Open Access Article
Open Access ArticleRetracted Article: FOXO4 overexpression suppresses hypoxia-induced-MCF-7 cell survival and promotes apoptosis through the HIF-2α/Bnip3 signal pathway
Yan Qiao,
Bin Wang,
Huimin Zhang,
Yu Yan and
Ligang Niu *
*
Department of Breast Surgery, The First Affiliated Hospital of Xi'an Jiao Tong University, No. 277 Yanta West Road, Xi'an 710061, China. E-mail: ligang_niu3@126.com; Fax: +86-029-85324605
First published on 19th August 2019
Abstract
Transcriptional regulator forkhead box O (FOXO) has implications in many diverse carcinomas and often acts as a tumour suppressor. Evidence suggests that FOXO4 may play a role in cancer cell proliferation and apoptosis; however, the function and mechanism of FOXO4 on breast cancer cell growth are still unknown. FOXO4 can respond to hypoxia and in the current study, our aim is to investigate the function and molecular mechanism of FOXO4 in hypoxia-induced MCF-7 cells. We first observed that hypoxia treatment reduced FOXO4 mRNA and protein expression in MCF-7 cells. Moreover, FOXO4 overexpression reversed hypoxia-induced MCF-7 cell survival. Hypoxia treatment markedly impeded MCF-7 cell apoptosis and inhibited caspase-3 activity, whereas FOXO4 overexpression promoted apoptosis and increased caspase-3 activity in hypoxia-induced MCF-7 cells. Further studies indicated that FOXO4 overexpression inhibited hypoxia-induced HIF-2α and Bnip3 expression in MCF-7 cells; moreover, FOXO4 suppressed Bnip3 expression, which is dependent on the low level of HIF-2α. Finally, we demonstrated that Bnip3 overexpression reversed the effects of FOXO4 overexpression on cell survival and apoptosis in hypoxia-induced MCF-7 cells. In conclusion, the present study suggests that FOXO4 overexpression mediated the HIF-2α/Bnip3 signal pathway, which has implications in cell survival and apoptosis in hypoxia-induced MCF-7 cells.
1. Introduction
Breast cancer is one of the most common carcinomas with high mortality and an increasing annual incidence worldwide.1,2 Accumulating evidence suggested that breast cancer could be classified into three major subtypes, namely, basal like, luminal and human epidermal growth factor receptor 2+ (HER2+) according to the results of comprehensive gene expression profiling.3,4 There are multiple influential factors in the therapeutic options of patients with heterogeneous breast cancer, including survival and physiological outcomes and a multitude of clinical outcomes.5 Moreover, because of the genetic alterations that act as the critical determinants and their involvement in the initiation and progression of breast cancer, breast cancer patients usually have different clinical outcomes.6 Genes-regulates genomic integrity and the novel molecular which plays roles in breast cancer are required to reveal.7Forkhead box (FOX) members have 19 sub-families of transcription factors, which share approximately 110 amino acids and have a highly conserved DNA-binding domain.8 The FOXO subgroup consists of four members, namely, FOXO1, FOXO3, FOXO4, and FOXO6.9,10 Evidence indicated that FOXO members act as important transcriptional regulators in different types of carcinomas, which are involved in cancer development and therapy.8 FOXO4 is involved in non-small-cell lung cancer,11 colon cancer,12 and bladder cancer.13 However, the function of FOXO4 in breast cancer cells is largely unknown.
Multiple solid tumors are always accompanied by hypoxia, which plays a role in tumor therapy resistance and aggressiveness.14 Hypoxia-inducible factors (HIFs) contribute to regulate gene expression in response to hypoxic conditions.15 HIF contains two hetero-dimeric subunits, one oxygen-sensitive α-subunit (HIF-1α, HIF-2α and HIF-3α) and one oxygen-insensitive β-subunit (HIFβ, also termed ARNT).15,16 HIF-2α, also named as a member of pas superfamily 2 (Mop2) and endothelial PAS domain protein-1 (EPAS1), was first identified in 1997.17,18 HIF-2α shares about 48% homology in amino-acid sequences with HIF-1, and hypoxic conditions also up-regulated the expression of HIF-2α.17,19 It is now well established that HIF-2α is universally expressed in a variety of cancer types, such as renal, glial, colon, head and neck, hepatocellular, ovarian, bladder, prostate and breast cancers.20–26 BCL-2/adenovirus E1B 19 kDa-interacting protein 3 (Bnip3), a gene positioned downstream of HIF, is a member of the Bcl-2 gene family, and it plays an important role in hypoxia-induced cell autophagy, necrosis and apoptosis. The current study aims to suggest the function of the HIF-2α/Bnip3 signal axis in FOXO4-mediated cell survival and apoptosis in hypoxia-induced MCF-7 cells.
