Open Access Article
Open Access ArticleIn situ formation of DNA-templated copper nanoparticles as fluorescent indicator for hydroxylamine detection†
Quanwei Song *ab,
Changzhao Chenab,
Wenhe Yuab,
Lixia Yangc,
Kunfeng Zhangab,
Jin Zhengab,
Xianyuan Duab and
Hongkun Chenab
*ab,
Changzhao Chenab,
Wenhe Yuab,
Lixia Yangc,
Kunfeng Zhangab,
Jin Zhengab,
Xianyuan Duab and
Hongkun Chenab
aState Key Laboratory of Petroleum Pollution Control, Beijing 102206, China. E-mail: song_quanwei@163.com; Tel: +86-10-80169547
bCNPC Research Institute of Safety and Environment Technology, Beijing 102206, China
cBeijing Key Laboratory of Metal Material Characterization, Central Iron and Steel Research Institute, Beijing 100081, China
First published on 20th August 2019
Abstract
Herein, we develop a facile method for selective and sensitive detection of hydroxylamine (HA) based on the in situ formation of DNA templated copper nanoparticles (DNA-CuNPs) as fluorescent probes. It is firstly found that HA as a reducing agent can play a key role in the in situ formation of fluorescent DNA-CuNPs. This special optical property of DNA-CuNPs with (λex = 340 nm, λem = 588 nm) with a mega-Stokes shifting (248 nm) makes it applicable for the turn-on detection of HA. In addition, this fluorescent method has several advantages such as being simple, rapid, and environmentally friendly, because it avoids the traditional organic dye molecules and complex procedures. Under optimized conditions, this platform achieves a fluorescent response for HA with a detection limit of 0.022 mM. Especially, successful detection capability in tap waters and ground waters exhibits its potential to be general method.
Introduction
Hydroxylamine (HA), NH2OH, is a potent reducing agent, and an important intermediate or raw material in industrial and pharmaceutical processes.1,2 In cellular metabolism, HA is well known as a nitric oxide (NO) donor, and an inhibitor for the release of insulin.3,4 Given the fact that HA itself is moderately toxic to humans and animals, HA residues in the ecosystem environment should be strictly controlled. The traditional analytical methods for the detection of HA includes acid–base titration, spectrophotometry, polarographic, and electrochemical methods.5,6 The above methods are low-sensitivity and poor reproducibility. In addition, HA after derived can also be analyzed by high-performance liquid chromatography (HPLC) and gas chromatography (GC).7,8 However, their operation requirements are complex and expensive, and their analytical procedures are laborious. Therefore, facile fluorescent strategies for HA sensing will be of great benefit, because fluorescent methods can partly avoid shortages of traditional assays and exhibit promising advantages, including high sensitivity, rapid analysis, and little damage to sample.9Currently, the majority of highly sensitive fluorescent methods frequently require an expensive fluorescent labeling process and complex chemical modification.10 Additionally, these methods usually suffer from inherent limitations, including relatively high toxicity, low emission intensity, and small Stokes shifting.11 To enhance the applicability, it will be desirable for fluorescent sensing systems by using environmental-friendly nano-dyes as alternatives of organic dye molecules.
In the recent years, DNA-templated copper nanoparticles (DNA-CuNPs) has opened up a new avenue and continually bring excellent features for label-free analysis in life science,12,13 thanks to its outstanding optical properties, mild synthesis conditions, good bio-compatibility, and sequence-specific manner. Compared to quantum dots and DNA-stabilized silver nanoclusters, the strong orange-red fluorescent emission with a mega-Stokes shifting (230–270 nm) can be easily observed in approximately 10 minutes under the room temperature by the reduction of ascorbate.14 Interestingly, the specific adenine–thymine (AT) rich regions of double-stranded DNA (dsDNA) or poly thymine can act as efficient templates for the in situ formation of CuNPs.15,16 Although fluorescent DNA-CuNPs are emerging as an attractive tools in label-free sensing systems for versatile analytical bio-applications,17,18 there is still great room for further investigation of its growth and long-term stability.19
In this study, it is firstly found that HA as a new reducing agent can also play a dominant role in the DNA-CuNPs formation. Subsequently, given the importance of HA residues detection in medicine and pesticide, DNA-CuNPs are employed to HA sensing. In our design, the optimal addition amount of HA can lead to fluorescence enhancement in the buffered solutions and real samples under the room temperature. Attractively, to the best of our knowledge, our proposed strategy is the first example of using environmental-friendly nanodyes as fluorescent indicator to detect HA.
