Open Access Article
Open Access ArticleModulation of JNK-1/ β-catenin signaling by Lactobacillus casei, inulin and their combination in 1,2-dimethylhydrazine-induced colon cancer in mice†
Mohammed S. Alia,
Rasha M. Hussein *ab,
Yasser Gaber
*ab,
Yasser Gaber bc,
Olfat A. Hammamd and
Mohamed A. Kandeile
bc,
Olfat A. Hammamd and
Mohamed A. Kandeile
aDepartment of Biochemistry, Faculty of Pharmacy, Beni-Suef University, Salah Salem Street, 62514, Beni-Suef, Egypt. E-mail: rasha.hussein@pharm.bsu.edu.eg; Tel: 002 1200136515
bDepartment of Pharmaceutics and Pharmaceutical Technology, College of Pharmacy, Mutah University, 61710, Al-Karak, Jordan
cDepartment of Microbiology and Immunology, Faculty of Pharmacy, Beni-Suef University, 62514, Beni-Suef, Egypt
dPathology Department, Theodor Bilharz Research Institute, 12411 Giza, Egypt
eDepartment of Biochemistry, Faculty of Veterinary Medicine, Beni-Suef University, Egypt
First published on 17th September 2019
Abstract
Colon cancer is a complex disease that involves numerous genetic alterations that change the normal colonic mucosa into invasive adenocarcinoma. In the current study, the protective effects of inulin (prebiotic), Lactobacillus casei (L. casei, probiotic) and their combination (synbiotic) on 1,2-dimethylhydrazine (DMH)-induced colon cancer in male Swiss mice were evaluated. Animals were divided into: Control group, DMH-treated group, DMH plus inulin, DMH plus L. casei and DMH plus inulin plus L. casei-treated groups. Fecal microbiome analysis, biochemical measurements, histopathological examination of the colon tissues, immunostaining and Western blotting analysis of β-catenin, GSK3β and JNK-1 were performed. The prebiotic-, probiotic- and synbiotic-treated groups showed decreased levels of carcinoembryonic antigen and a lower number of aberrant crypt foci compared to the DMH-treated group with the synbiotic group exhibiting a superior effect. Furthermore, all treatments showed a body weight-reducing effect. Administration of inulin, L. casei or their combination increased the expression level of phospho-JNK-1 while they decreased the expression level of β-catenin and phospho-GSK3β. Remarkably, L. casei treatment resulted in enrichment of certain beneficial bacterial genera i.e. Akkermansia and Turicibacter. Therefore, administration of L. casei and inulin as a synbiotic combination protects against colon cancer in mice.
1. Introduction
Colon cancer is the second cause of cancer-related deaths.1 It is a complex process which involves numerous genetic mutations that eventually change normal colonic mucosa into invasive adenocarcinoma.1,2 Colon cancer is caused by many predisposing factors such as microbial exposure, imbalanced diet, inflammation, intestinal epithelial damage and, importantly, normal gut microbiota imbalance.3–5 1,2-Dimethylhydrazine (DMH) is a chemical compound widely used to induce colon cancer in experimental animals since it shows high pathological similarity with human colon cancer.6,7Probiotics are live microorganisms that have beneficial effects to the host such as immunomodulation and improvement of gut microbiota through improving gut homeostasis and alleviating the inflammation caused by pathogenic strains.8–11 Interestingly, the administration of probiotics is increasingly used to prevent various human diseases. For example, it was found that administration of a probiotic mixture could prevent the conversion of liver cirrhosis to hepatocellular carcinoma.12 Also, a mixture of Bifidobacterium and Lactobacillus species could alleviate chronic kidney failure by reducing the plasma levels of urea, indoxyl sulfate and p-cresol.13 More relevant to this study, probiotics were found to inhibit the development of colon cancers through inducing apoptosis and modulating the cellular defensive mechanisms.14
Prebiotics are bioactive substances, fermented by the normal gut microflora to beneficial products such as butyrate and propionate which inhibit colon cancer biomarkers.15 Inulin is one of the prebiotics which contains a polymer of fructooligosaccharides linked together by β-(2-1) bond and it previously showed an important role in decreasing the pre-cancerous lesions of colon cancer.16
Interestingly, a combination of probiotic and prebiotic is known as synbiotic and it provides a synergistic effect that improves growth of the beneficial microbiome along with supplying new ones into the colon.17 Therefore, administration of synbiotics can restore the normal microbiome balance, immunity and consequently reduce the severity of cancer lesions.18,19
β-Catenin signaling plays an important role in colon carcinogenesis and its inhibition is viewed as a good target for the action of many chemotherapeutic drugs.20 More than 90% of the inherited colon cancer cases showed a mutation in the canonical Wnt/β-catenin signaling pathway.21 Importantly, JNK-1 is a member of Mitogen activated protein kinase (MAPK) family and it negatively regulates the β-catenin expression through its effect on glycogen synthase kinase 3 beta (GSK3β) phosphorylation.22 Therefore, it was found that some drugs such as sulindac and Pizotifen combat the prostate and colon cancer respectively via decreasing β-catenin expression levels.23,24
In the current study, we investigated the possible protective effects of inulin (prebiotic), Lactobacillus casei (probiotic), and their combination (synbiotic) on DMH-induced colon cancer in male Swiss mice and the role of JNK-1/β-catenin signaling pathway in this process.
2. Materials and methods
2.1. Chemicals
1,2-Dimethylhydrazine dihydrochloride (C2H10Cl2N2) and inulin from chicory were purchased from Acros Organics, Thermo Fisher Scientific, Belgium. Lactobacillus casei DSM 20011 (ATCC 393) was purchased from the German collection of microorganisms and cell cultures (DSMZ) Germany. MRS Broth was purchased from the Foreign Trade Association, Egypt.2.2. Animals
Forty male Swiss mice (6 weeks old, weighing about 25 g) were obtained from Animal House Colony, Pharmacology and Chemistry Research Center, Misr University and Technology Park, Egypt. Mice were kept in plastic cages (8 mice per cage) under the following standard conditions: 12 h light/dark cycles, 25 ± 2 °C temperature, and humidity 48 ± 10% at the Animal House Laboratory at Faculty of Pharmacy, Beni-Suef University. Mice had free access to water and standard pellet diet composed of 14.5% protein, 4.8% fat and 59.4% carbohydrates. All the experimental procedures performed in this study were in agreement with the guidelines of National Institutes of Health guide for use and care of laboratory animals revised in 1978. The study protocol was approved by the Animal Ethics Committee at Beni-Suef University (Approval number: 018-56).2.3. Bacterial culture and viable counting
Lactobacillus casei DSM 20011 were grown on MRS media adjusted to pH 5.5 and incubated at 30 °C. The grown cells were collected under cooling centrifugation at 1700 × g for 15 min. The cells were washed with saline and resuspended in saline to a final a concentration of 2 × 109 CFU/0.3 ml. Optical turbidity was recorded at wavelength 600 nm as a measure of the concentration of bacterial suspension. A calibration curve was constructed between the bacterial optical density and the viable counting to facilitate dose adjustment.2.4. In vivo study
Control (untreated) group. Mice received SC. injections of 1 mM EDTA, pH 6.5 once per week for 20 weeks.
DMH-treated group. Mice received SC. injections of DMH as described above.
DMH + inulin (prebiotic) treated group. Mice received SC. injections of DMH as DMH-treated group plus inulin (5 mg/0.3 ml/animal) by oral gavage daily from the start of the experiment till animal sacrifice. The dose of inulin was selected based on the previously published study.17
DMH + L. casei (probiotic) treated group. Mice received SC. injections of DMH as DMH-group plus L. casei (2 × 109 CFU/0.3 ml per animal) 3 times per week by oral gavage from the start of the experiment till animal sacrifice. The selected bacterial concentration was based on ref. 25.
DMH + inulin + L. casei (synbiotic) treated group. Mice received SC. injections of DMH as DMH-group plus inulin (5 mg/0.3 ml per animal) via oral gavage daily plus L. casei (2 × 109 CFU/0.3 ml per animal) 3 times per week by oral gavage from the start of the experiment till animal sacrifice.