2. Methods
2.1. Cell culture, treatment and transfection
Human breast cancer cell line (MCF-7) was obtained from ATCC (Manassas, VA, USA). Cells were maintained in DMEM supplemented with 10% FBS and cultured at 37 °C in an atmosphere of 5% CO2 and 21% O2. MCF-7 cells were transfected with pcDNA3.1-FOXO4 or co-transfected with pcDNA3.1-FOXO4 and pcDNA3.1-HIF-2α or pcDNA3.1-FOXO4 and pcDNA3.1-Bnip3 for 48 h and then cultured under normoxia (21% O2; 5% CO2) or hypoxia (1% O2; 5% CO2) for 24 h.2.2. CCK-8 assay
MCF-7 cell viability was determined using a cell counting kit-8 (CCK-8) assay (Dojindo Molecular Technologies, Inc., Kumamoto, Japan) according to the manufacturer's instructions. MCF-7 cells were transfected and treated as described earlier. The cells were seeded into 96-well plates at a density of 3 × 104 cells per well. CCK-8 (10 μL per well) solution was added to each well for another 2 h for measuring the cell viability. The absorbance of the cells was measured at 450 nm.2.3. Cell apoptosis
Cell apoptosis was analyzed using a cell death detection enzyme-linked immunosorbent assay (ELISA) kit (Roche, Mannheim, Germany) according to the manufacturer's protocol. The DNA fragments present in the supernatant of MCF-7 cell lysate were quantified.2.4. Caspase-3 activity
MCF-7 cells were transfected with pcDNA3.1-FOXO4 or co-transfected with pcDNA3.1-FOXO4 and pcDNA3.1-HIF-2α or pcDNA3.1-FOXO4 and pcDNA3.1-Bnip3 for 48 h and then cultured under normoxia (21% O2; 5% CO2) or hypoxia (1% O2; 5% CO2) for 24 h. The caspase-3 activity assay was performed using a commercial kit (APOPCYTO Caspase-3 Colorimetric Assay Kit, Medical and Biological Laboratories, Japan) according to the manufacturer's instructions.2.5. Quantitative real-time PCR
Total cellular RNA was isolated from MCF-7 cells using TRIzol Reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions. A Takara reverse transcription kit (Takara Bio, Otsu, Japan) was used to synthesize total cDNA. RT-qPCRs were performed using a SYBR Premix Ex Taq kit (Takara Bio) and the ABI ViiA 7 Real-Time PCR System (Applied Biosystems, USA). The PCR conditions are as follows: 95 °C for 5 min, then 30 cycles at 95 °C for 1 min, followed by 58 °C for 40 s and 72 °C for 1 min. To quantify the relative mRNA levels of FOXO4, the 2-ΔΔCt method was used, and data were normalized to β-actin.2.6. Western blot analysis
Total protein was isolated from MCF-7 cells using a RIPA lysis buffer (Millipore). Equal amounts of the extracts (40 μg per lane) were separated on a 10% SDS-PAGE gel. FOXO4, HIF-2α and Bnip3 protein expression were assayed by Western blotting using antibodies directed against FOXO4 (ab63254, 1:500, Abcam), HIF-2α (ab199, 1:500, Abcam), Bnip3 (ab109362, 1:1000, Abcam) and β-actin (ab8226, 1:5000, Abcam). Protein bands were detected using an ECL kit according to the manufacturer's recommendations (Amersham Corp., Buckinghamshire, UK) and captured using the G:BOX chemiXR5 Gel Imaging System. β-Actin acts as the control.2.7. Statistical analysis