Experimental
Materials and instruments
All agents used in this work were of analytical grade and do not require purification. 3-(N-Morpholino) propanesulfonic acid (MOPS), NaCl, MgCl2, CuSO4, ethylenediamine, aniline, sulfanilamide, acrylamide, glutamic acid, and HA hydrochloride were purchased from Sigma-Aldrich Chemical Co., (St, Louis, MO, USA). Solutions were prepared by using ultra-pure water with an electric resistance > 18.2 MΩ based on a Milli-Q water purification system (Millipore, USA). The HPLC-purified and MS-verified oligonucleotides were synthesized and purchased from Shanghai Sangon Biotechnology Co. Ltd. (Beijing, China). The stock solutions of oligonucleotides were prepared in the MOPS buffered solutions (10 mM MOPS, 2 mM MgCl2, 150 mM NaCl, pH 7.5). All the specific AT-rich regions of dsDNA in this work were given in the ESI.†High-resolution transmission electron microscopy (HR-TEM) images of DNA-CuNPs were collected on a JEOL 2100F microscope operating at 200 kV. In this study, all of fluorescence emission measurements were obtained on a Shimadzu RF-5301PC under an excitation wavelength of 340 nm, and both excitation and emission slits were set on 5 nm. Time-resolved fluorescence decay curves were carried out based on an Edinburgh Analytical Instrument OB920 single-photon counting fluorometer with a pulsed microsecond nitrogen lamp as excitation source.
HA assay
HA assay was carried out in the 0.5 mL of MOPS buffered solutions (10 mM MOPS, 2 mM MgCl2, 150 mM NaCl, pH 7.5) and real samples. First, different concentrations of HA were pre-incubated with DNA templates (500 nM) at room temperatures for 1 min. Subsequently, final concentration of Cu2+ (0.2 mM) was introduced, and then the synthesis of fluorescent CuNPs were finished within 10 min incubation time. All of experimental processes can be achieved by the temperature regulating device of PCR machine. Finally, fluorescence signal of this sensing system was recorded by the formation of DNA-CuNPs as judged by fluorescence spectroscopy. Details experimental information were located in the ESI† section.Results and discussion
Feasibility verification of the fluorescent method
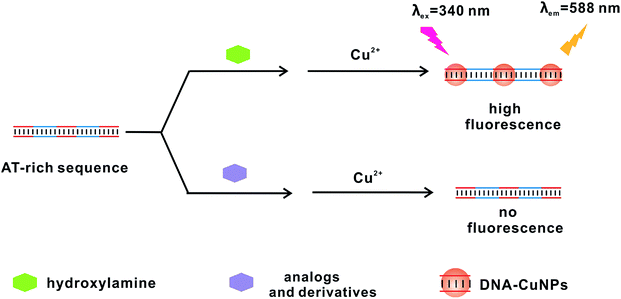
The proposed fluorescent method is shown in Scheme 1. A number of sensing systems exhibit specific dependence on target compounds, which, in turn, provide an efficient fluorescent method for developing selective platforms for these compounds. For example, a label-free and turn-on fluorescent strategy is achieved for the determination of Cu2+ ions based on the poly thymine templated CuNPs by He and co-workers.20 In light of mild reduction performance of HA, we discover that the addition of HA can effectively initiate the in situ formation of DNA-CuNPs, resulting in a strong fluorescence emission. On the contrary, in the absence of HA, fluorescent signal will be hardly observed owing to the lack of reducing agents. Therefore, by virtue of these unique feature, the turn-on sensing platform for the detection of HA will be established by collecting the variation of fluorescence intensity.Characterization of the DNA-CuNPs
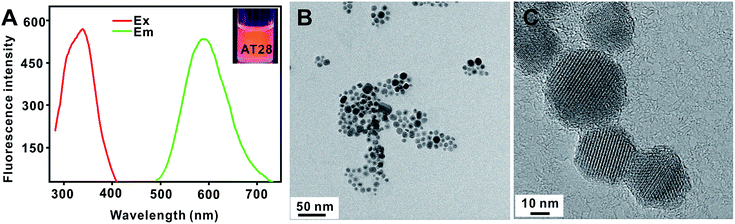
