of the young shoots. The metabolic profiles of the polar and non-polar metabolites in the shoots under Y2O3 NP stress were detected and analyzed by combining the 1H NMR-based metabonomics technique and multivariable data analysis. The main metabolic pathways are depicted and the corresponding metabolic regularity discussed. This study helps elucidate the response of crop seed germination and shoots to Y2O3 NP stress, and aids the understanding of the potential environmental risk of RENPs.
Materials and methods
Experimental materials
Y2O3 NPs, with the mean size of about 30 nm and purity of 99.9% and without any other modifiers on the surface, were purchased from Beijing DK Nano Technology Co., Ltd. Maize seeds (Zea mays L. cv. Zhengdan 958) were purchased from the Chinese Academy of Agricultural Sciences, Beijing, China. Heavy water, chloroform-D (containing 0.03% tetramethylsilane (TMS)), and 3-trimethylsilyl[2,2,3,3-D4]propionate (TSP) were all purchased from J&K Scientific Co., Ltd. All other chemical reagents were analytically pure and purchased from China Sinopharm Group. All the used water was ultrapure water.Characterization of the Y2O3 NPs
The size and morphology of the Y2O3 NPs were determined by transmission electron microscopy (TEM, JEM 200CX, Japan) and scanning electron microscopy (SEM, SSX-550, Shimadzu, Japan). The crystal structure of the Y2O3 NPs was detected by X-ray diffractometry (XRD, X'pert PRO MPD, Holland). 100 mg L−1 of Y2O3 NPs suspension after ultrasonic dispersion for 1 h was used to detect the hydrodynamic sizes, zeta potential and polydispersity index (PDI) using a Nano ZS90 Zeta Potential/Particle System (Malvern Panalytical, UK).Seed germination and Y2O3 NPs use
First, the uniform seeds were selected. Then, they were sterilized in 10% fresh sodium hypochlorite solution for 15 min, and rinsed with ultrapure water five times. Second, the Y2O3 NPs were sterilized by ultraviolet irradiation for 1 h, followed by 30 min of ultrasonic dispersion before use. One concentration gradient with seven concentrations (0, 10, 30, 50, 100, 300, and 500 mg L−1) was set. Third, the NPs were added into the sterilized plastic tubes with five germination holes, and each hole was inoculated with two seeds. Each gradient had 10 parallels and 4 repetitions. Finally, each germination hole was covered with a piece of thin plastic film with pinholes. These tubes loaded with 500 mL of Y2O3 NPs suspension were put in a constant-temperature shaker, and the seeds were germinated in the dark under 25 °C. Precipitation of the Y2O3 NPs was prevented by reciprocal shaking to ensure the seeds stayed in full contact with the NPs. After 6 days exposure, seed germination was terminated. Pictures were taken. At the same time, the germination rate, germination potential, germination index, vigor index, and number of roots were calculated. The lengths of the root and shoot were measured using a meter ruler.| Germination rate (%) = (number of germinated seeds/number of total testing seeds) × 100% | (1) |
| Germination potential (%) = (number of germinated seeds in 5 d/number of total testing seeds) × 100% | (2) |
| Germination index (GI) = ∑Gt/Dt, | (3) |
| Vigor index (VI) = GI × S, | (4) |
Antioxidant enzyme activity and malondialdehyde (MDA) content
The seedlings were rinsed with ultrapure water thoroughly after 6 days exposure. In an ice bath, 0.5 g of shoots was homogenated in phosphate buffer solution (PBS, 50 mmol L−1, pH 7.8, containing 1% polyvinylpyrrolidone for protecting the enzyme activity). Second, the mixture was centrifuged for 10 min at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g under 4 °C. The supernatants were preserved for the determination of the enzyme activities and MDA contents. The supernatants were used to measure POD and CAT activities as previously described by Wu and Von.30 POD activity was measured by following the formation of guaiacol dehydrogenation products, as determined by an increase in absorbance at 450 nm. CAT activity was assayed by measuring the change in absorbance at 240 nm that accompanied the consumption of H2O2. MDA content in shoots was measured by the TBARS (thiobarbituric acid reactive substances) assay as described by Chaoui et al.31
000g under 4 °C. The supernatants were preserved for the determination of the enzyme activities and MDA contents. The supernatants were used to measure POD and CAT activities as previously described by Wu and Von.30 POD activity was measured by following the formation of guaiacol dehydrogenation products, as determined by an increase in absorbance at 450 nm. CAT activity was assayed by measuring the change in absorbance at 240 nm that accompanied the consumption of H2O2. MDA content in shoots was measured by the TBARS (thiobarbituric acid reactive substances) assay as described by Chaoui et al.31
1H NMR detection and metabolites
The selected shoot samples of the low-concentration group (L, 10 mg L−1 Y2O3 NPs) and high-concentration group (H, 500 mg L−1 Y2O3 NPs) were detected by 1H NMR with the control group (CK, without Y2O3 NPs). Preparation of the samples and NMR detection were accomplished according to a method by Sun et al.32 First, 0.5 g of shoots was quickly ground into powder in liquid nitrogen, to which was added 3 mL pre-cooled methanol–water mixture (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) and 3 mL chloroform. The mixture was then transferred to a pre-cooled centrifuge tube, followed by vortex processing for 1 min and ultrasonic treatment for 1 min in an ice bath. Subsequently, the samples were centrifuged for 20 min at 10
1) and 3 mL chloroform. The mixture was then transferred to a pre-cooled centrifuge tube, followed by vortex processing for 1 min and ultrasonic treatment for 1 min in an ice bath. Subsequently, the samples were centrifuged for 20 min at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g under 4 °C. This step was repeated three times. The polar and non-polar parts were combined and collected, respectively. Methanol was eliminated from the polar samples under vacuum conditions. The supernatant was frozen for at least 24 h under −80 °C and then processed to a brown powder by a vacuum freeze-drier. The non-polar samples were vacuum dried in a rotary vacuum evaporator. Then, 650 μL of 100% D2O, 130 μL of 0.1 mol L−1 PBS (pH 7.0) containing 10% D2O, and 0.02 mmol L−1 TSP were added into the polar samples. While, 1 mL chloroform-D solution containing 0.1% TMS was added into the non-polar samples. TSP and TMS were used as internal standards, respectively. These polar and non-polar samples were transferred to centrifuge tubes and centrifuged for 5 min at 10
000g under 4 °C. This step was repeated three times. The polar and non-polar parts were combined and collected, respectively. Methanol was eliminated from the polar samples under vacuum conditions. The supernatant was frozen for at least 24 h under −80 °C and then processed to a brown powder by a vacuum freeze-drier. The non-polar samples were vacuum dried in a rotary vacuum evaporator. Then, 650 μL of 100% D2O, 130 μL of 0.1 mol L−1 PBS (pH 7.0) containing 10% D2O, and 0.02 mmol L−1 TSP were added into the polar samples. While, 1 mL chloroform-D solution containing 0.1% TMS was added into the non-polar samples. TSP and TMS were used as internal standards, respectively. These polar and non-polar samples were transferred to centrifuge tubes and centrifuged for 5 min at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g. For each sample, 600 μL supernatant was transferred into an NMR sample tube. Four repetitions of each treatment were used for the 1H NMR analysis.