After 24 weeks, animals were overnight fasting, and blood samples were collected in the morning in heparinized tubes. Plasma was separated by centrifugation at 3000 rpm for 20 min and stored at −80 °C for biochemical measurements. Animals were then anesthetized by diethyl ether and killed by cervical decapitation. The colons were dissected, opened longitudinally, washed with saline and dried on filter paper. A small part of the distal colon was excised and immersed into a protease inhibitor buffer and stored at −80 °C for Western blotting analysis. The remaining part of the colon was kept in 10% buffered formalin solution for histopathology and immunostaining analysis.
2.5. Biochemical measurement in plasma
The concentrations of glucose, triglyceride and total cholesterol were measured in the plasma samples by commercial kits (glucose – TR, triglycerides – LQ and cholesterol assay kits, SPINREACT, Spain respectively) according to the provided instructions.2.6. Histopathological examination
The number of goblet cells was quantified in the colonic tissues using a computer controlled microscope (Zeiss Axioscope microscope, Germany) and the image analysis was performed using the software program Axio vision 4.8 which allows for the determination of the colored compound in the goblet cells stained with methylene blue. The number of goblet cells was analyzed for each colon/group under a light microscope at 400×.
2.7. Immunohistochemical analysis of the carcinoembryonic antigen (CEA)
Immunostaining of the colon tissues with CEA was performed as follows: the slides were deparaffinized, hydrated, blocked and then stained by CEA monoclonal antibody solution (sc-55547, Santa Cruz Biotechnology, Inc, USA) 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50 dilution for 20 min. The slides were washed followed by applying the secondary antibody; HRP Envision kit, DAKO, Agilent, US (no dilution) for 20 min and applying 3,3′-diaminobenzidine (DAB, Sigma Fast DAB tablets, Sigma-Aldrich, St. Louis MO) for 20 min as previously mentioned.31 The slides were finally counterstained with EnVision FLEX hematoxylin for 5 min, dehydrated, cleared and covered by a slip. The colonic sections were examined using a Zeiss light microscope (Oberkochen, Germany). The brownish-color was considered as a positive expression of CEA in the cells. Immunostaining expression was classified according to its distribution into either cytoplasmic type, in which immunostaining was homogeneously distributed in the cytoplasm of neoplastic cells or an apical type in which, there was predominant immunostaining on the cytoplasmic edge closest to the lumen of the neoplastic tissue cells.32 The positively stained cells were recorded within ten successive fields in each sample and quantified using Leica application suite for image analysis at 400× magnification.
50 dilution for 20 min. The slides were washed followed by applying the secondary antibody; HRP Envision kit, DAKO, Agilent, US (no dilution) for 20 min and applying 3,3′-diaminobenzidine (DAB, Sigma Fast DAB tablets, Sigma-Aldrich, St. Louis MO) for 20 min as previously mentioned.31 The slides were finally counterstained with EnVision FLEX hematoxylin for 5 min, dehydrated, cleared and covered by a slip. The colonic sections were examined using a Zeiss light microscope (Oberkochen, Germany). The brownish-color was considered as a positive expression of CEA in the cells. Immunostaining expression was classified according to its distribution into either cytoplasmic type, in which immunostaining was homogeneously distributed in the cytoplasm of neoplastic cells or an apical type in which, there was predominant immunostaining on the cytoplasmic edge closest to the lumen of the neoplastic tissue cells.32 The positively stained cells were recorded within ten successive fields in each sample and quantified using Leica application suite for image analysis at 400× magnification.
2.8. Western blotting analysis
Thirty mg of each colon tissue sample was homogenized in 400 μl ice-cold RIPA buffer. The homogenate was put on a pre-cooled microcentrifuge tube and agitated at 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g for 30 min at 4 °C then spun for 20 min. The supernatant was then collected, and the protein concentration was determined by Bradford assay. 20 μg of protein was mixed with an equal volume of Laemmli buffer and boiled for 5 min at 95 °C to run SDS–PAGE at 100–150 V for 1 h. The proteins were then transferred from the gel to PVDF membrane at 25 V. The blots were blocked with 3% BSA in TBST for 1 h at room temperature. The blots were incubated separately overnight at 4 °C with the following primary antibodies:
000 × g for 30 min at 4 °C then spun for 20 min. The supernatant was then collected, and the protein concentration was determined by Bradford assay. 20 μg of protein was mixed with an equal volume of Laemmli buffer and boiled for 5 min at 95 °C to run SDS–PAGE at 100–150 V for 1 h. The proteins were then transferred from the gel to PVDF membrane at 25 V. The blots were blocked with 3% BSA in TBST for 1 h at room temperature. The blots were incubated separately overnight at 4 °C with the following primary antibodies:
P-JNK monoclonal antibody (G-7: sc-6254, Santa Cruz Biotechnology, Inc, Oregon, USA), β-catenin polyclonal antibody (H-102: sc-7199, Santa Cruz Biotechnology, Inc, Oregon, USA), P-GSK-3β polyclonal antibody (Ser 9: sc-11757, Santa Cruz Biotechnology, Inc, Oregon, USA) (dilution 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500). The membranes were washed thoroughly and incubated with the secondary conjugated antibody poly HRP anti-rabbit IgG (31460, Thermo Fisher Scientific, USA) (dilution 1
500). The membranes were washed thoroughly and incubated with the secondary conjugated antibody poly HRP anti-rabbit IgG (31460, Thermo Fisher Scientific, USA) (dilution 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200) for 1 h at room temp. The membranes were re-washed and the signal was detected by a chemiluminescent substrate, Thermo Fisher Scientific, USA. The images were captured using a CCD camera-based imager and the band intensity of each protein was determined using Image J analysis software (Bitplane, USA).