Firstly, in order to confirm the effect of HA for the formation of DNA-CuNPs, AT-rich sequences in MOPS buffered conditions were carefully investigated (10 mM MOPS, 2 mM MgCl2, 150 mM NaCl, pH 7.5), because it was vital to select efficient DNA templates for preparing DNA-CuNPs. In the view of previous reports, fluorescence intensity could be used to evaluate AT-rich sequences as templates for CuNPs formation.16,21 As shown in Fig. 1(A), the AT28-templated CuNPs (AT28-CuNPs) were strongly emissive with excitation (Ex) and emission (Em) bands at 340 nm and 588 nm. As shown in Fig. S1,† with the increase of the length of AT-rich sequences from 16 bp to 28 bp, the fluorescent intensity of DNA-CuNPs was clearly improved under the same conditions. As shown in Fig. 1(B) and (C), further examination by HR-TEM images exhibited that the average size of AT28-CuNPs was 24 ± 8 nm in the presence of 2 mM HA, and the crystal lattice structure indicated that AT28 could support the formation of fluorescent DNA-CuNPs. We could infer that HA was able to facilitate the reduction of Cu2+ ions, and to further accumulate CuNPs from Cu0 clusters localized on the dimension of major groove of dsDNA. Therefore, our results indicated that the role of HA was consistent with ascorbate in the process of the reduction of Cu2+ ions to generate CuNPs.To confirm this speculation, CuNPs stabilized by these AT-rich sequences were identified by the time-correlated single-photon counting (TCSPC) technique. As shown in Fig. S2,† the average fluorescence lifetime of AT16-CuNPs, AT22-CuNPs, AT28-CuNPs was calculated to be approximately 21.2 μs, 31.5 μs, and 55.1 μs, respectively. According to previous reports, electron energy relaxation of fluorescent DNA-CuNPs was size-dependent, and the size of DNA-CuNPs was responsible for its long fluorescence lifetime in the microsecond range.16,22 Hence, in accordance with above results, we could infer that these DNA-CuNPs were also sequence-dependent and size-dependent.
Optimization of the fluorescent method
The above results demonstrated that our proposed fluorescent strategy for HA assay was feasible. Thus, a series of measurements were optimized in this work.Firstly, the addition amount of Cu2+ ions and HA was carefully investigated with the template of AT28, because previous reports indicated that nucleic acid backbone could be gradually damaged by hydroxyl radicals following reducing reactions.14 Fig. S3† suggested that the final concentration of 0.2 mM Cu2+ ions and the final concentration of 2 mM HA were the optimum signal-to-background ratio of fluorescence emission at room temperature. Therefore, these mole radio of Cu2+ ions and HA were finally chosen in following tests.
To improve the accuracy, the stability of fluorescent signal of AT28-CuNPs was further explored. As shown in Fig. S4,† the fluorescence emission maximum of CuNPs (λex = 340 nm, λem = 588 nm) could be observed at incubation time of 10 min, and then the fluorescent signal could maintain 40 min at least, which suggested that AT-rich sequences can provide advantageous microenvironments for the formation of fluorescent CuNPs. Hence, DNA-CuNPs by using HA reduction systems had the relative longer-term stability, which was not so far different from using ascorbate.23,24
Finally, in the view that the formation of DNA-CuNPs was strongly sequence-dependent and length-dependent, the relatively longer AT-rich templates with the length of 34 and 40 bp were also studied. As shown in Fig. S5,† the trend of fluorescence enhancement became slow when the length of AT pairs reached 28. Although the results indicated that the longer AT-rich templates might probably provide a relatively higher sensitivity and broader dynamic range, taking into consideration of the balance between fluorescence enhancement and cost efficiency, the AT28 was selected as the most suitable substrate DNA for the following sensing methods.