000g. For each sample, 600 μL supernatant was transferred into an NMR sample tube. Four repetitions of each treatment were used for the 1H NMR analysis.
High-resolution 1H NMR spectra were screened using a Bruker AVIII 600 nuclear magnetism spectrometer (Bruker Biospin, Germany), thus producing polar and non-polar metabolic profiles. The processing, automatization, and collection of samples were controlled by Topspin 2.1 software (Bruker Biospin). The standard 1H 90° pulse sequences were used for the polar and non-polar samples, and residual water resonance was suppressed in the polar samples. After introducing the probe, the samples were allowed to balance for 1 min. Each spectrum was gained as the 32 K data point at a spectral width of 16 ppm, and as the sum of 128 transients with a relaxation delay of 2 s. The phase and baseline of the 1H NMR spectra were corrected automatically, and the internal standard TSP and TMS were used as the reference peaks for the chemical shift at δH 0.00 ppm. Metabolite resonances were assigned according to the public databases (e.g., Biological Magnetic Resonance Data Bank, Spectral Database for Organic Compounds, and Madison Metabolomics Consortium Database) and previous studies.32
Data and statistical analysis
Overall variances of the different Y2O3 NPs treatments in terms of seed germination and physiological indexes were analyzed by one-way ANOVA using SPSS 17.0 software. Significant differences among the treatment groups were analyzed using LSD and Duncan tests. For all, the statistical significance was set at P < 0.05.1H NMR data were processed by MestReC software. Spectral intensities were scaled to TSP (polar extracts) and to TMS (non-polar extracts). Spectral intensities were decreased to integrated regions with an equal width (0.01 ppm) corresponding to the δ 10.00 to δ −0.05 region (non-polar phase) or δ 9.00 to δ −0.05 region (polar phase). For the polar phase, the regions of δ 5.00 to δ 4.70 and δ 3.38 to δ 3.30 were eliminated from the analysis due to residual signals of water and methanol. For the non-polar phase, the region of δ 7.40 to δ 7.20 was excluded from the analysis due to residual signals of chloroform. Subsequently, the normalized NMR spectral data were imported into the SIMCA-P+ software package (Version 14.0, Umetrics AB, Umea, Sweden) for principal component analysis (PCA). The PCA results were visualized with score plots. The overall variances among the metabolites of CK, L, and H were analyzed by one-way ANOVA using SPSS 17.0 software. For all, the statistical significance was set at P < 0.05. The main metabolic pathways of the shoots in response to Y2O3 NP stress were depicted by VANTED software (Version 2.6.5, Germany). Correlation analyses between all the metabolite pairs were performed using Pearson's correlation in VANTED software.
Results
Physicochemical properties of the Y2O3 NPs
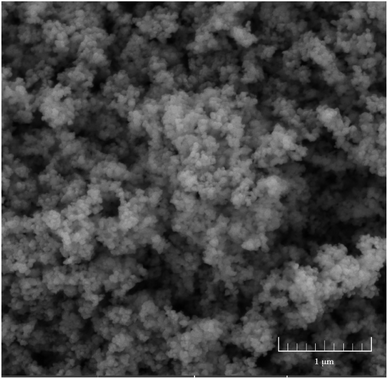
The used commercial Y2O3 NPs sized about 30 nm were nearly spherical (Fig. 1 and S1†). Because of their minute size, large superficial area, and highly active surface, the NPs tended to aggregate. After ultrasonic treatment, the hydrodynamic size, zeta-potential, and PDI of the Y2O3 NPs dispersed in ultrapure water were 300.5 ± 14.1 nm, 5.27 ± 0.03 mV, and 0.489 ± 0.008, respectively. It was found that their hydrodynamic size was far larger than their particle size, possibly due to particle aggregation and there existing a hydration layer on the particles. The XRD pattern of the Y2O3 NPs showed a cubic crystal structure (Fig. S2†).Responses of seed germination to Y2O3 NP stress
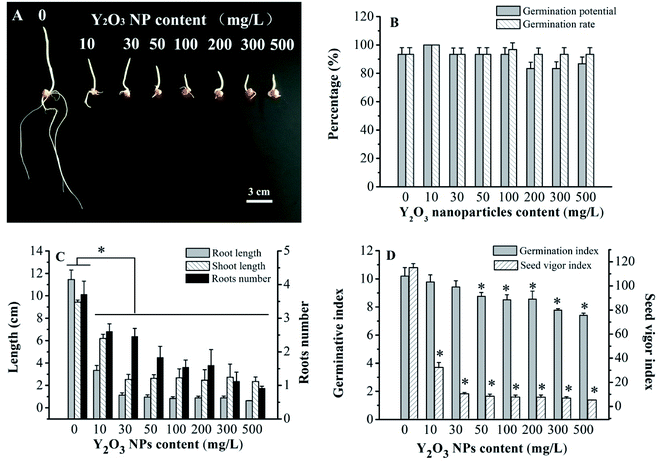