200) for 1 h at room temp. The membranes were re-washed and the signal was detected by a chemiluminescent substrate, Thermo Fisher Scientific, USA. The images were captured using a CCD camera-based imager and the band intensity of each protein was determined using Image J analysis software (Bitplane, USA).
2.9. Microbiome determination and analysis
Fecal pellets were collected from each animal, the collected samples within the same group were, pooled and kept at −80 °C freezer until DNA extraction.33,34 The DNA extraction was done using the homogenized pooled samples and using the GeneJet Genomic DNA purification Kit (Thermo Scientific, cat. no. K0721). The DNA concentration and quality were checked using PicoGreen, Qubit® fluorimeter.2.10. Statistical analysis
SPSS software version 22 (SPSS Inc., Chicago, Illinois) was used for the statistical analysis of the results. One way analysis of variance (ANOVA) followed by post hoc Tukey's test was used to examine for the statistical significance among groups. A p-value < 0.05 was considered to be statistically significant. Data were represented as mean ± SE.3. Results
3.1. Effect of pre-, pro- and synbiotic treatments on the mice body weight
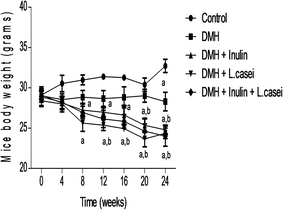
We followed the change in the mice body weight throughout the experimental period to evaluate the possible effects of our studied pre-, pro- and synbiotic treatments. The results showed a significant difference between the body weight of the control group and the DMH-treated group only at week 24 of the experiment at which DMH administration significantly decreased the mice body weight (p < 0.01). Interestingly, administration of prebiotic decreased the body weight significantly at weeks 8, 12, 16, 20 and 24 compared to the control group (p < 0.01) and at weeks 20 and 24 compared to the DMH group at p < 0.01. In addition, administration of probiotic decreased the body weight significantly at weeks 8,12,16, 20 and 24 compared to the control group (p < 0.01) and at weeks 12, 16, 20 and 24 compared to DMH group (p < 0.01). Similarly, the mice body weights of the synbiotic were decreased significantly at weeks 8, 12, 16, 20 and 24 compared to the control group and at weeks 20 and 24 compared to the DMH group (p < 0.01). These results indicate that administration of the prebiotic, probiotic or both treatments has a prominent body weight reducing effect (Fig. 1).3.2. Effect of pre-, pro- and synbiotic treatments on the biochemical measurements
To explain the mechanism by which our treatments reduced the animals' body weights, we measured the blood glucose, triglycerides and total cholesterol levels at the end of the experiment. The results showed no significant difference in the blood glucose, total cholesterol or triglycerides concentrations between the DMH-treated and the control groups. However, the blood glucose and total cholesterol levels were significantly reduced in the prebiotic, probiotic and synbiotic treated groups compared to the control or DMH-treated groups at p < 0.05. In addition, the blood triglycerides level was reduced significantly in the symbiotic-treated group compared to both the control and DMH-treated groups at p < 0.05 (Table 1). These results indicate the potential hypoglycemic and the lipid-lowering effect of our treatments that ultimately lead to a reduction in body weight.| Blood glucose (mg dl−1) | Total cholesterol (mg dl−1) | Triglycerides (mg dl−1) | |
|---|---|---|---|
| a Significantly different from the control group (p < 0.05).b Significantly different from the control group (p < 0.01).c Significantly different from the DMH group (p < 0.05). | |||
| Control | 108.83 ± 22.80 | 103.22 ± 15.51 | 86.33 ± 18.57 |
| DMH | 85.01 ± 20.78 | 92.52 ± 14.98 | 87.42 ± 28.01 |
| DMH + inulin | 58.12 ± 23.67b | 66.44 ± 27.78a | 53.87 ± 16.06 |
| DMH + L. casei | 58.66 ± 34.04b | 60.88 ± 24.18a,c | 72.12 ± 35.23 |
| DMH + inulin + L. casei | 53.71 ± 21.19b | 56.28 ± 21.95b,c | 46.42 ± 10.58a,c |
3.3. Effect of pre-, pro- and synbiotic treatments on the colon macroscopic and microscopic architecture
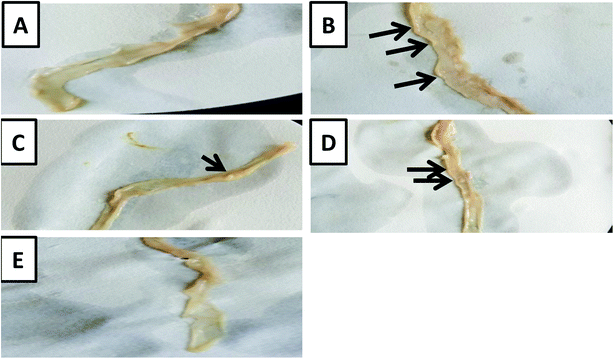
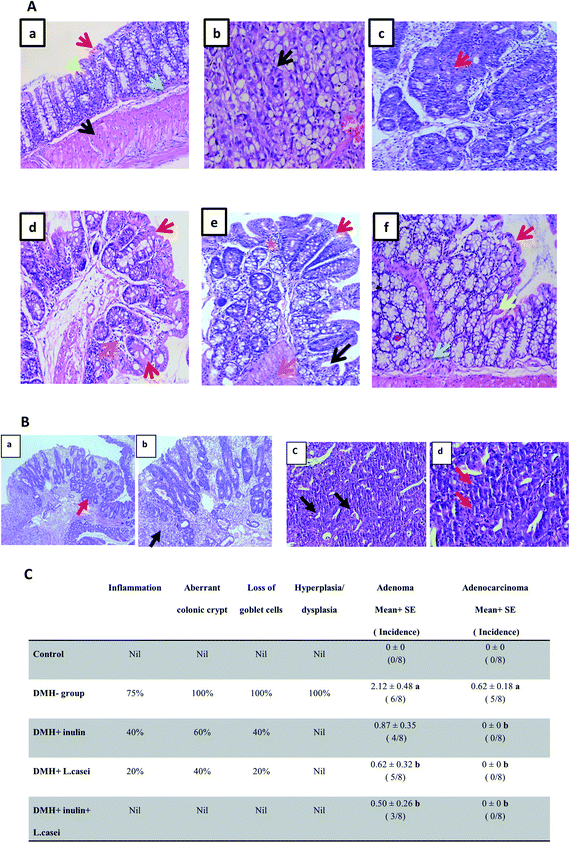
The macroscopic examination of the entire colon showed a formation of multi-plaque lesions in the DMH-treated group compared to the control group. However, there was an apparent decrease in the number of plaque lesions in the prebiotic treated group and probiotic treated group with a complete absence of plaque lesions in the synbiotic-treated group (Fig. 2).Further examination of the colon tissues at the microscopic scale using H&E staining showed that 75% of the examined samples of DMH-treated group exhibited signs of both inflammation and adenoma development compared to the control group. Moreover, 100% of the examined samples of DMH-treated group exhibited severe loss of goblet cells, aberrant colonic crypt, hyperplasia, and dysplasia compared to the control group. 62.5% of the DMH group showed adenocarcinoma. Remarkably, the prebiotic treated group showed a decrease in the degree of inflammation and loss of the goblet cells to 40%, in addition, the formation of aberrant colonic crypts decreased to 60% with no signs of dysplasia, hyperplasia or adenocarcinoma. Similar observations were recorded in the case of probiotic-treated group where the signs of inflammation and loss of goblet cells decreased to 20%, the aberrant colonic crypts were reduced to 40% with no signs of dysplasia or adenocarcinoma. In the synbiotic treated group, the results showed no signs of inflammation, aberrant colonic crypt, loss of the goblet cells, dysplasia or adenocarcinoma (Fig. 3A–C).
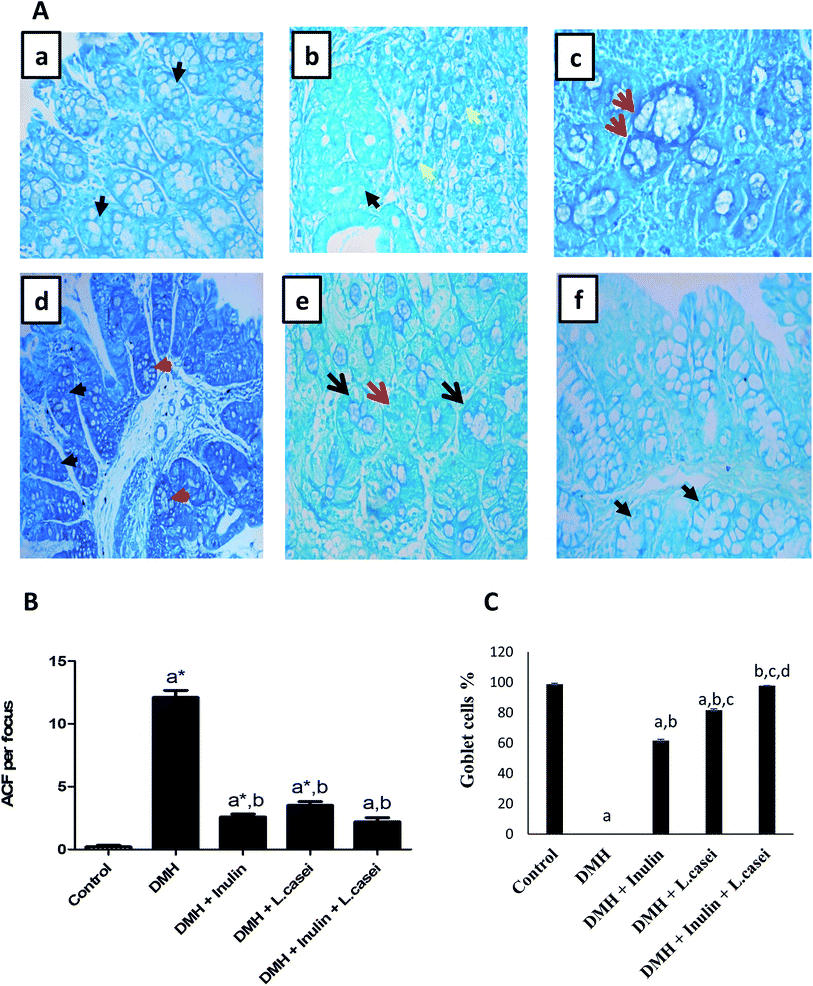
Interestingly, estimating the number of ACF in each colon/group by methylene blue staining revealed that, in the control group, there were normal crypts without ACF formation. Although the DMH treated group showed an increased number of ACF that reached 12.1 ± 0.56 ACF per focus. Importantly, administration of probiotic, prebiotic and synbiotic significantly decreased the number of ACF by three, five and six times respectively compared to the DMH-treated group (p < 0.001) (Fig. 4A and B). Additionally, the percentages of goblet cells in pre-, pro- and synbiotic groups were estimated as 61.7%, 81.7% and 97.7% respectively compared with the DMH group that showed 0% of goblet cells (Fig. 4C).