Detection performance of the fluorescent method
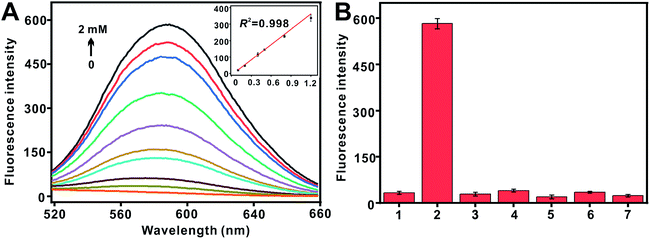
In order to evaluate the sensitivity of the proposed analysis method, an increasing amount of HA from 0 to 2 mM were introduced under the optimized conditions. As shown in Fig. 2(A), the fluorescence intensity gradually increased when the concentrations of HA was raised, thus suggesting that the formation of CuNPs was highly rely on the additive concentration of HA. The good linear range from 0.1 to 1.2 mM with an R2 of 0.998 was obtained. The detection limit was determined to be 0.022 mM (3σ/slope, where σ was the standard deviation in blank samples, n = 11). Thus, our proposed fluorescent assay was relative sensitive among a group of reported traditional methods by derivative reaction, and the detailed comparison was shown in Table S2.†For an fluorescent method, selectivity was also an important matter. As the proof-of-concept, the selectivity of this sensing strategy toward HA was explored by comparing with its several analogs and derivatives, including ethylenediamine, aniline, sulfanilamide, acrylamide, and glutamic acid. As shown in Fig. 2(B), only the solution including HA offered a bright fluorescent emission, and however the others exhibited almost few emission even when present at higher concentrations (10 mM) compared to HA (2 mM). The increase of fluorescence emission suggested that HA could specifically make the in situ formation of DNA-CuNPs, which caused that it was difficult to exclude the interference from other reducing agents, especially ascorbate. Hence, DNA-CuNPs could be as a strong application potential for fluorescence probe for the sensitive and selective determination of HA in its its analogs and derivatives.
Application of the fluorescent strategy
For evaluating the practicality of this strategy, standard recovery experiments were performed in the tap water, the ground water, and the river water. As listed in Table S1,† the average recovery range was between 95.9%, 93.8%, and 95.6%. Besides, the relative standard deviation (RSD) was generally preferable. Thus, there was little inference from substrates in environmental samples, and the capability of our proposed approach to the HA analysis was feasible.It could be concluded that this ideal analytical performance may depend on these factors: (i) the fluorescence emission of AT28-CuNPs (λex = 340 nm, λem = 588 nm) with a mega-Stokes shifting (248 nm) was successfully eliminate the stronger background interference in optical analysis to some extent; (ii) this design of label-free strategy offered an efficient platform for the in situ formation of DNA-CuNPs; (iii) this turn-on method for the detection of HA effectively decreased the false positive possibilities.
Conclusion
In conclusion, we firstly found that HA could be efficient in creating the in situ formation of fluorescent DNA-CuNPs with (λex = 340 nm, λem = 588 nm) with a mega-Stokes shifting (248 nm). Subsequently, a turn-on fluorescent strategy for highly sensitive and selective detection of HA was developed. AT-rich sequence templated CuNPs as fluorescent probe were more environmental-friendly and low-cost, and the homogenous “mix-and-detect” fashion contributed to more easily accessible fluorescent emission signal. In addition, the good detection performance showed potential applications in complex environmental samples. As a part of our interest, our future work will focus on the further exploration of long-term stability of DNA-CuNPs and their applications in environmental research.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors gratefully acknowledge the support from the CNPC Key Technologies for Safety and Environmental Protection Funds (Grant number 2019D-4624, 2018D-4616, 2017D-4613), and the National Science and Technology Major Project (Grant number 2016ZX05040002-001-004).Notes and references
- M. Kumasaki, Y. Fujimoto and T. Ando, J. Loss Prev. Process Ind., 2003, 16, 507 CrossRef.
- Y. Geng, E. Ko, V. Tran, W. S. Chung, C. H. Park, M. K. Kim, G. H. Jin and G. H. Seong, Anal. Sci., 2017, 9, 993 CrossRef PubMed.