Data in this study are expressed as means ± SD from three independent experiments. One-way ANOVA was used to perform statistical comparisons followed by the least significant difference (LSD) test. P < 0.05 was accepted as statistically significant.3. Results
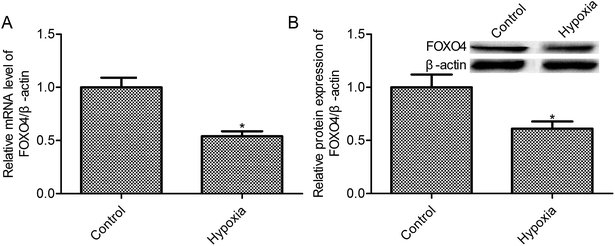
3.1. Hypoxia treatment elevated FOXO4 mRNA and protein expression
Evidences indicated that oxygen tension in solid tumors probably between 0.1 and 2%,27 and 1% oxygen concentration may be the appropriate concentration in cancer cell studies under hypoxic conditions. According to this, we have chosen 1% oxygen concentration to mimic hypoxic conditions of MCF-7 cells. First, we observed that the exposure to 1% O2 tension for 24 h reduced the FOXO4 mRNA level when compared with the control group (21% O2, p < 0.05, Fig. 1A). Moreover, hypoxia treatment (1% O2) markedly suppressed FOXO4 protein expression in MCF-7 cells (p < 0.05, Fig. 1B)3.2. High expression of FOXO4-impeded hypoxia-induced MCF-7 cell survival
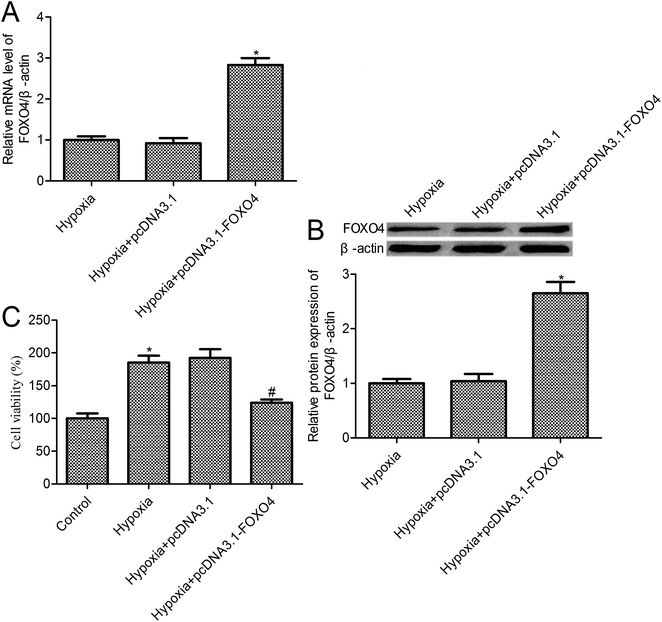
To further investigate the effects of FOXO4 on hypoxia-induced MCF-7 cell biology, FOXO4 overexpression was established in MCF-7 cells (Fig. 2A and B). Cell survival was determined using a CCK-8 assay. The survival of the control cells under normoxic conditions was set as 100%, and relative cell survival in the experimental group was quantified. Compared with the control group, we observed that hypoxia treatment increased MCF-7 cell survival to 185.3% ± 10.42% (p < 0.05). In addition, a 123.8% ± 5.14% cell survival rate was observed for FOXO4 overexpression in MCF-7 cells (p < 0.05), whereas there was no significant difference between the pcDNA3.1 group and the hypoxia group (p > 0.05).3.3. FOXO4 overexpression promoted MCF-7 cell apoptosis under hypoxic conditions
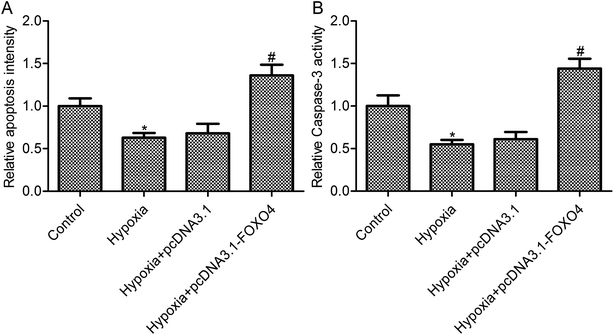
As shown in Fig. 3A, hypoxia treatment leads to an obvious decrease in cell apoptosis in MCF-7 cells (p < 0.05). FOXO4 overexpression promoted cell apoptosis in MCF-7 cells under hypoxic conditions when compared with the hypoxia group (Fig. 3A, p < 0.05), whereas there was no significant differences between the control vector pcDNA 3.1 group and the hypoxia group (Fig. 3A, p > 0.05). To further confirm the effects of FOXO4 overexpression on MCF-7 cell apoptosis, caspase-3 activity was assayed. Hypoxia treatment decreased caspase-3 activity in MCF-7 cells compared with the control group (Fig. 3B, p < 0.05). In addition, FOXO4 overexpression increased caspase-3 activity in MCF-7 cells compared with the hypoxia group (Fig. 3B, p < 0.05).3.4. FOXO4 overexpression inhibited hypoxia-induced HIF-2α and Bnip3 expression