3.4. Effect of pre-, pro- and synbiotic treatments on CEA expression
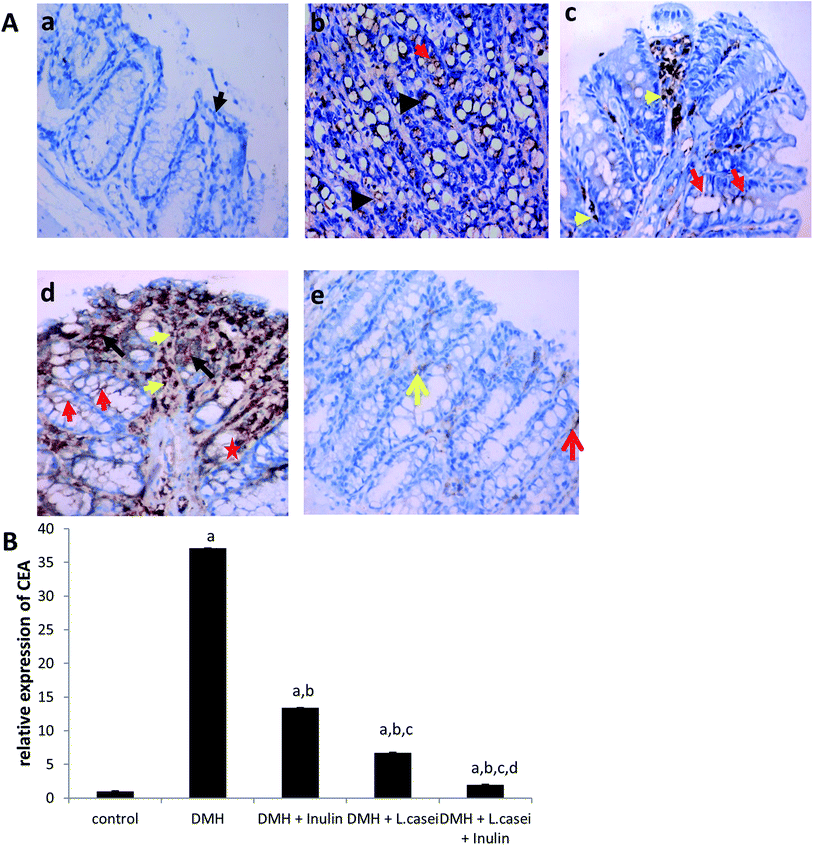
To confirm the histopathology findings, we immuno-stained the colon tissues with CEA antibody. Our results showed an increased expression of CEA in the DMH-treated group by 37 folds compared to the control group (p < 0.001). However, the prebiotic, the probiotic and synbiotic groups showed a significant reduction in the CEA expression estimated by six, three and 19 folds respectively, compared to the DMH-treated group at p < 0.001 (Fig. 5A and B).3.5. Effect of pre-, pro- and synbiotic treatments on the expression of p-JNK-1, p-GSK3-β and β-catenin proteins
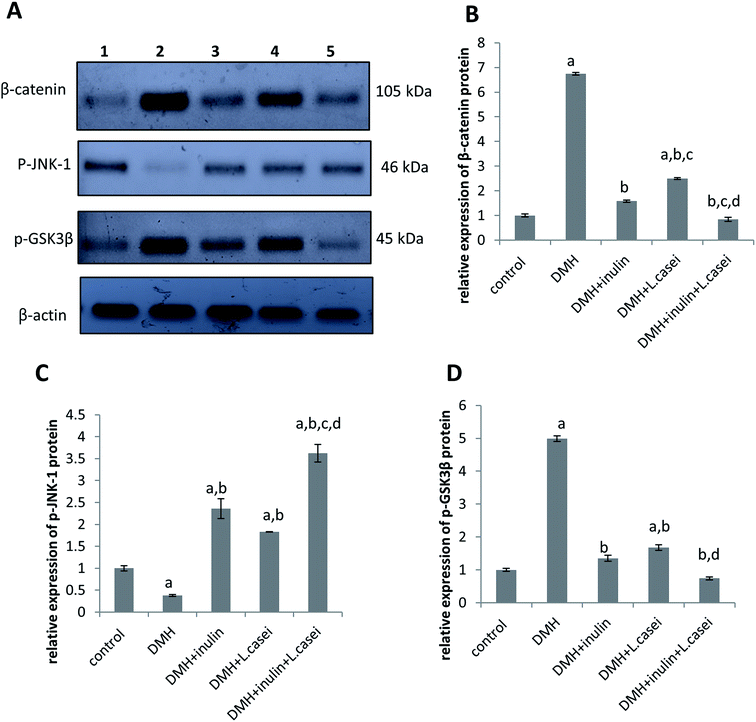
To identify the molecular pathway targeted by our treatments that may explain their protective effects against colon carcinogenesis, we measured the protein expression level of β-catenin, the master gene in regulating the colon cancer development, with its regulatory proteins; p-JNK-1, and p-GSK3β. The findings demonstrated that DMH-treated group showed a significant decrease in the expression of p-JNK-1 by two and a half folds but an increase in the expression of β-catenin and p-GSK3β proteins by seven and five folds, respectively compared to the control group at p < 0.05 (Fig. 6). Interestingly, administration of prebiotic resulted in a significant increase in the expression level of p-JNK-1 protein by six folds while decreasing the expression level of both β-catenin and p-GSK3β by four folds compared to the DMH-treated group at p < 0.05. In the same way, administration of probiotic increased the expression level of p-JNK-1 protein by five folds while decreasing the expression of β-catenin and p-GSK3β by three folds compared to the DMH-treated group at p < 0.05. Interestingly, the synbiotic-treated group showed an increased p-JNK-1 expression level by nine folds with a decreased expression of β-catenin and p-GSK3β proteins by eight and seven folds respectively compared to the DMH-treated group at p < 0.05 (Fig. 6).3.6. Microbiome analysis
High-throughput sequencing resulted in 585![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 554 reads, average 117
554 reads, average 117![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 110 reads per sample, more detailed results are found in ESI Tables S1–S7.† The data filtration resulted in reduction in the reads count, the highest reads count recorded was 114
110 reads per sample, more detailed results are found in ESI Tables S1–S7.† The data filtration resulted in reduction in the reads count, the highest reads count recorded was 114![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 847 in DMH treated group, while the lowest reads count was 57
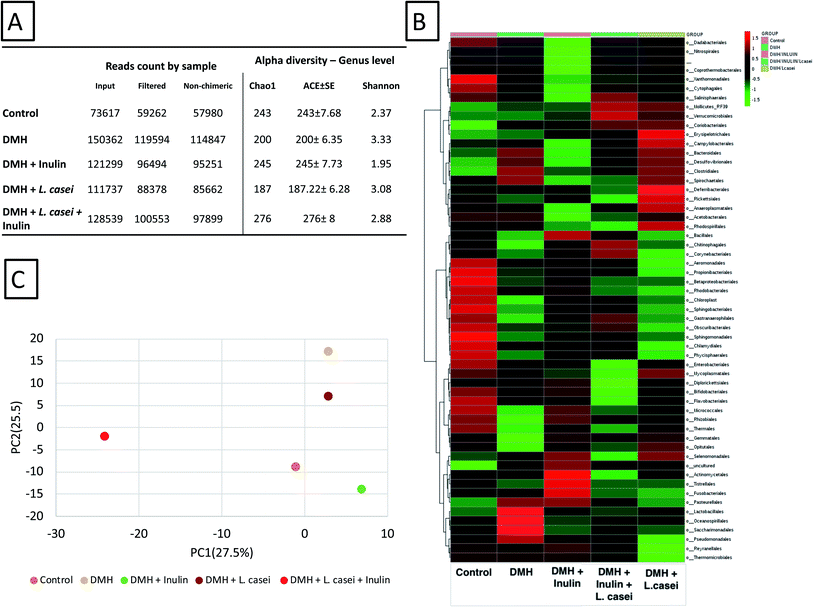
847 in DMH treated group, while the lowest reads count was 57![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 980 in the control group (Fig. 7A). The total reads obtained were classified into 23 phyla, 40 classes, 103 orders, 201 families and 464 genera. Alpha diversity of the groups was calculated including Chao1, ACE and Fisher indices (Fig. 7A). The Shannon and Chao1 indices were highest at the synbiotic-treated group. Fig. 7B shows the heatmap representation calculated at the order level of the classified reads obtained. Importantly, the principal component analysis among the five tested groups showed distinct separation of the synbiotic-treated group compared to all other sample groups (see also ESI Fig. S1†). The PC1 was computed at 27.5% while the PC2 was computed at 25.5% (Fig. 7C).