Visibly, compared to CK, the Y2O3 NPs treatments of 10–500 mg L−1 had strong inhibition effects on seed germination (Fig. 2A). With the increasing concentration of Y2O3 NPs, young roots became shorter and swollen. According to the statistical analysis, the germination rates (>93%) and germination potentials (>83%) of all the treatments showed no significant difference (P > 0.05, Fig. 2B), but the shoot length, root length, number of roots, and vigor index in the range of 10–500 mg L−1 of Y2O3 NPs were significantly decreased compared to CK (P < 0.05, Fig. 2C and D). The inhibition effect of the Y2O3 NPs on root elongation was much stronger than that on shoot elongation. After the Y2O3 NPs concentration reached 50 mg L−1, the germination index was significantly lowered compared to CK (P < 0.05, Fig. 2D), indicating the delay of seed germination. On the whole, Y2O3 NPs treatment could reduce seed vigor, delay germination, and inhibit seedling growth in a dose-dependent manner.Physiological and biochemical responses to Y2O3 NP stress
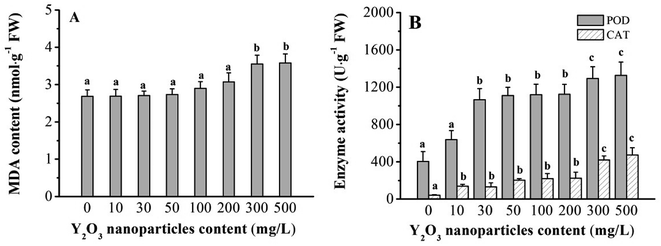
Adversity stress can influence plant growth and trigger physiological and biochemical changes. MDA is often used to measure the peroxidation degree of plasma membrane under adversity stress.33,34 It was found here that the Y2O3 NPs slightly increased MDA content in the shoots in the range of 10–200 mg L−1, and then significantly (P < 0.05) increased it at ≥300 mg L−1 of Y2O3 NPs compared to CK (Fig. 3A). Compared to CK, the POD activity (10–200 mg L−1 of Y2O3 NPs) and CAT activity (30–200 mg L−1 of Y2O3 NPs) in shoots significantly increased (Fig. 3B, P < 0.05). After the NPs reached 300 mg L−1, both further significantly increased.Metabolic responses of maize shoot to Y2O3 NP stress
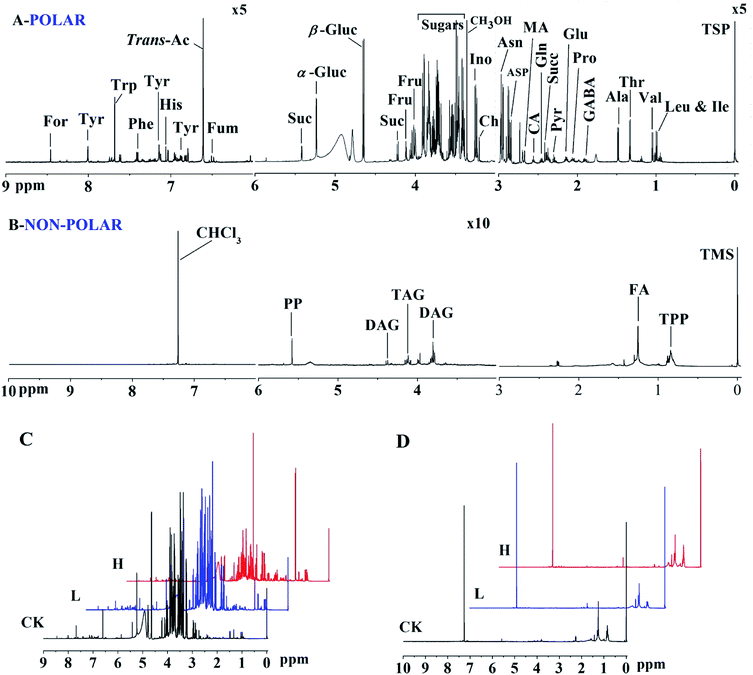
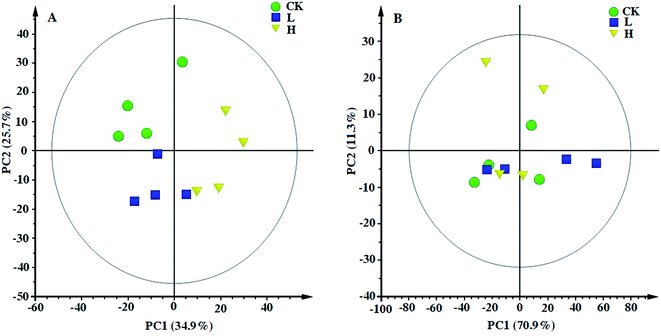
The typical 1H NMR spectra of the polar and non-polar metabolites in maize shoots are shown in Fig. 4A and B, respectively. Compared with the 1H NMR spectra of the CK, L, and H groups, some bin areas of polar metabolites increased in the presence of Y2O3 NPs, indicating that Y2O3 NP stress improved the level of some polar metabolites (Fig. 4C). However, almost no difference was found among the non-polar metabolites of the three groups (Fig. 4D).Principal component analysis (PCA) was performed to provide a general overview of the trends, grouping, and outliers in the 1H NMR data.23 PCAs of the polar and non-polar metabolite profiles were extracted from maize shoot samples. The PCA score plots showed different patterns of variations between the polar and non-polar metabolites (Fig. 5). For the polar metabolites, the score plots reflected that there was no intersection of CK, L, and H, along with an evident separation trend (Fig. 5A). H was clearly separated from CK and L by PC1. This indicated that their polar metabolic profiles differed and responded in a dose-dependent manner. However, the non-polar metabolites among the three groups exhibited an intersection, without a clear separation trend (Fig. 5B), indicating that the non-polar metabolic profiles had no significant response to stress.
Differences in the polar and non-polar metabolites among L, H, and CK were further analyzed by ANOVA (Table 1, P < 0.05). For sugars, compared to CK, sucrose in L and H was upregulated the most, followed by fructose, while sucrose and fructose from L to H did not increase significantly. All the amino acids in L and H were upregulated with the aggravation of Y2O3 NP stress, but the fold changes varied greatly. Among all the amino acids, proline (4.70-fold) in L was upregulated the most compared to CK, whereas alanine upregulated the most (15.11-fold) in H. Only tyrosine levels in H was almost unchanged compared to L. Except for aconitate and fumarate, the other organic acids in L and H were upregulated compared to CK. Pyruvate (2.61-fold) in L was upregulated the most, but it was citrate (3.90-fold) in H. Compared to CK, choline increased significantly with the increasing Y2O3 NP stress. All the non-polar metabolites in L and H showed no significant difference compared to CK. These results confirmed that most of the polar metabolites in the maize shoots were markedly upregulated by the Y2O3 NPs, while there was no significant alteration in the non-polar metabolites.