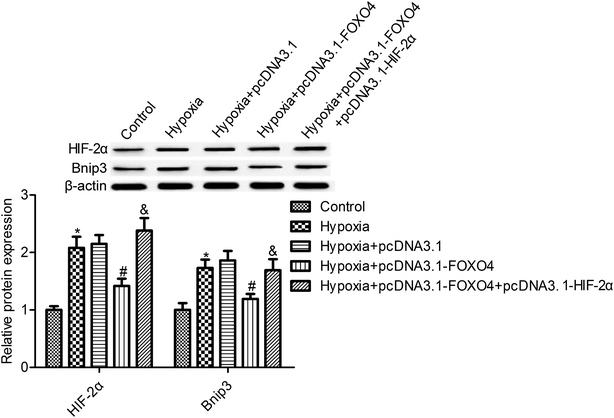
We observed that the hypoxic condition resulted in an elevation of HIF-2α protein expression in MCF-7 cells compared with the control normoxia group (Fig. 4, p < 0.05). Moreover, Bnip3 expression, the downstream gene of HIF, was also determined. As shown in Fig. 4, the protein expression of Bnip3 was also significantly accumulated in MCF-7 cells under hypoxic conditions. Moreover, our study suggests that FOXO4 overexpression markedly reduced the protein expression of HIF-2α and Bnip3 in MCF-7 cells compared with the hypoxia group (Fig. 4, p < 0.05). To further reveal the function of HIF-2α in FOXO4-induced Bnip3 inhibition under hypoxic conditions, HIF-2α and FOXO4 overexpression was established in the cells. HIF-2α overexpression increased the protein expression of HIF-2α and reversed the inhibition effects of FOXO4 on Bnip3 expression. These results indicated that FOXO4-induced Bnip3 expression inhibition was dependent on the low level of HIF-2α in MCF-7 cells under hypoxic conditions (Fig. 4, p < 0.05).3.5. Bnip3 overexpression reversed the effects of FOXO4 on cell survival and apoptosis in hypoxia-induced MCF-7 cells
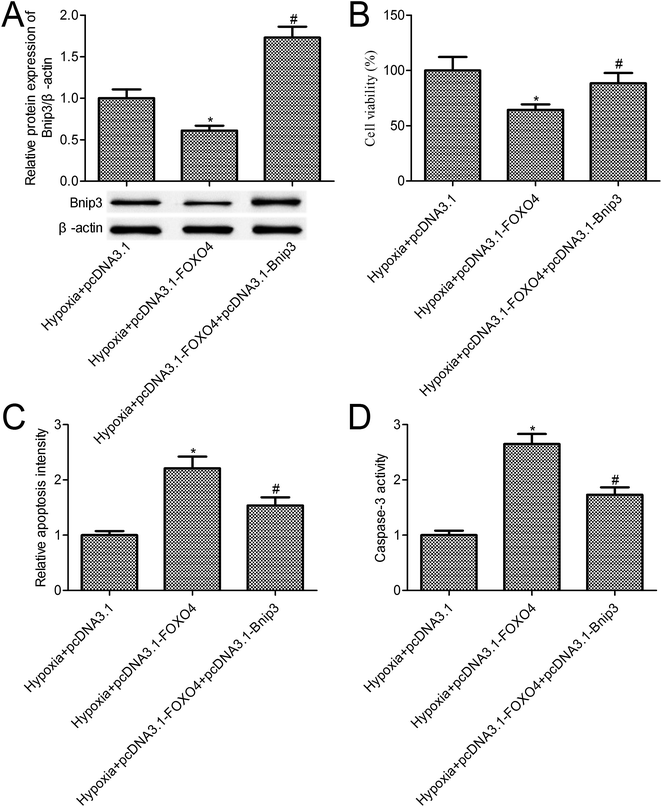
The above-mentioned results confirmed that FOXO4 overexpression inhibited Bnip3 expression. To further reveal the function of Bnip3 in hypoxia-induced MCF-7 cells, we constructed MCF-7 cells co-transfected with pcDNA3.1-FOXO4 and pcDNA3.1-Bnip3. Western blot results indicated that Bnip3 overexpression significantly increased Bnip3 levels when compared with the pcDNA3.1-FOXO4 group (Fig. 5A, p < 0.05). Compared with the only FOXO4 overexpression group, Bnip3 overexpression significantly promoted MCF-7 cell survival (Fig. 5B, p < 0.05). Further studies confirmed that Bnip3 overexpression reversed the effects of FOXO4 overexpression on cell apoptosis and caspase-3 activity in MCF-7 cells treated with 1% O2 tension for 24 h (Fig. 5C and D, p < 0.05).4. Discussion
Breast cancer is one of the most common cancers, and 30% of new cancer cases among women are breast cancer cases.2 The altered expression of genes may act as a biomarker for the early detection of breast cancer. Evidence indicated that hypoxic stress contributes to the change in tumor cell biology, including increasing tumor heterogeneity, promoting cell differentiation, helping cells in gaining new functional properties, controlling tumor immunogenicity, and causing the expansion of immune-suppressive stromal cells.14 Moreover, hypoxia always contributes to breast cancer development.28 In the