980 in the control group (Fig. 7A). The total reads obtained were classified into 23 phyla, 40 classes, 103 orders, 201 families and 464 genera. Alpha diversity of the groups was calculated including Chao1, ACE and Fisher indices (Fig. 7A). The Shannon and Chao1 indices were highest at the synbiotic-treated group. Fig. 7B shows the heatmap representation calculated at the order level of the classified reads obtained. Importantly, the principal component analysis among the five tested groups showed distinct separation of the synbiotic-treated group compared to all other sample groups (see also ESI Fig. S1†). The PC1 was computed at 27.5% while the PC2 was computed at 25.5% (Fig. 7C).
Fig. 8 shows the reads counts recorded for the highly represented bacterial phyla among the five tested groups. Interestingly the Verrucomicrobia phylum was enriched in animal groups treated with L. casei (probiotic group and synbiotic treated group). The major contribution to the phylum Verrucomicrobia is due to the genus Akkermansia, the only identified genus from this phylum. Bacteroidetes were found at highest abundance in the DMH treated group, while the lowest abundance was at the prebiotic treated group. On the contrary, Firmicutes were found at the lowest abundance at the DMH-treated group, while the highest abundance was found in the prebiotic treated groups.
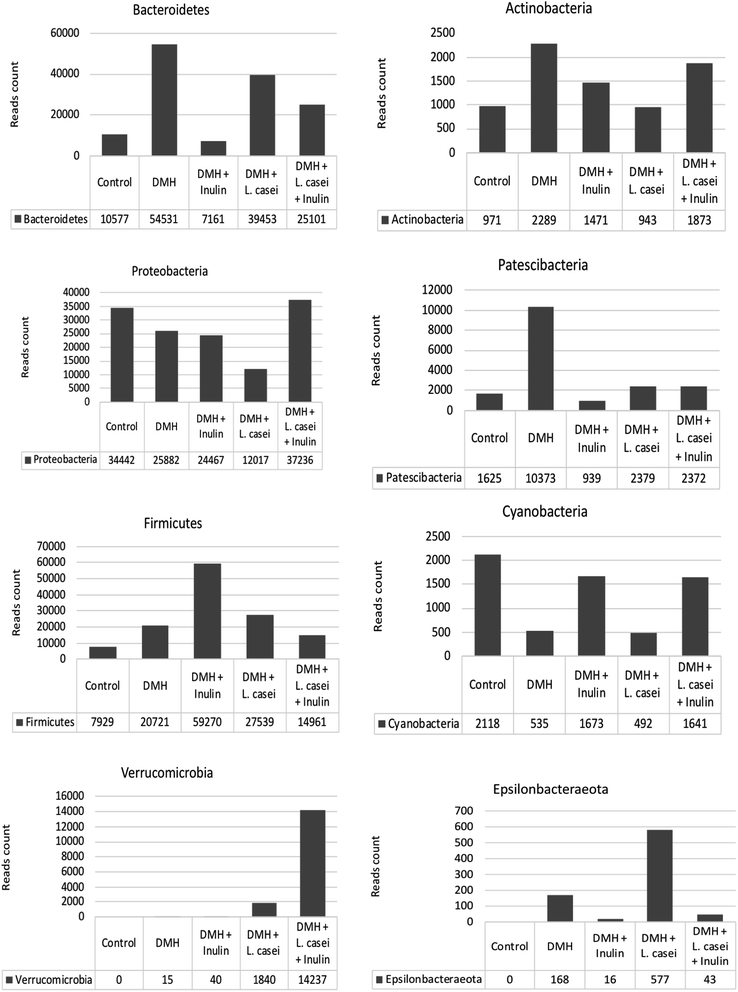 | ||
| Fig. 8 Reads count for major phyla and genera. The reads count for the major phyla recorded among the different groups. | ||
The genus Akkermansia reads count were 1821 and 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 231 in the case of probiotic and synbiotic treated groups respectively compared to zero and 40 in the non-L. casei treated groups (DMH) and prebiotic respectively (Table 2). Similarly, the genus Turicibacter reads count were 1094 and 1841 in probiotic and synbiotic groups respectively compared to 681 and 465 reads count in the DMH and prebiotic treated groups respectively. Based on these observations, the genera Akkermansia and Turicibacter were significantly enriched in response to L. casei treatment (Table 2, p < 0.05).
231 in the case of probiotic and synbiotic treated groups respectively compared to zero and 40 in the non-L. casei treated groups (DMH) and prebiotic respectively (Table 2). Similarly, the genus Turicibacter reads count were 1094 and 1841 in probiotic and synbiotic groups respectively compared to 681 and 465 reads count in the DMH and prebiotic treated groups respectively. Based on these observations, the genera Akkermansia and Turicibacter were significantly enriched in response to L. casei treatment (Table 2, p < 0.05).
| Non-L. casei | L. casei | Total | P value | |
|---|---|---|---|---|
| Genus Akkermansia | ||||
| DMH | 0 | 1821 | 1821 | 0.0199 |
| DMH + inulin | 40 | 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 231 231 |
14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 271 271 |
|
| Total | 40 | 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 052 052 |
16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 092 092 |
|
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||
| Genus Turicibacter | ||||
| DMH | 681 | 1094 | 1775 | <0.00001 |
| DMH + inulin | 465 | 1841 | 2306 | |
| Total | 1146 | 2935 | 4081 | |
| Non-inulin | Inulin | Total | P value | |
|---|---|---|---|---|
| Genus Novosphingobium | ||||
| DMH | 8210 | 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 920 920 |
29![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 130 130 |
<0.00001 |
| DMH + L. casei | 9750 | 33![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 073 073 |
42![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 823 823 |
|
| Total | 17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 960 960 |
53![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 993 993 |
71![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 953 953 |
|
4. Discussion
In the current study, we found that co-administration of L. casei and inulin as a synbiotic exhibited a superior protective effect against a DMH-induced colon cancer in male Swiss mice through affecting JNK-1/β-catenin signaling pathway as well as enriching beneficial bacteria in the colon such as Akkermansia and Turicibacter.Our results showed no change in the body weight between the DMH-treated group and the control group except after 24 weeks of the experimental period which agrees with the previously published reports.42 In addition, DMH showed no significant effect on calorie intake between groups.43 However, in the current study, administration of inulin, L. casei or inulin + L. casei together with DMH significantly decreased the body weight compared to either the control or the DMH groups. It is worthy to mention that the effect of administration of pre-, pro- or synbiotic formulations on the body weight in literature is controversial so far. For instance, it was observed that ingestion of inulin or fructo-oligosaccharides for long periods reduces the body weight.44–47 Nevertheless, in other studies inulin administration had no effect or even increased the body weight of experimental animals.48–50 On the same way, administration of different probiotics either showed a weight loss51 or no effect on the body weight of mice.52,53 Interestingly, it was found that administration of a synbiotic mixture (inulin plus Lactobacillus acidophilus, Lactobacillus bulgaricus, Bifidobacterium bifidum, Bifidobacterium longum, and Streptococcus thermophilus) for 5 weeks resulted in a decrease in the body weight of animals consistently with our findings.54 Although, administration of Bifidobacterium longum BL999 with a prebiotic mixture (fructo-oligosaccharides and galacto-oligosaccharides) to infants from 14th to 112th day after birth resulted in a weight gain.55
In the current study, we did not record the food intake of animals throughout the experimental period. In literature, the administration of inulin in a model of DMH-induced colon cancer, showed no significant effect on food intake of animals although there was a significant reduction in the body weight compared to that of the normal rats.56 On the same way, there was no significant difference in the food and water intake between mice given Lactobacillus casei and control mice although the L. casei group showed less blood glucose level and more protection from the diabetes incidence.57,58 In an attempt to explain the body weight reducing effect, Drissi et al., determined the differences in the genomes of some Lactobacillus species that have either weight protection or weight gain effect and they found that weight protecting Lactobacillus strains developed mechanisms for increased glycolysis and antioxidant defense.59 In addition, Lactobacillus species decreased body weight through decreasing fatty acid synthase activity and hence white adipose tissue mass without any effect on energy intake.60 These studies supporting our results that, the decrease in the blood glucose and lipids is partly responsible for the body weight reduction. Our results are also compatible with previous studies showing the reducing effect on inulin on blood glucose, total cholesterol and triglycerides levels in mice and humans.50,61 On the same way, administration of Lactobacillus casei CCFM419 to mice with type 2 diabetes mellitus decreased the blood glucose level.53 Interestingly, administration of a synbiotic formula to women suffering from polycystic ovary syndrome for 12 weeks resulted in decreased levels of plasma glucose, triglyceride, and cholesterol levels.62
Importantly, it was previously found that certain Lactobacillus strains isolated from human breast milk could combat cervical cancer via inducing apoptosis and inhibiting proliferation.63 In particular, L. casei potentiated the antiproliferative effect of geniposide through the production of β-glucosidase in oral squamous cell carcinoma.64 Remarkably, the anti-cancer activity of inulin was previously elucidated against melanoma as well as the transplanted mammary tumors in rats.65 Moreover, inulin was found to decrease the preneoplastic lesions and multiplicity in colon cancer.19 It acts through enhancing the healthy microflora which prevents the pathogenic bacteria from producing toxins or carcinogenic substances.66 Our results support the previous studies and provide evidence that co-administration of L. casei and inulin exhibits a superior protective effect against colon cancer compared to the administration of either L. casei or inulin alone. This observation was seen in a significant decrease in CEA levels, ACF number as well as colon cancer morphological and histological changes in the synbiotic treated group compared to either pre- or probiotic treated groups.