| Metabolites | L/CK | H/CK | H/L | Metabolites | L/CK | H/CK | H/L |
|---|---|---|---|---|---|---|---|
| The bolder parts in the table are the different categories of metabolites. Metabolites with a significant change among CK, L, and H are indicated in bold and italics (P < 0.05). CK, L, and H stand for 0, 10, and 500 mg L−1 of Y2O3 NPs, respectively. | |||||||
| Sugars | Amino acids | ||||||
| Sucrose | 3.12 | 3.63 | 1.61 | GABA | 2.29 | 4.81 | 2.11 |
| Glucose | 1.43 | 1.13 | 0.79 | Organic acids | |||
| Fructose | 2.32 | 2.37 | 1.02 | Formate | 2.07 | 2.34 | 1.13 |
| Amino acids | Aconitate | 0.98 | 0.93 | 0.95 | |||
| Tryptophan | 1.08 | 1.55 | 1.44 | Fumarate | 0.78 | 0.70 | 0.90 |
| Histidine | 1.52 | 1.76 | 1.16 | Malate | 1.42 | 2.65 | 1.86 |
| Phenylalanine | 2.61 | 3.33 | 1.28 | Citrate | 1.72 | 3.90 | 2.27 |
| Tyrosine | 3.40 | 3.50 | 1.03 | Succinate | 1.50 | 2.35 | 1.57 |
| Valine | 2.88 | 3.75 | 1.30 | Pyruvate | 2.61 | 3.85 | 1.48 |
| Isoleucine | 2.82 | 3.98 | 1.41 | Others | |||
| Leucine | 2.35 | 4.51 | 1.92 | Choline | 2.31 | 3.32 | 1.44 |
| Alanine | 3.97 | 15.11 | 3.81 | Inositol | 1.34 | 1.18 | 0.88 |
| Threonine | 2.11 | 3.39 | 1.61 | Non-polar metabolites | |||
| Asparagine | 3.77 | 4.37 | 1.16 | Polyphenols | 1.51 | 1.24 | 0.81 |
| Aspartate | 1.09 | 2.40 | 2.20 | Triacylglyceride | 1.12 | 1.10 | 0.98 |
| Glutamine | 3.44 | 4.25 | 1.23 | Diacylglyceride | 1.62 | 1.46 | 0.90 |
| Glutamate | 3.60 | 7.01 | 1.95 | Fatty acid | 1.00 | 1.01 | 1.00 |
| Proline | 4.70 | 6.19 | 1.32 | Triterpenoids | 1.01 | 1.01 | 1.00 |
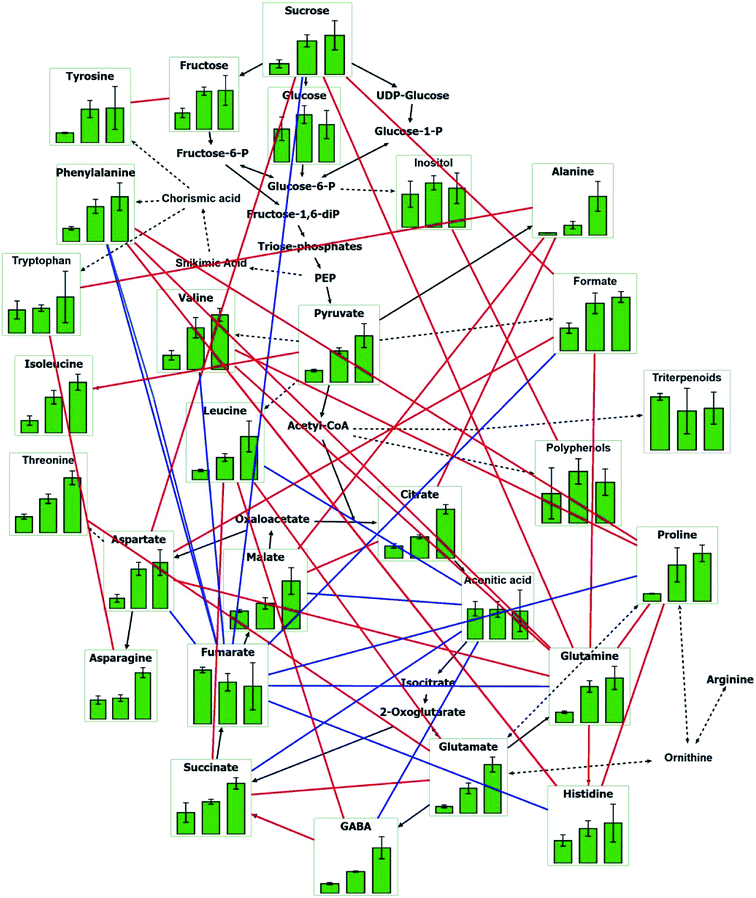
In order to better understand the responses of young shoots to Y2O3 NP stress as a whole, the major pathways were depicted by VANTED software, including the carbohydrate metabolism, amino acid synthesis, and tricarboxylic acid cycle (TCA) (Fig. 6). Correlations among the different metabolites were further analyzed. Except for aconitate and fumarate, there was a positive correlation between most metabolites.
Discussions
Response of maize seed germination to Y2O3 NP stress
Seed germination is considered to start with water absorption and end with the radicle breaking the surrounding structure through a series of metabolic processes.35 In this process, seeds are sensitive to environmental stresses, and respond accordingly. In the present study, the germination rates of maize seeds exposed to all treatments of Y2O3 NPs showed no significant difference compared to CK (P > 0.05, Fig. 2A and B), which was in agreement with seeds exposed to other nanoparticles.9,34,36 However, some other nanoparticles have exhibited either the promotion37 or inhibition38 of the seed germination rate. These contradictory results indicated that the toxicity of nanoparticles to seed germination is related to the concentration, type, size, and morphology of the nanoparticles and the plant species.39 Interestingly, although Y2O3 NPs had no effect on the germination rate of maize seeds, the germination indexes significantly decreased at ≥50 mg L−1 of Y2O3 NPs compared to CK (P < 0.05, Fig. 2D), indicating a slowdown of the emergence speed and the delay of seed germination. At ≥10 mg L−1 of Y2O3 NPs, the seed vigor indexes significantly decreased, indicating the decline of seed vitality (Fig. 2D). Seed germination is a physiological process of water absorption and saturation.40 The seed germination speed depends on its water absorption rate. Generally, water can pass through the dense seed coat freely, but particles like Y2O3 NPs (about 300 nm in hydrodynamic size) are difficult to enter. Y2O3 NPs in the environment may be wrapped in an outer layer of the seed coat by adsorption, which reduces the speed and amount of water absorption of the seeds, resulting in the reduction of free water content, the hindrance of cell expansion, and the inhibition of metabolic activity (substance transport, enzyme activation, signal transduction, etc.) in cells, further causing the decline of seed vigor and the delay of seed germination.41,42After seed germination, Y2O3 NPs (≥10 mg L−1) altered the morphology of early seedlings; for example, Y2O3 NPs caused the shoot and root elongation, reduction of lateral roots, and hardening and swelling of the main roots. Compared with the case of shoot elongation, the Y2O3 NPs had a much stronger inhibition effect on root elongation (Fig. 2A and C), which was consistent with other RENPs.3,9,10,34 Roots were directly exposed to NPs after breaking through the seed coat. The NPs could easily be absorbed on the root surface due to the large amounts of mucilage secreted by the root tips and hairs.9,34 Chen et al. reported that Y2O3 nanotubes could be taken up by roots but were not found in the cabbage shoot mainly due to their accumulation at the primary-lateral-root junction.26 However, some NPs (e.g., Fe3O4 NPs,43 multiwalled carbon nanotubes,44 CeO2 NPs45) could be transported up through the xylem vessels with water and nutrients to the stems and leaves of plants.46,47 This indicates that whether NPs can be transported upward to the stems and leaves is related to various factors (such as particle type, size, morphology, and plant species). Compared with Y2O3 nanotubes,26 it was unclear whether our used Y2O3 NPs with a smaller size, nearly spherical shape, and a little agglomeration were more conducive to upward transport. Moreover, large numbers of the suspended NPs accumulated on the root surfaces could damage the roots. For instance, CeO2 NPs could reduce meristematic cells in root tips of asparagus lettuce, leading to retarding cell division and hindering root growth.34 Damage to the meristematic cells could affect the synthesis of cytokinins. A recent report showed that La2O3 NPs could accelerate the development of apoplastic barriers in roots, leading to a decrease in water uptake.48
Physiological and biochemical response of maize shoots to Y2O3 NP stress
The phytotoxicity of NPs has been attributed to the excess generation of reactive oxygen species (ROS).49,50 ROS can cause lipid peroxidation, osmotic changes, protein oxidation, and metabolic hindrance.51 MDA is a major cytotoxic product of membrane lipid peroxidation, which can reflect the damage degree of plants under adverse stress.34 To protect cells from the cytotoxicity of ROS, plants develop various defense mechanisms to scavenge ROS caused by damage. The enzyme-antioxidant system is one of the protective mechanisms against ROS and works through a series of ROS scavenging enzymes, such as SOD, POD, and CAT.52 In this study, compared to CK, the activities of POD and CAT in maize shoots were increased significantly in the range of 30 to 500 mg L−1 of Y2O3 NPs (Fig. 3B). However, the MDA level did not increase significantly until the NPs concentration was greater than or equal to 300 mg L−1 of Y2O3 NPs (Fig. 3A). This indicated that ROS accumulation and the ROS scavenging system could maintain a dynamic balance at less than 300 mg L−1 of Y2O3 NPs, but this balance was broken at greater than or equal to 300 mg L−1 and then excess ROS could not be cleared timely, resulting in oxidative damage to the membrane system and an increase in cell membrane permeability.34,53 Similar results were reported by previous studies on the induction of antioxidative enzymes activity in plants exposed to other nanoparticles.50,54,55Metabolic response of maize shoots to Y2O3 NPs stress