current study, we observe that FOXO4 was significantly reduced in breast cancer cells under hypoxic conditions. Thus, we speculated that FOXO4 overexpression is involved in MCF-7 cell biology under hypoxic conditions. We found that hypoxia treatment markedly promoted MCF-7 cell survival and suppressed its apoptosis. FOXO4 overexpression reversed the effects of hypoxia on MCF-7 cell survival and apoptosis. Finally, we confirmed that FOXO4 overexpression plays a role in hypoxia-induced MCF-7 cells via reducing hypoxia-responsive HIF-2α and Bnip3 gene expressions.FOXO1, FOXO3a, FOXO4 and FOXO6 transcription factors are members of the Forkhead box O (FOXO) family, which act as tumor suppressors and play important inhibitory roles in cancer cell proliferation and promotive roles in cell apoptosis.8 Accumulating evidence suggested that FOXO1 plays a role in breast cancer cell proliferation, migration, and invasion.29 Park S. H. et al. confirmed that FOXO3a activation suppresses triple-negative breast cancer cell survival and proliferation, thereby inhibiting tumor growth in vivo.30 Lallemand F. and his colleague also confirmed that FOXO6 transcriptional factor has implications in the carcinogenesis of breast cancer.31 Thus, we hypothesized that FOXO4 may also play a role in breast cancer cell survival and apoptosis. A recent study has also confirmed that the down-regulation of FOXO4 was associated with high histological grade of breast cancer patients.32 Moreover, evidence suggested that FOXO4 leads to the inhibition of various responses to hypoxia.33 Consistent with these reports, our study indicates that FOXO4 overexpression suppressed hypoxia-induced MCF-7 cell survival and promoted its apoptosis.
HIF-2α activates and accumulates in cancer cells exposed to hypoxic stress, and are involved in cancer development.18 Helczynska K. and his colleague suggested that HIF-2α was associated with the poor outcome and distant recurrence in invasive breast cancer.34 Moreover, evidences confirmed that both the transcription factors HIF-1 and HIF-2 correlated with the poor prognosis and metastasis of breast cancers and indicated high-grade breast cancer types.26 HIF-2α is involved in the carcinogenesis and breast cancer progression via mediating inflammation, tissue remodeling, cell survival and death.26 In agreement with the previous reports, our study indicates that hypoxia treatment up-regulated the expression of HIF-2α in MCF-7 cells. The down-regulation of the expression of HIF-2α via FOXO4 overexpression impeded hypoxic stress-induced cell survival and promoted MC-7 cell apoptosis. In the present study, we also demonstrate that Bnip3, which is downstream of HIF-2α, plays essential roles in the effects of FOXO4 on hypoxia-treated MCF-7 cells.