There are several types of JNK proteins such as JNK-1, JNK-2, and JNK-3 involved in the regulation of various biological processes.67,68 Although JNK-3 is only expressed in the brain and testis, both JNK-1 and JNK-2 proteins are ubiquitously expressed. JNK-1 was found to suppress the development of colon cancer through inhibiting the proliferation of intestinal epithelial cells and inducing apoptosis.68–70 β-Catenin is a multifunctional overexpressed molecule in colorectal cancers.71 Mutations in either β-catenin or adenomatous polyposis coli (APC) genes are commonly associated with the development of various cancer types especially, colon cancer.72 Interestingly, β-catenin is regulated through its phosphorylation by GSK3β whereas, after its phosphorylation, a degradation complex is formed followed by ubiquitination and destruction by the proteasome.73 Phosphorylation of GSK3β on Ser-9 leads to its inactivation which in turn prevents β-catenin degradation. Interestingly, JNK-1 activation decreases the phosphorylation of GSK3β.73 Fang et al. 2010 described a reverse correlation between JNK-1 and β-catenin since JNK-1 decreases the activity of β-catenin.20 Our novel findings indicate that administration of L. casei, inulin or both exhibit protective effects against DMH-induced colon cancer through increasing the expression of phosphorylated JNK-1 while decreasing the expression of phosphorylated GSK3β and ultimately β-catenin expression.
The bacterial flora of the gut is found in a dynamic balance with the host, and their composition is considered an indicator for the health status. It is worth mentioning that previous studies aimed to characterize the gut microbiome dysbiosis in relation to colon cancer showed conflicting results and no specific bacterial populations have been consistently linked to colon cancer.74,75 We aimed in our microbiome analysis to trace the alteration in the bacterial taxa in response to different treatments. The results obtained revealed significant bacterial enrichment of the genera Akkermansia and Turicibacter to the animal groups treated with L. casei compared to those not treated with L. casei. Similarly, administration of inulin has resulted in significant enrichment of the genus Novosphingobium compared to animal groups not treated with inulin. Akkermansia muciniphila is anaerobic bacteria that was recently identified in the intestinal flora as a specialized mucin degrader.76 Everard et al., 2013 reported on the beneficial effect of Akkermansia muciniphila as a probiotic in which impairment of the mucin production by intestinal cells was related to colon cancer and can rational the reduction of Akkermansia in our DMH-treated group.77 Derrien et al., 2011 reported on the immune modulating effect of Akkermansia muciniphila in mice.76 In addition, our results agree with the data obtained by Irecta-Nájera et al., 2017, as they showed in their study the protective effect of L. casei on DMH-treated mice.78 Previous studies reported on the beneficial effect of Turicibacter enrichment in the gut. Turicibacter has been correlated to the production of butyric acid, a short chain fatty acid, that act as an immunomodulators.79 Moreover, an anti-inflammatory activity of Turicibacter bacteria has been suggested.80,81 In the light of 95% similarity of the microbiome functional properties of the mouse and the human, based on Kyoto Encyclopedia of Genes and Genomes (KEGG) orthologous groups, the beneficial effect of the synbiotic treatment should be applicable to humans.82
5. Conclusion
Our novel findings indicate that administration of L. casei, inulin or their combination effectively protect from the development of colon cancer in male Swiss mice through targeting the JNK-1/β-catenin axis with the synbiotic treatment showing a superior effect. In addition, enhancing Akkermansia and Turicibacter beneficial bacteria in the colon was achieved. However, further studies are needed to elucidate if our pre-, pro- or synbiotic treatments have the same effect if used after the induction of colon cancer. In addition, it will be more informative to identify the main bacterial metabolites responsible for the protective effect against colon cancer.Funding
This study did not receive a specific grant.Conflicts of interest
Authors declare no conflict of interests.Acknowledgements
Microbiome experiments and related data analysis were done by The Foundation for the Promotion of Health and Biomedical Research of Valencia Region, FISABIO, under supervision of Dr Giuseppe D'Auria, Valencia, Spain.References
- S. Roy and T. Chakraborty, Deciphering the molecular mechanism and apoptosis underlying the in vitro and in vivo chemotherapeutic efficacy of vanadium luteolin complex in colon cancer, Cell Biochem. Funct., 2018, 36, 116–128 CrossRef CAS PubMed.
- N. Sangeetha, S. Aranganathan and N. Nalini, Silibinin ameliorates oxidative stress induced aberrant crypt foci and lipid peroxidation in 1,2 dimethylhydrazine induced rat colon cancer, Invest. New Drugs, 2010, 28, 225–233 CrossRef CAS PubMed.
- M. C. S. Mendes, et al., Microbiota modification by probiotic supplementation reduces colitis associated colon cancer in mice, World J. Gastroenterol., 2018, 24, 1995 CrossRef CAS PubMed.
- C. Liu, et al., Phytic acid improves intestinal mucosal barrier damage and reduces serum levels of proinflammatory cytokines in a 1,2-dimethylhydrazine-induced rat colorectal cancer model, Br. J. Nutr., 2018, 120, 121–130, DOI:10.1017/s0007114518001290.
- G. Y. Koh, et al., Parabacteroides distasonis attenuates toll-like receptor 4 signaling and Akt activation and blocks colon tumor formation in high-fat diet-fed azoxymethane-treated mice, Int. J. Cancer, 2018, 143, 1797–1805 CrossRef CAS PubMed.
- H. Svitina, et al., Transplantation of placenta-derived multipotent cells in rats with dimethylhydrazine-induced colon cancer decreases survival rate, Oncol. Lett., 2018, 15, 5034–5042 Search PubMed.
- H. Svitina, et al., Placenta-derived multipotent cells have no effect on the size and number of DMH-induced colon tumors in rats, Exp. Ther. Med., 2017, 14, 2135–2147 CrossRef CAS PubMed.
- E. Talero, et al., Inhibition of chronic ulcerative colitis-associated adenocarcinoma development in mice by VSL# 3, Inflamm. Bowel Dis., 2015, 21, 1027–1037 CrossRef PubMed.
- M.-T. Liong, Roles of probiotics and prebiotics in colon cancer prevention: postulated mechanisms and in vivo evidence, Int. J. Mol. Sci., 2008, 9, 854–863 CrossRef CAS PubMed.
- M. Kumar, et al., Probiotic metabolites as epigenetic targets in the prevention of colon cancer, Nutr. Rev., 2013, 71, 23–34 CrossRef PubMed.
- A. Tiptiri-Kourpeti, et al., Lactobacillus casei exerts anti-proliferative effects accompanied by apoptotic cell death and up-regulation of TRAIL in colon carcinoma cells, PLoS One, 2016, 11, e0147960 CrossRef PubMed.
- M. L. Wan and H. El-Nezami, Targeting gut microbiota in hepatocellular carcinoma: probiotics as a novel therapy, Hepatobiliary Surg. Nutr., 2018, 7, 11 CrossRef PubMed.