The external influences of Y2O3 NPs on seed germination and seedling growth overall reflected in the internal metabolic changes. Polar metabolites in shoots mainly respond to Y2O3 NPs stress, involving the metabolic pathways of glycolysis, tricarboxylic acid cycle, and amino acid synthesis (Table 1 and Fig. 6). Sugars are important participants in the glycolytic pathway (EMP), TCA, and pentose phosphate pathway (PPP) during seed germination and young seedling growth.56 Sucrose, among the soluble sugars, had the strongest response to Y2O3 NPs stress (Table 1). The accumulation of sucrose plays an important role in lowering cell osmotic potential and protecting the cell molecular structure and membrane stability.57 Moreover, sucrose is more effective than other oligosaccharides in maintaining the stability of biomembrane systems and biomacromolecules.58 Similarly, low temperature stress promoted the accumulation of soluble sugars in chickpea, especially the accumulation of sucrose was found to be much greater than that of glucose and fructose.59 In addition, the soluble sugars can act as antioxidants directly to regulate plant growth and to cope with environmental stress.60 They have dual effects on the accumulated ROS in plants, which can participate not only in ROS production pathways, but also in the NADPH production pathway, thus promoting ROS removal.61 It has been reported that sucrose as a signal molecule may play an important role in the activation of antioxidant-related genes, such as CAT.62 Therefore, soluble sugars, especially sucrose, may play multiple important roles in coping with Y2O3 NPs stress at the early stage of a maize seedling.TCA is the major pathway of energy source for life activities, which is the central link for the mutual transformation of different types of organic matters in plants. With the aggravation of Y2O3 NPs stress, the key intermediates of TCA are accumulated significantly, especially pyruvate, citrate, and malate (Table 1). Pyruvate is an important link between the glycolysis pathway and TCA. Malate can maintain the osmotic pressure and charge balance of cells under adversity stress.63 Water stress, mineral nutrient shortage, and salt stress all can induce citrate accumulation in plants.64 Too much ROS can make cis-aconitase irreversibly inactivated under adversity stress, resulting in a decrease in electron flow in the mitochondrial electron-transport chain and a rise of the citrate level, thus alleviating oxidative stress.65 Moreover, citrate, as an allosteric inhibitor of phosphofructokinase, can slow down glycolysis to achieve feedback regulation for plant respiration.66 Therefore, the upregulation of these key intermediates indicated that maize seedlings attempted to accumulate energy and resist the Y2O3 NPs stress at the cost of slowing down glycolysis and TCA, so as to maintain normal physiological processes or to enhance their defense-related activities.25
Free amino acids are produced by the decomposition of proteins and the transformation of sugars in plants, which can be a response to environmental stress directly or indirectly.67 It was found from Table 1 that free amino acids had the strongest responses to Y2O3 NPs stress among all the metabolites. The amino acids showed a dose-dependent increase in response to the stress (Table 1). Proline in all the metabolites was upregulated most at 10 mg L−1 of Y2O3 NPs, whereas alanine upregulated the most at 500 mg L−1 of Y2O3 NPs, indicating that the transformation of glutamic acid to proline was weakened and the pathway of pyruvate to alanine was enhanced with the increase in Y2O3 NPs.68 Both are important organic osmotic regulators in plants. Proline is also an effective antioxidant, which can regulate ROS balance in cells by synergizing with antioxidant enzymes (SOD, POD, and CAT) and non-enzymatic peroxide systems (e.g., ascorbic acid, vitamin E, reducibility glutathione).69 Glutamic acid is the precursor of some characteristic compounds (alanine, GABA) of the anaerobic metabolism.70 Under anoxia stress, a reversible reaction catalyzed by alanine aminotransferase promotes the amino transfer of glutamic acid and pyruvate to produce alpha-ketoglutarate and alanine to maintain carbon and nitrogen balance.71 To a certain extent, this reduces the loss of carbon sources, thus enabling alpha-ketoglutarate to enter TCA and produce ATP through a substrate-level phosphorylation.72 Mustroph et al. also found that alanine accumulated in roots and shoots during anoxia.73 Under anoxia stress, the increased proportion of alanine was significantly higher than that of GABA,74 which was consistent with our results. We speculated that with the increase of Y2O3 NPs, the aerobic respiration of seedlings would decrease, while the anaerobic respiration would increase, resulting in a significant increase in alanine accumulation. Moreover, GABA and asparagine also showed a marked response to Y2O3 NPs stress. GABA, as a stress indicator, is a non-protein amino acid, in which the content in plants is increased by several or even dozens of times upon environmental stresses (e.g., anoxia stress, cold stress, thermal stress, salt stress, drought stress, and mechanical damages).75 It can also relieve ROS damage and increase antioxidant enzyme activity. Asparaginate accumulation may be a direct response to the stress for maintaining osmotic pressure, or an indirect result of restricting protein synthesis.76 In addition, as aromatic amino acids, phenylalanine and tyrosine increased markedly in the presence of Y2O3 NPs compared to CK (Table 1). Both are the precursors of the general phenylpropanoid pathway that produces lots of secondary metabolites based on the few intermediates of the shikimate pathway as the core unit.77 The upregulation of both in maize shoots under Y2O3 NPs stress may serve as an increased supply for phenylpropanoid synthesis, indicating that the plant defense system was activated to scavenge excessive ROS and to protect the cells from damage.25 Therefore, these accumulated free amino acids under Y2O3 NPs stress were involved in osmotic regulation and ROS scavenging to maintain the stability of the proteins and enzymes in maize shoots.78
Formate accumulated significantly in a dose-dependent manner under Y2O3 NPs stress (Table 1). The one-carbon metabolism that formate participates in is essential for plants. Formate provides a C1 unit for the synthesis of nucleotides, mitochondria, chloroplast protein, and methyl, as well as for amino acid metabolism and vitamin metabolism.79 Hence, formate accumulation indicated a suppression of the one-carbon metabolism in maize shoots under Y2O3 stress.