5. Conclusion
In conclusion, our study has suggested that FOXO4 plays a pivotal role in the survival and apoptosis of MCF-7 cells exposed to hypoxia, and the HIF-2α/Bnip3 axis is involved in this process. However, additional cell and animal studies are needed to confirm if FOXO4 has any roles in breast cancer.Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
This study was supported by the project of the First Affiliated Hospital of Xi'an Jiaotong University (No. XJTU1AF-CRF-2018-001) in China.References
- A. Jemal, F. Bray, M. M. Center, J. Ferlay, E. Ward and D. Forman, Ca-Cancer J. Clin., 2011, 61, 69–90 CrossRef PubMed.
- M. Seifi-Alan, R. Shamsi, S. Ghafouri-Fard, R. Mirfakhraie, D. Zare-Abdollahi, A. Movafagh, M. H. Modarressi, G. Kazemi, L. Geranpayeh and M. Najafi-Ashtiani, Asian Pac. J. Cancer Prev., 2014, 14, 6625–6629 CrossRef PubMed.
- T. Sorlie, C. M. Perou, R. Tibshirani, T. Aas, S. Geisler, H. Johnsen, T. Hastie, M. B. Eisen, M. van de Rijn, S. S. Jeffrey, T. Thorsen, H. Quist, J. C. Matese, P. O. Brown, D. Botstein, P. E. Lonning and A. L. Borresen-Dale, Proc. Natl. Acad. Sci. U. S. A., 2001, 98, 10869–10874 CrossRef CAS PubMed.
- C. M. Perou, T. Sorlie, M. B. Eisen, M. van de Rijn, S. S. Jeffrey, C. A. Rees, J. R. Pollack, D. T. Ross, H. Johnsen, L. A. Akslen, O. Fluge, A. Pergamenschikov, C. Williams, S. X. Zhu, P. E. Lonning, A. L. Borresen-Dale, P. O. Brown and D. Botstein, Nature, 2000, 406, 747–752 CrossRef CAS PubMed.
- K. Polyak, J. Clin. Invest., 2011, 121, 3786–3788 CrossRef CAS PubMed.
- C. DeSantis, J. Ma, L. Bryan and A. Jemal, Ca-Cancer J. Clin., 2014, 64, 52–62 CrossRef PubMed.
- T. Walsh and M. C. King, Cancer Cell, 2007, 11, 103–105 CrossRef CAS PubMed.
- A. Coomans de Brachene and J. B. Demoulin, Cell. Mol. Life Sci., 2016, 73, 1159–1172 CrossRef CAS PubMed.
- Z. Fu and D. J. Tindall, Oncogene, 2008, 27, 2312–2319 CrossRef CAS PubMed.
- H. Huang and D. J. Tindall, J. Cell Sci., 2007, 120, 2479–2487 CrossRef CAS PubMed.
- H. Li, R. Ouyang, Z. Wang, W. Zhou, H. Chen, Y. Jiang, Y. Zhang, M. Liao, W. Wang, M. Ye, Z. Ding, X. Feng, J. Liu and B. Zhang, Sci. Rep., 2016, 6, 39001–39011 CrossRef CAS PubMed.
- I. K. Kwon, R. Wang, M. Thangaraju, H. Shuang, K. Liu, R. Dashwood, N. Dulin, V. Ganapathy and D. D. Browning, Oncogene, 2010, 29, 3423–3434 CrossRef CAS PubMed.
- Y. Wang, X. L. Kang, F. C. Zeng, C. J. Xu, J. Q. Zhou and D. N. Luo, Pathol., Res. Pract., 2017, 213, 766–772 CrossRef CAS PubMed.
- S. Terry, R. Faouzi Zaarour, G. Hassan Venkatesh, A. Francis, W. El-Sayed, S. Buart, P. Bravo, J. Thiery and S. Chouaib, Int. J. Mol. Sci., 2018, 19, E3044–E3062 CrossRef PubMed.
- G. L. Semenza, Cell, 2012, 148, 399–408 CrossRef CAS PubMed.