- R. A. B. Fagundes, T. F. Soder, K. C. Grokoski, F. Benetti and R. H. Mendes, Probiotics in the treatment of chronic kidney disease: a systematic review, J. Bras. Nefrol., 2018, 40, 278–286 CrossRef PubMed.
- C.-C. Chen, et al., Oral inoculation of probiotics Lactobacillus acidophilus NCFM suppresses tumour growth both in segmental orthotopic colon cancer and extra-intestinal tissue, Br. J. Nutr., 2012, 107, 1623–1634 CrossRef CAS PubMed.
- T. R. Qamar, et al., Novel Combination of Prebiotics Galacto-Oligosaccharides and Inulin-Inhibited Aberrant Crypt Foci Formation and Biomarkers of Colon Cancer in Wistar Rats, Nutrients, 2016, 8, 465 CrossRef PubMed.
- A. Verma and G. Shukla, Administration of prebiotic inulin suppresses 1,2-dimethylhydrazine dihydrochloride induced procarcinogenic biomarkers fecal enzymes and preneoplastic lesions in early colon carcinogenesis in Sprague Dawley rats, J. Funct. Foods, 2013, 5, 991–996 CrossRef CAS.
- A. Verma and G. Shukla, Synbiotic (Lactobacillus rhamnosus + Lactobacillus acidophilus + inulin) attenuates oxidative stress and colonic damage in 1,2-dimethylhydrazine dihydrochloride-induced colon carcinogenesis in Sprague–Dawley rats: a long-term study, Eur. J. Cancer Prev., 2014, 23, 550–559 CrossRef CAS PubMed.
- L. B. Bindels, et al., Synbiotic approach restores intestinal homeostasis and prolongs survival in leukaemic mice with cachexia, ISME J., 2016, 10, 1456 CrossRef CAS PubMed.
- B. L. Pool-Zobel, Inulin-type fructans and reduction in colon cancer risk: review of experimental and human data, Br. J. Nutr., 2005, 93, S73–S90 CrossRef CAS PubMed.
- W. Fang, A. Han, X. Bi, B. Xiong and W. Yang, Tumor inhibition by sodium selenite is associated with activation of c-Jun NH2-terminal kinase 1 and suppression of β-catenin signaling, Int. J. Cancer, 2010, 127, 32–42 CrossRef CAS PubMed.
- R. H. Giles, J. H. van Es and H. Clevers, Caught up in a Wnt storm: Wnt signaling in cancer, Biochim. Biophys. Acta, Rev. Cancer, 2003, 1653, 1–24 CrossRef CAS.
- D. Hu, X. Bi, W. Fang, A. Han and W. Yang, GSK3β is involved in JNK2-mediated β-catenin inhibition, PLoS One, 2009, 4, e6640 CrossRef PubMed.
- J. Du, et al., Anticancer activities of sulindac in prostate cancer cells associated with c-Jun NH2-terminal kinase 1/beta-catenin signaling, Oncol. Lett., 2014, 8, 313–316 CrossRef CAS PubMed.
- S. S. Piao and B. Shang, Pizotifen Inhibits the Proliferation and Migration of Colon Cancer HCT116 Cells by Down-regulating Wnt Signaling Pathway, Ann. Clin. Lab. Sci., 2019, 49, 183–188 Search PubMed.
- J.-H. Chang, Y. Y. Shim, S.-K. Cha, M. J. Reaney and K. M. Chee, Effect of Lactobacillus acidophilus KFRI342 on the development of chemically induced precancerous growths in the rat colon, J. Med. Microbiol., 2012, 61, 361–368 CrossRef CAS PubMed.
- J. D. Bancroft and M. Gamble, Theory and practice of histological techniques, Elsevier Health Sciences, 2008 Search PubMed.
- T. K. Motawi, S. A. El-Maraghy, A. N. ElMeshad, O. M. Nady and O. A. Hammam, Cromolyn chitosan nanoparticles as a novel protective approach for colorectal cancer, Chem.-Biol. Interact., 2017, 275, 1–12 CrossRef CAS PubMed.
- B. Shpitz, et al., Proliferating cell nuclear antigen as a marker of cell kinetics in aberrant crypt foci, hyperplastic polyps, adenomas, and adenocarcinomas of the human colon, Am. J. Surg., 1997, 174, 425–430 CrossRef CAS.
- L. O. Whiteley, L. Hudson Jr and T. P. Pretlow, Aberrant crypt foci in the colonic mucosa of rats treated with a genotoxic and nongenotoxic colon carcinogen, Toxicol. Pathol., 1996, 24, 681–689 CrossRef CAS PubMed.
- Y. Jia, et al., Metformin prevents DMH-induced colorectal cancer in diabetic rats by reversing the Warburg effect, Toxicol. Pathol., 2015, 4, 1730–1741 CAS.
- S.-M. Hsu, L. Raine and H. Fanger, Use of avidin–biotin–peroxidase complex (ABC) in immunoperoxidase techniques: a comparison between ABC and unlabeled antibody (PAP) procedures, J. Histochem. Cytochem., 1981, 29, 577–580 CrossRef CAS PubMed.
- J. Waisberg, et al., Padrão da distribuição tecidual do CEA no carcinoma colo-retal: relação com o nível sérico do CEA e classificação de Dukes, Rev. Bras. Coloproctol., 2002, 22, 7 Search PubMed.
- S. Yamada, et al., Bile acid metabolism regulated by the gut microbiota promotes non-alcoholic steatohepatitis-associated hepatocellular carcinoma in mice, Oncotarget, 2018, 9, 9925 Search PubMed.
- Y. Gendo, et al., Dysbiosis of the Gut Microbiota on the Inflammatory Background due to Lack of Suppressor of Cytokine Signalling-1 in Mice, Inflamm. Intest. Dis., 2018, 3, 145–154 CrossRef PubMed.
- A. Klindworth, et al., Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies, Nucleic Acids Res., 2013, 41, e1 CrossRef CAS PubMed.
- R. Schmieder and R. Edwards, Quality control and preprocessing of metagenomic datasets, Bioinformatics, 2011, 27, 863–864 CrossRef CAS PubMed.
- R. C. Team, A language and environment for statistical computing R Foundation for Statistical Computing, Vienna, Austria, 2012, ISBN 3-900051-07-0 Search PubMed.
- J. G. Caporaso, et al., QIIME allows analysis of high-throughput community sequencing data, Nat. Methods, 2010, 7, 335 CrossRef CAS PubMed.
- B. J. Callahan, et al., DADA2: high-resolution sample inference from Illumina amplicon data, Nat. Methods, 2016, 13, 581 CrossRef CAS PubMed.
- C. Quast, et al., The SILVA ribosomal RNA gene database project: improved data processing and web-based tools, Nucleic Acids Res., 2012, 41, D590–D596 CrossRef PubMed.
- B. D. Ondov, N. H. Bergman and A. M. Phillippy, Interactive metagenomic visualization in a Web browser, BMC Bioinf., 2011, 12, 385 CrossRef PubMed.
- V. Chithra and S. Leelamma, Coriandrum sativum—effect on lipid metabolism in 1, 2-dimethyl hydrazine induced colon cancer, J. Ethnopharmacol., 2000, 71, 457–463 CrossRef CAS PubMed.
- A. Lopez-Mejia, et al., Chemopreventive effect of Callistemon citrinus (Curtis) Skeels against colon cancer induced by 1,2-dimethylhydrazine in rats, J. Cancer Res. Clin. Oncol., 2019, 145, 1417–1426 CrossRef CAS PubMed.
- C. Morris and G. A. Morris, The effect of inulin and fructo-oligosaccharide supplementation on the textural, rheological and sensory properties of bread and their role in weight management: A review, Food Chem., 2012, 133, 237–248 CrossRef CAS PubMed.
- N. M. Delzenne and P. D. Cani, Nutritional modulation of gut microbiota in the context of obesity and insulin resistance: Potential interest of prebiotics, Int. Dairy J., 2010, 20, 277–280 CrossRef CAS.
- P. D. Cani, et al., Gut microbiota fermentation of prebiotics increases satietogenic and incretin gut peptide production with consequences for appetite sensation and glucose response after a meal, Am. J. Clin. Nutr., 2009, 90, 1236–1243 CrossRef CAS PubMed.