Correlation analysis of the metabolites revealed that there were positive correlations between the main metabolites related with antioxidation and osmotic regulation (Fig. 6). For instance, sucrose was significantly positively correlated with aspartate and glutamine. Glutamine was significantly positively correlated with proline. Alanine presented significantly positive correlations with tryptophan, citrate, and malate. These main metabolites synergistically coped with Y2O3 NPs stress by adjusting the carbohydrate metabolism, TCA, and amino acid synthesis. Moreover, their upregulation also indicated that the synthesis of proteins and other organics may be retarded by Y2O3 NPs stress, leading to the inhibition of seedling growth.
Conclusions
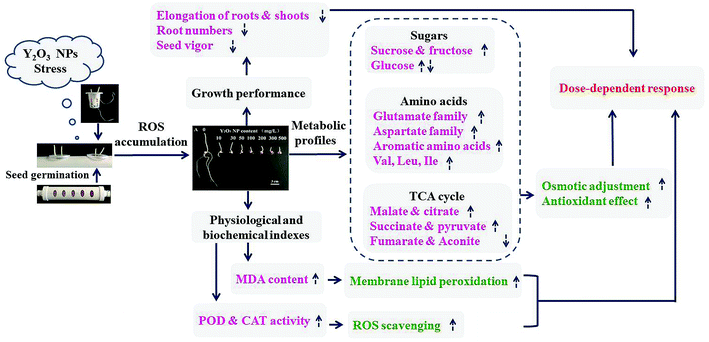
Based on maize germination as well as the physiological and biochemical properties and metabolic profiles of shoots under Y2O3 NPs stress, a hypothetic model for adaptation was proposed (Fig. 7). With the aggravation of Y2O3 NPs stress, seed germination is delayed, seed vitality declined, and the inhibition of seedling growth enhanced, accompanied by the intensification of oxidative damage and osmotic pressure change. Meanwhile, the activities of the antioxidant enzymes (POD and CAT) and the levels of the metabolites related with the antioxidant response and osmotic adjustment showed a dose-dependent increase in response to the stress as a whole. Besides, carbohydrate metabolism, TCA, and amino acid synthesis were the major metabolic pathways in response to the stress. There may be cross-talks between these metabolites and antioxidant gene expression. However, when the stress was aggravated to some extent, the increased antioxidant level could not completely offset the excessive ROS produced by this stress, causing the imbalance of osmotic pressure, the decrease in energy metabolism, and the synthesis hindrance of proteins and other organics in cells. These hindered effects were finally manifested by inhibiting the apparent growth of seedlings. The findings provide an idea to alleviate the toxicity of Y2O3 NPs to plants by adding extra sucrose, amino acids, or simple organic acids (e.g., malate and citrate) into the matrix in the environment.Conflicts of interest
No conflict of interest exits in the submission of this manuscript, and manuscript is approved by all authors for publication.Acknowledgements
This work was supported by National Training Program of Innovation and Entrepreneurship for Undergraduates (No. 201810145206) and the Fundamental Research Funds for the Central Universities (No. N182410001). We would also like to acknowledge Shanghai Sensichip Biotech Co., Ltd for the help in multivariate analysis.References
- R. F. Service, Science, 2003, 300, 243 CrossRef PubMed.
- F. Gottschalk, T. Y. Sun and B. Nowack, Environ. Pollut., 2013, 181, 287–300 CrossRef CAS PubMed.
- C. X. Ma, J. C. White, O. P. Dhankher and B. S. Xing, Environ. Sci. Technol., 2015, 49, 7109–7122 CrossRef CAS PubMed.
- F. Schwab, G. S. Zhai, M. Kern, A. Turner, J. L. Schnoor and M. R. Wiesner, Nanotoxicology, 2016, 10, 257–278 CAS.
- S. L. Gai, C. X. Li, P. P. Yang and J. Lin, Chem. Rev., 2014, 114, 2343–2389 CrossRef CAS PubMed.
- L. Yue, C. Ma, X. Zhan, J. C. White and B. Xing, Environ. Sci.: Nano, 2017, 4, 843–855 RSC.
- P. Eriksson, A. A. Tal, A. Skallberg, C. Brommesson, Z. Hu, R. D. Boyd, W. Olovsson, N. Fairley, I. A. Abrikosov, X. Zhang and K. Uvdal, Sci. Rep., 2018, 8, 6999 CrossRef PubMed.
- R. La Torre Roche, A. Servin, J. Hawthorne, B. Xing, L. A. Newman, X. Ma, G. Chen and J. C. White, Environ. Sci. Technol., 2015, 49, 11866–11874 CrossRef PubMed.
- Y. H. Ma, L. L. Kuang, X. He, W. Bai, Y. Y. Ding, Z. Y. Zhang, Y. L. Zhao and Z. F. Chai, Chemosphere, 2010, 78, 273–279 CrossRef CAS PubMed.