- G. L. Semenza, Annu. Rev. Phytopathol., 2014, 9, 47–71 CrossRef CAS PubMed.
- H. Tian, S. L. McKnight and D. W. Russell, Genes Dev., 1997, 11, 72–82 CrossRef CAS PubMed.
- J. Zhao, F. Du, G. Shen, F. Zheng and B. Xu, Cell Death Dis., 2015, 6, e1600 CrossRef CAS PubMed.
- J. D. Gordan and M. C. Simon, Curr. Opin. Genet. Dev., 2007, 17, 71–77 CrossRef CAS PubMed.
- K. L. Talks, H. Turley, K. C. Gatter, P. H. Maxwell, C. W. Pugh, P. J. Ratcliffe and A. L. Harris, Am. J. Pathol., 2000, 157, 411–421 CrossRef CAS PubMed.
- G. Bangoura, Z. S. Liu, Q. Qian, C. Q. Jiang, G. F. Yang and S. Jing, World J. Gastroenterol., 2007, 13, 3176–3182 CrossRef CAS PubMed.
- G. Xia, Y. Kageyama, T. Hayashi, S. Kawakami, M. Yoshida and K. Kihara, Cancer, 2001, 91, 1429–1436 CrossRef CAS PubMed.
- T. Imamura, H. Kikuchi, M. T. Herraiz, D. Y. Park, Y. Mizukami, M. Mino-Kenduson, M. P. Lynch, B. R. Rueda, Y. Benita, R. J. Xavier and D. C. Chung, Int. J. Cancer, 2009, 124, 763–771 CrossRef CAS PubMed.
- S. C. Winter, K. A. Shah, C. Han, L. Campo, H. Turley, R. Leek, R. J. Corbridge, G. J. Cox and A. L. Harris, Cancer, 2006, 107, 757–766 CrossRef CAS PubMed.
- Y. Guan, K. R. Reddy, Q. Zhu, Y. Li, K. Lee, P. Weerasinghe, J. Prchal, G. L. Semenza and N. Jing, Mol. Ther., 2010, 18, 188–197 CrossRef CAS PubMed.
- S. Påhlman, L.R. Lund and A. Jögi, PLoS One, 2015, 10, e0125771 CrossRef PubMed.
- A. Valli, M. Rodriguez, L. Moutsianas, R. Fischer, V. Fedele, H. L. Huang, R. Van Stiphout, D. Jones, M. Mccarthy, M. Vinaxia, K. Igarashi, M. Sato, T. Soga, F. Buffa, J. Mccullagh, O. Yanes, A. Harris and B. Kessler, Oncotarget, 2015, 6, 1920–1941 CrossRef PubMed.
- Y. Cui, Y. Huang, X. Wu, M. Zheng, Y. Xia, Z. Fu, H. Ge, S. Wang and H. Xie, J. Cell. Physiol., 2018, 26, 27533 Search PubMed.
- D. Z. Liu, B. Chang, X. D. Li, Q. H. Zhang and Y. H. Zou, Clin. Transl. Oncol., 2017, 19, 1133–1140 CrossRef CAS PubMed.
- S. H. Park, Y. M. Chung, J. Ma, Q. Yang, J. S. Berek and M. C. Hu, Oncotarget, 2016, 7, 42110–42125 Search PubMed.
- F. Lallemand, A. Petitalot, S. Vacher, L. de Koning, K. Taouis, B. S. Lopez, S. Zinn-Justin, N. Dalla-Venezia, W. Chemlali, A. Schnitzler, R. Lidereau, I. Bieche and S. M. Caputo, Oncotarget, 2017, 9, 7464–7475 Search PubMed.
- A. Rehman, Y. Kim, H. Kim, J. Sim, H. Ahn, M. S. Chung, S. J. Shin and K. Jang, J. Clin. Pathol., 2018, 71, 806–813 CrossRef CAS PubMed.
- T. T. Tang and L. A. Lasky, J. Biol. Chem., 2003, 278, 30125–30135 CrossRef CAS PubMed.
- K. Helczynska, A. M. Larsson, L. Holmquist Mengelbier, E. Bridges, E. Fredlund, S. Borgquist, G. Landberg, S. Påhlman and K. Jirström, Cancer Res., 2008, 68, 9212–9220 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2019 |