- K. R. Pandey, S. R. Naik and B. V. Vakil, Probiotics, prebiotics and synbiotics-a review, J. Food Sci. Technol., 2015, 52, 7577–7587 CrossRef CAS PubMed.
- S. Tan and J. A. Caparros-Martin, Isoquercetin and inulin synergistically modulate the gut microbiome to prevent development of the metabolic syndrome in mice fed a high fat diet, Sci. Rep., 2018, 8, 10100 CrossRef PubMed.
- C. Andrieux, S. Lory, C. Dufour-Lescoat, R. De Baynast and O. Szylit, Physiological effects of inulin in germ-free rats and in heteroxenic rats inoculated with a human flora, Food Hydrocolloids, 1991, 5, 49–56 CrossRef CAS.
- Q. Zhang, et al., Inulin-type fructan improves diabetic phenotype and gut microbiota profiles in rats, PeerJ, 2018, 6, e4446 CrossRef PubMed.
- M. Million, et al., Comparative meta-analysis of the effect of Lactobacillus species on weight gain in humans and animals, Microb. Pathog., 2012, 53, 100–108 CrossRef PubMed.
- S. Yun, H. Park and J. Kang, Effect of Lactobacillus gasseri BNR17 on blood glucose levels and body weight in a mouse model of type 2 diabetes, J. Appl. Microbiol., 2009, 107, 1681–1686 CrossRef CAS PubMed.
- X. Li, et al., Effects of Lactobacillus casei CCFM419 on insulin resistance and gut microbiota in type 2 diabetic mice, Benefic. Microbes, 2017, 8, 421–432 CrossRef CAS PubMed.
- S.-C. Yang, et al., Effect of synbiotics on intestinal microflora and digestive enzyme activities in rats, World J. Gastroenterol., 2005, 11, 7413 CrossRef CAS PubMed.
- G. Puccio, et al., Clinical evaluation of a new starter formula for infants containing live Bifidobacterium longum BL999 and prebiotics, Nutrition, 2007, 23, 1–8 CrossRef CAS PubMed.
- T. Qamar, et al., Novel combination of prebiotics galacto-oligosaccharides and inulin-inhibited aberrant crypt foci formation and biomarkers of colon cancer in wistar rats, Nutrients, 2016, 8, 465 CrossRef PubMed.
- T. Matsuzaki, et al., Effect of oral administration of Lactobacillus casei on alloxan-induced diabetes in mice, APMIS, 1997, 105, 637–642 CrossRef CAS PubMed.
- H. Yadav, S. Jain and P. Sinha, Antidiabetic effect of probiotic dahi containing Lactobacillus acidophilus and Lactobacillus casei in high fructose fed rats, Nutrition, 2007, 23, 62–68 CrossRef PubMed.
- F. Drissi, et al., Comparative genomics analysis of Lactobacillus species associated with weight gain or weight protection, Nutr. Diabetes, 2014, 4, e109 CrossRef CAS PubMed.
- T. Arora, S. Singh and R. K. Sharma, Probiotics: interaction with gut microbiome and antiobesity potential, Nutrition, 2013, 29, 591–596 CrossRef CAS PubMed.
- C. M. Williams, Effects of inulin on lipid parameters in humans, J. Nutr., 1999, 129, 1471S–1473S CrossRef CAS PubMed.
- M. Samimi, A. Dadkhah and H. Haddad Kashani, The Effects of Synbiotic Supplementation on Metabolic Status in Women With Polycystic Ovary Syndrome: a Randomized Double-Blind Clinical Trial, Probiotics Antimicrob. Proteins, 2018, 10, 1–7 CrossRef PubMed.
- M. S. R. Rajoka, et al., Anticancer potential against cervix cancer (HeLa) cell line of probiotic Lactobacillus casei and Lactobacillus paracasei strains isolated from human breast milk, Food Funct., 2018, 9, 2705–2715 RSC.
- Y. Qian, et al., Lactobacillus casei Strain Shirota Enhances the In Vitro Antiproliferative Effect of Geniposide in Human Oral Squamous Carcinoma HSC-3 Cells, Molecules, 2018, 23, 1069 CrossRef PubMed.
- H. S. Taper and M. Roberfroid, Influence of inulin and oligofructose on breast cancer and tumor growth, J. Nutr., 1999, 129, 1488S–1491S CrossRef CAS PubMed.
- K. Kalyani Nair, S. Kharb and D. Thompkinson, Inulin dietary fiber with functional and health attributes—a review, Food Rev. Int., 2010, 26, 189–203 CrossRef CAS.
- A. Saadeddin, R. Babaei-Jadidi, B. Spencer-Dene and A. S. Nateri, The links between transcription, β-catenin/JNK signaling, and carcinogenesis, Mol. Cancer Res., 2009, 7, 1189–1196 CrossRef CAS.
- L. Hui, K. Zatloukal, H. Scheuch, E. Stepniak and E. F. Wagner, Proliferation of human HCC cells and chemically induced mouse liver cancers requires JNK1-dependent p21 downregulation, J. Clin. Invest., 2008, 118, 3943–3953 CrossRef CAS.
- C. Tong, et al., c-Jun NH2-terminal kinase 1 plays a critical role in intestinal homeostasis and tumor suppression, Am. J. Pathol., 2007, 171, 297–303 CrossRef CAS.
- Q.-B. She, N. Chen, A. M. Bode, R. A. Flavell and Z. Dong, Deficiency of c-Jun-NH2-terminal kinase-1 in mice enhances skin tumor development by 12-O-tetradecanoylphorbol-13-acetate, Cancer Res., 2002, 62, 1343–1348 CAS.
- T. Brabletz, et al., Variable β-catenin expression in colorectal cancers indicates tumor progression driven by the tumor environment, Proc. Natl. Acad. Sci. U. S. A., 2001, 98, 10356–10361 CrossRef CAS.
- B. Javan, F. Atyabi and M. Shahbazi, Hypoxia-inducible bidirectional shRNA expression vector delivery using PEI/chitosan-TBA copolymers for colorectal Cancer gene therapy, Life Sci., 2018, 202, 140–151 CrossRef CAS.
- D. Hu, et al., c-Jun N-terminal kinase 1 interacts with and negatively regulates Wnt/β-catenin signaling through GSK3β pathway, Carcinogenesis, 2008, 29, 2317–2324 CrossRef CAS PubMed.
- H.-M. Chen, et al., Decreased dietary fiber intake and structural alteration of gut microbiota in patients with advanced colorectal adenoma, Am. J. Clin. Nutr., 2013, 97, 1044–1052 CrossRef CAS.
- T. Wang, et al., Structural segregation of gut microbiota between colorectal cancer patients and healthy volunteers, ISME J., 2012, 6, 320–329 CrossRef CAS.
- M. Derrien, M. C. Collado, K. Ben-Amor, S. Salminen and W. M. de Vos, The Mucin degrader Akkermansia muciniphila is an abundant resident of the human intestinal tract, Appl. Environ. Microbiol., 2008, 74, 1646–1648 CrossRef CAS.
- A. Everard, et al., Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 9066–9071 CrossRef CAS.
- C. A. Irecta-Nájera, M. del Rosario Huizar-López, J. Casas-Solís, P. Castro-Félix and A. Santerre, Protective effect of Lactobacillus casei on DMH-induced colon carcinogenesis in mice, Probiotics Antimicrob. Proteins, 2017, 9, 163–171 CrossRef.
- Y. Zhong, M. Nyman and F. Fåk, Modulation of gut microbiota in rats fed high-fat diets by processing whole-grain barley to barley malt, Mol. Nutr. Food Res., 2015, 59, 2066–2076 CrossRef CAS.
- J. S. Suchodolski, Fecal Microbiome in Dogs with Acute Diarrhea and Idiopathic Inflammatory Bowel Disease, Encyclopedia of Metagenomics: Environmental Metagenomics, 2015, pp. 183–186 Search PubMed.
- T. Werner, et al., Depletion of luminal iron alters the gut microbiota and prevents Crohn's disease-like ileitis, Gut, 2011, 60, 325–333 CrossRef CAS.
- L. Xiao, et al., A catalog of the mouse gut metagenome, Nat. Biotechnol., 2015, 33, 1103 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9ra04388h |
| This journal is © The Royal Society of Chemistry 2019 |