- P. Zhang, Y. H. Ma, Z. Y. Zhang, X. He, Z. Guo, R. Z. Tai, Y. Y. Ding, Y. L. Zhao and Z. F. Chai, Environ. Sci. Technol., 2012, 46, 1834–1841 CrossRef CAS PubMed.
- Q. Wang, X. M. Ma, W. Zhang, H. C. Pei and Y. S. Chen, Metallomics, 2012, 4, 1105–1112 RSC.
- C. M. Rico, M. I. Morales, A. C. Barrios, R. McCreary, J. Hong, W. Y. Lee, J. Nunez, J. R. Peralta-Videa and J. L. Gardea-Torresdey, J. Agric. Food Chem., 2013, 61, 11278–11285 CrossRef CAS PubMed.
- P. Zhang, Y. H. Ma, Z. Y. Zhang, X. He, J. Zhang, Z. Guo, R. Z. Tai, Y. L. Zhao and Z. F. Chai, ACS Nano, 2012, 6, 9943–9950 CrossRef CAS PubMed.
- P. Zhang, Y. Ma, Z. Zhang, X. He, Y. Li, J. Zhang, L. Zheng and Y. Zhao, Nanotoxicology, 2015, 9, 1–8 CrossRef CAS PubMed.
- F. Schwabe, S. Tanner, R. Schulin, A. Rotzetter, W. Starkb, A. von Quadtc and B. Nowack, Metallomics, 2015, 7, 466–477 RSC.
- J. Nordmann, S. Buczka, B. Voß, M. Haasea and K. Mummenhoff, J. Mater. Chem. B, 2015, 3, 144–150 RSC.
- D. J. Oliver, B. Nikolau and E. S. Wurtele, Metab. Eng., 2002, 4, 98–106 CAS.
- O. Fiehn, Plant Mol. Biol., 2002, 48, 155–171 CrossRef CAS PubMed.
- A. A. Keller, A. S. Adeleye, J. R. Conway, K. L. Garner, L. Zhao, G. Cherr, J. Hong, J. L. Gardea-Torresdey, H. Godwin and S. Hanna, NanoImpact, 2017, 7, 28–40 CrossRef.
- L. Zhao, Y. Huang, J. Hu, H. Zhou, A. S. Adeleye and A. A. Keller, Environ. Sci. Technol., 2016, 50, 2000–2010 CrossRef CAS PubMed.
- L. Zhao, Y. Huang, A. S. Adeleye and A. A. Keller, Environ. Sci. Technol., 2017, 51, 10184–10194 CrossRef CAS PubMed.
- L. Zhao, Y. Huang and A. A. Keller, J. Agric. Food Chem., 2018, 66, 6628–6636 CAS.
- R. Marangoni, D. Paris, D. Melck, L. Fulgentini, G. Colombetti and A. Motta, Biophys. J., 2011, 100, 215–224 CrossRef CAS PubMed.
- R. B. Li, Z. X. Ji, C. H. Chang, D. R. Dunphy, X. M. Cai, H. Meng, H. Y. Zhang, B. B. Sun, X. Wang, J. Y. Dong, S. J. Lin, M. Y. Wang, Y. P. Liao, C. J. Brinker, A. Nel and T. Xia, ACS Nano, 2014, 8, 1771–1783 CrossRef CAS PubMed.
- R. Srinivasan, N. R. Yogamalar, J. Elanchezhiyan, R. J Joseyphus and A. C. Bose, J. Alloys Compd., 2010, 496, 472–477 CrossRef CAS.
- Y. Y. Chen, C. Sanchez, Y. Yue, M. D. Almeida, J. M. González, D. Y. Parkinson and H. Liang, J. Nanobiotechnol., 2016, 14, 23 CrossRef PubMed.
- J. Strable and M. J. Scanlon, Cold Spring Harb. Protoc., 2009, 10, pdb.emo132 Search PubMed.
- L. Rajjou, M. Duval, K. Gallardo, J. Catusse, J. Bally, C. Job and D. Job, Annu. Rev. Plant Biol., 2012, 63, 507–533 CrossRef CAS PubMed.
- K. Weitbrecht, K. Müller and G. Leubner-Metzger, J. Exp. Bot., 2011, 62, 3289–3309 CrossRef CAS PubMed.
- X. Y. Wu and T. A. Von, Environ. Pollut., 2002, 116, 37–47 CrossRef PubMed.
- A. Chaoui, S. Mazhoudi, M. H. Ghorbal and E. El Ferjani, Plant Sci., 1997, 127, 139–147 CrossRef CAS.
- C. Sun, X. Gao, J. Fu and J. Zhou, Plant Soil, 2015, 388, 99–117 CrossRef CAS.
- D. Liu, X. Wang, Y. Lin, Z. Chen, H. Xu and L. Wang, Environ. Sci. Pollut. Res., 2012, 19, 3282–3291 CrossRef CAS PubMed.
- D. Cui, P. Zhang, Y. Ma, X. He, Y. Li, J. Zhang, Y. Zhao and Z. Zhang, Environ. Sci.: Nano, 2014, 1, 459–465 RSC.
- J. D. Bewley, K. J. Bradford, H. W. M. Hilorst and H. Nonogaki, Physiology of Development, Germination and Dormancy, Springer, New York, 3rd edn, 2013 Search PubMed.
- D. Lin and B. Xing, Environ. Pollut., 2007, 150, 243–250 CrossRef CAS PubMed.
- M. H. Siddiqui and M. H. Alwhaibi, Saudi J. Biol. Sci., 2014, 21, 13–17 CrossRef CAS PubMed.
- L. C. Woo, M. Shaily, Z. Katherine, D. Li, Y. Tsai, J. Braam and P. J. J. Alvarez, Environ. Toxicol. Chem., 2010, 29, 669–675 CrossRef PubMed.
- V. Aruoja, H. C. Dubourguier, K. Kasemets and A. Kahru, Sci. Total Environ., 2009, 407, 1461–1468 CrossRef CAS PubMed.
- M. Wierzbicka and J. Obidzińska, Plant Sci., 1998, 137, 155–171 CrossRef CAS.
- A. E. Rubio-Casal, J. M. Castillo, C. J. Luque and M. E. Figueroa, J. Arid Environ., 2003, 53, 145–154 CrossRef.
- P. Schopfer, Am. J. Bot., 2006, 93, 1415–1425 Search PubMed.
- H. Zhu, J. Han, J. Q. Xiao and Y. Jin, J. Environ. Monit., 2008, 10, 713–717 RSC.