- M. H. Antoine, R. Ouedraogo, J. Sergooris, M. Hermann, A. Herchuelz and P. Lebrun, Eur. J. Pharmacol., 1996, 313, 229 CrossRef CAS PubMed.
- P. G. Wang, M. Xian, X. P. Tang, X. J. Wu, Z. Wen, T. W. Cai and A. J. Janczuk, Chem. Rev., 2002, 102, 1091 CrossRef CAS PubMed.
- A. C. Sedgwick, R. Chapman, J. E. Gardiner, L. R. Peacock, G. Kim, J. Yoon, S. D. Bull and T. D. James, Chem. Commun., 2017, 75, 10441 RSC.
- C. Zhang, G. Wang, M. Liu, Y. Feng, Z. Zhang and B. Fang, Electrochim. Acta, 2010, 55, 2835 CrossRef CAS.
- W. D. Korte, J. Chromatogr., 1992, 603, 145 CrossRef CAS.
- Y. Seike, R. Fukumori, Y. Senga, H. Oka, K. Fujinaga and M. Okumura, Anal. Sci., 2004, 20, 139 CrossRef CAS PubMed.
- S. K. Krishnan, E. Singh, P. Singh, M. Meyyappan and H. S. Nalwa, RSC Adv., 2019, 16, 423 Search PubMed.
- L. Luo, F. Xu, H. Shi, X. He, T. Qing, Y. Lei, J. Tang, D. He and K. Wang, Talanta, 2017, 169, 57 CrossRef CAS PubMed.
- Z. Qing, X. He, T. Qing, K. Wang, H. Shi, D. He, Z. Zou, L. Yan, F. Xu, X. Ye and Z. Mao, Anal. Chem., 2013, 85, 12138 CrossRef CAS PubMed.
- R. Liu, C. Wang, J. Hu, Y. Su and Y. Lv, TrAC, Trends Anal. Chem., 2018, 105, 436 CrossRef CAS.
- Z. Qing, A. Bai, S. Xing, Z. Zou, X. He, K. Wang and R. Yang, Biosens. Bioelectron., 2019, 137, 96 CrossRef CAS PubMed.
- A. Rotaru, S. Dutta, E. Jentzsch, K. Gothelf and A. Mokhir, Angew. Chem., Int. Ed., 2010, 49, 5665 CrossRef CAS PubMed.
- Z. Qing, X. He, D. He, K. Wang, F. Xu, T. Qing and X. Yang, Angew. Chem., Int. Ed., 2013, 52, 9719 CrossRef CAS PubMed.
- Q. Song, S. Yu, D. He, S. Xu and J. Ouyang, Chem.–Eur. J., 2015, 21, 2417 CrossRef CAS PubMed.
- H. Wang, H. Zhang, Y. Chen and Y. Liu, Biosens. Bioelectron., 2015, 74, 581 CrossRef CAS PubMed.
- H. Wang, H. Zhang, Y. Chen, Y. Li and T. Gan, RSC Adv., 2015, 5, 77906 RSC.
- Y. Guo, F. Cao, X. Lei, L. Mang, S. Cheng and J. Song, Nanoscale, 2016, 8, 4852 RSC.
- Z. Qing, L. Zhu, S. Yang, Z. Cao, X. He, K. Wang and R. Yang, Biosens. Bioelectron., 2016, 78, 471 CrossRef CAS PubMed.
- Q. Song, L. Yang, H. Chen, R. Zhang, N. Na and J. Ouyang, Talanta, 2017, 174, 444 CrossRef CAS PubMed.
- T. Ye, C. Y. Li, S. Chen, X. H. Ji, J. Zheng, P. Tinnefeld and Z. K. He, Chem. Commun., 2015, 51, 8644 RSC.
- H. Wang, Y. Chen, Y. Li, H. Zhang and J. Cao, RSC Adv., 2015, 5, 94099 RSC.
- H. Wang, H. Zhang, Y. Chen, K. Huang and Y. Liu, Sens. Actuators, B, 2015, 220, 146 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9ra04476k |
| This journal is © The Royal Society of Chemistry 2019 |