- S. Lin, J. Reppert, Q. Hu, J. S. Hudson, M. L. Reid, T. A. Ratnikova, A. M. Rao, H. Luo and P. C. Ke, Small, 2009, 5, 1128–1132 CrossRef CAS PubMed.
- L. V. Nhan, C. Ma, Y. Rui, S. Liu, X. Li, B. Xing and L. Liu, Sci. Rep., 2015, 5, 11618 CrossRef CAS PubMed.
- Y. Q. Deng, J. C. White and B. S. Xing, J. Zhejiang Univ., Sci., A, 2014, 15, 552–572 CrossRef CAS.
- G. Zhai, K. S. Walters, D. W. Peate, P. J. J. Alvarez and J. L. Schnoor, Environ. Sci. Technol. Lett., 2014, 1, 146–151 CrossRef CAS PubMed.
- L. Yue, F. Chen, K. Yu, Z. Xiao, X. Yu, Z. Wang and B. Xing, Sci. Total Environ., 2019, 653, 675–683 CrossRef CAS PubMed.
- W. Du, W. Tan, J. R. Peralta-Videa, J. L. Gardea-Torresdey, R. Ji, Y. Yin and H. Guo, Plant Physiol. Biochem., 2017, 110, 210–225 CrossRef CAS PubMed.
- B. Ahmed, M. S. Khan and J. Musarrat, Environ. Pollut., 2018, 240, 802–816 CAS.
- J. T. Li, Z. B. Qiu, X. W. Zhang and L. S. Wang, Acta Physiol. Plant., 2011, 33, 835–842 CrossRef CAS.
- S. Garcia-Sanchez, I. Bernales and S. Cristobal, BMC Genomics, 2015, 16, 341 CrossRef PubMed.
- P. M. G. Nair and I. M. Chung, 3 Biotech, 2017, 7, 293 CrossRef PubMed.
- A. Cox, P. Venkatachalam, S. Sahi and N. Sharma, Plant Physiol. Biochem., 2016, 107, 147–163 CrossRef CAS PubMed.
- M. Tiwari, N. C. Sharma, P. Fleischmann, J. Burbage, P. Venkatachalam and S. V. Sahi, Front. Plant Sci., 2017, 8, 633 CrossRef PubMed.
- T. Boriboonkaset, C. Theerawitaya, N. Yamada, A. Pichakum, K. Supaibulwatana, S. Cha-Um, T. Takabe and C. Kirdmanee, Protoplasma, 2013, 250, 1157–1167 CrossRef CAS PubMed.
- K. E. Koch, Curr. Opin. Plant Biol., 2004, 7, 235–246 CrossRef CAS PubMed.
- J. Buitink, M. A. Hemming and F. A. Hoekstra, Plant Physiol., 2000, 122, 1217–1224 CrossRef CAS PubMed.
- H. Nayyar, T. S. Bains and S. Kumar, Environ. Exp. Bot., 2005, 54, 275–285 CrossRef CAS.
- Z. Liu, J. P. Yan, D. K. Li, Q. Luo, Q. J. Yan, Z. B. Liu, L. M. Ye, J. M. Wang, X. F. Li and Y. Yang, Plant Physiol., 2015, 167, 1659–1670 CrossRef CAS PubMed.
- A. Matros, D. Peshev, M. Peukert, H. P. Mock and W. Van den Ende, Plant J., 2015, 82, 822–839 CrossRef CAS PubMed.
- A. L. Contento, S. J. Kim and D. C. Bassham, Plant Physiol., 2004, 135, 2330–2347 CrossRef CAS PubMed.
- V. Emmerlich, N. Linka, T. Reinhold, M. A. Hurth, M. Traub, E. Martinoia and H. E. Neuhaus, Proc. Natl. Acad. Sci. U. S. A., 2003, 100, 11122–11126 CrossRef CAS PubMed.
- U. Luettge, Plant, Cell Environ., 1990, 13, 977–982 CrossRef CAS.
- D. F. Klessig, J. Duner, R. Noad, D. A. Navarre, D. Wendehenne, D. Kumar, J. M. Zhou, J. Shah, S. Zhang, P. Kachroo, Y. Trifa, D. Pontier, E. Lam and H. Silva, Proc. Natl. Acad. Sci. U. S. A., 2000, 97, 8849–8855 CrossRef CAS PubMed.
- E. Martinoia and D. Rentsch, Annu. Rev. Plant Biol., 1994, 45, 447–467 CrossRef CAS.
- M. Ashraf and P. J. C. Harris, Plant Sci., 2004, 166, 3–16 CrossRef CAS.
- R. Majumdar, B. Barchi, S. A. Turlapati, M. Gagne, R. Minocha, S. Long and S. C. Minocha, Front. Plant Sci., 2016, 7, 78 Search PubMed.
- R. Rusty and R. Regina, Proc. Natl. Acad. Sci. U. S. A., 2005, 102, 3175–3176 CrossRef PubMed.
- R. Reggiani and A. Bertani, Russ. J. Plant Physiol., 2003, 50, 733–736 CrossRef CAS.
- M. Rocha, F. Licausi, W. L. Araujo, A. Nunes-Nesi, L. Sodek, A. R. Fernie and J. T. van Dongen, Plant Physiol., 2010, 152, 1501–1513 CrossRef CAS PubMed.
- J. Bailey-Serres, T. Fukao, D. J. Gibbs, M. J. Holdsworth, S. C. Lee, F. Licausi, P. Perata, L. A. Voesenek and J. T. van Dongen, Trends Plant Sci., 2012, 17, 129–138 CrossRef CAS PubMed.
- A. Mustroph, G. A. Jr Barding, K. A. Kaiser, C. K. Larive and J. Bailey-Serres, Plant, Cell Environ., 2014, 37, 2366–2380 CAS.
- C. A. F. De Sousa and L. Sodek, Environ. Exp. Bot., 2003, 50(1), 1–8 CrossRef CAS.
- A. M. Kinnersley and F. J. Turano, Crit. Rev. Plant Sci., 2000, 19, 479–509 CrossRef CAS.
- P. J. Lea, L. Sodek, M. A. J. Parry, P. R. Shewry and N. G. Halford, Ann. Appl. Biol., 2007, 150, 1–26 CrossRef CAS.
- T. Vogt, Mol. Plant, 2010, 3, 2–20 CrossRef CAS PubMed.
- A. J. Keutgen and E. Pawelzik, Food Chem., 2008, 111, 642–647 CrossRef CAS.
- D. R. Appling, FASEB J., 1991, 5, 2645–2651 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9ra04672k |
| This journal is © The Royal Society of Chemistry 2019 |