Open Access Article
Open Access ArticleIncreased expression of core-fucosylated glycans in human lung squamous cell carcinoma
Tianran Maa,
Yan Wanga,
Liyuan Jiaa,
Jian Shua,
Hanjie Yua,
Haoqi Dua,
Jiajun Yanga,
Yiqian Liangb,
Mingwei Chen*b and
Zheng Li *a
*a
aLaboratory for Functional Glycomics, College of Life Sciences, Northwest University, No. 229 Taibai Beilu, Xi'an 710069, China. E-mail: zhengli@nwu.edu.cn; Fax: +86-29-88303572; Tel: +86-29-88304104
bDepartment of Respiratory and Critical Care Medicine, The First Affiliated Hospital of School of Medicine of Xi'an, Jiaotong University, Xi'an 710061, China. E-mail: chenmw36@163.com
First published on 16th July 2019
Abstract
Lung cancer is the most frequent cancer and the leading cause of cancer around the world. As one of the major types of lung cancer, lung squamous cell carcinoma (LUSC) is closely associated with smoking and shows poor sensitivity to therapy and prognosis. Although alteration of glycopatterns are reliable indicators of cancer, little is known about the alterations of protein glycosylation related to LUSC. In this study, we compared the differential expression levels of glycopatterns in seven pairs of LUSC tissues and normal pericarcinomatous tissues (PCTs) using lectin microarrays. Fluorescence-based lectin histochemistry and lectin blotting were utilized to validate and assess the expression and distribution of certain glycans in LUSC tissues and PCTs. And we further analyzed their total N-linked glycans using MALDI-TOF/TOF-MS to provide more information about the aberrant glycopatterns. The results showed that the expression level of the core fucosylation recognized by Pisum sativum agglutinin (PSA) and Lens culinaris agglutinin (LCA) was significantly increased in LUSC tissues compared with PCTs. There were 10 and 15 fucosylated N-linked glycans that were detected in PCTs and LUSC tissues respectively, 10 fucosylated N-glycans were common, while five fucosylated N-glycans were unique to LUSC tissues. And the abundance of the fucosylated N-glycans was increased from 40.9% (PCTs) to 48.3% (LUSC). These finding is helpful to elucidate the molecular mechanisms underlying the lung diseases and develop new treatment strategies.
1. Introduction
Lung cancer is the major malignant tumor with the highest incidence and mortality in both sexes around the world; there were about 2.1 million new lung cancer cases and 1.8 million deaths predicted in 2018.1 Lung cancer can be classified into four major histologic types of lung squamous cell carcinoma (LUSC), adenocarcinoma, small cell carcinoma, and large cell carcinoma.2,3 As the major types of lung cancer, LUSC is associated with smoking and represents approximately 30% of all cases of non-small cell lung cancer.4,5 Although kinase inhibitors (targeting epidermal growth factor receptor, anaplastic lymphoma kinase and c-ros oncogene 1) and immune checkpoint inhibitors (anti-programmed cell death 1 antibodies, anti-programmed cell death ligand 1 antibodies, atezolizumab and durvalumab) have been widely used in clinical practices according to the aberrant status of the cancer, it still a quite limited number of LUSC cases that were actually suitable and efficient for these treatment strategies.6–10 It is still an urgent requirement to investigate the definite molecular mechanism and antitumor treatment strategies of LUSC.11,12 Therefore, identification of the difference between tumor tissues and adjacent tissues is very meaningful for contributing to the molecular mechanism research.Glycosylation is one of the most ubiquitous and crucial post-translational modifications, occurring on virtually all known proteins in mammalian cells and involving in almost all important biological processes.13 The alteration of the glycosylation occupancy or specific glycan structures are often detected during inflammation, infection, neoplastic transformation and cancer.13,14 One of the most important types of glycosylation, core fucosylation is only catalyzed by fucosyltransferase 8 (FUT8).15,16 Evidences showed that the increased expression level of the core fucosylation is detected in variety of disease, such as chronic pancreatitis, hepatocellular carcinoma and inflammatory bowel disease.17–19 And the up-regulation of FUT8, also has been observed in several cancers including colorectal cancers, brain cancer, and lung cancer.20–22 Although the FUT8 as a functional regulator of non-small cell lung cancer is revealed,22 the correlation between aberrant glycans and LUSC is still no clear. Therefore, exploring the glycopatterns of LUSC tissues is also necessary to research the pathogenesis of LUSC.
In this study, the glycopatterns of seven pairs of lung tissues including tumor and normal pericarcinomatous tissues (PCTs) obtained from LUSC patients were detected using the lectin microarrays. The results were verified by fluorescence-based lectin blotting and histochemistry. Furthermore, the total N-linked glycans were identified using MALDI-TOF/TOF-MS to provide more information about the aberrant glycopatterns between PCTs and LUSC tissues. This study provides the comprehensive information of glycopatterns from lung LUSC tissues and PCTs, which is helpful to elucidate the molecular mechanisms underlying the lung diseases and develop new treatment strategies.
2. Materials and methods
2.1 Clinical specimens
The collection and use of all human pathological specimens for the research presented here were approved by the Ethical Committee of the First Affiliated Hospital of Xi'an Jiao Tong University and Northwest University in Xi'an, China. All patients enrolled in the study provided written informed consent. This study was conducted in accordance with the ethical guidelines of the Declaration of Helsinki.Seven pairs of lung tissues including normal PCTs and tumor tissues were obtained from seven LUSC patients, who had no history of treatment before their operation in the First Affiliated Hospital of Xi'an Jiao Tong University (Xi'an, China). Detailed information on patients is described in Table 1. The lung tissues were obtained during surgical resection and were snap-frozen in liquid nitrogen before being stored at −80 °C until required for use. PCTs were collected at a distance of at least 5 cm from the cancer lesions. All the tissues were histologically examined and were confirmed by experienced pathologists. Tissue microarrays which comprised of paraffin-embedded LUSC tissues and PCTs were purchased from Shanghai Outdo Biotech Co. Ltd. (Shanghai, China).
2.2 Tissue protein extraction
The total protein of lung tissue was extracted with T-PER (Thermo Scientific, Herts, UK) according to the manufacturer's instructions. Approximately 30 mg of lung tissue was homogenized in a mixer on ice in 1 mL of T-PER containing a 1% protease inhibitor cocktail (complete, EDTA-free, Roche, Basel, Switzerland). Following centrifugation at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 5 min, the supernatant was immediately transferred for use or stored at −80 °C. The protein concentration was determined using a BCA assay.
000g for 5 min, the supernatant was immediately transferred for use or stored at −80 °C. The protein concentration was determined using a BCA assay.
2.3 Lectin microarrays and data analysis
The lectin microarrays were produced using 37 lectins with different binding preferences covering N- and O-linked glycans. The Cy3-labeled proteins were incubated in the lectin microarrays to detect the different glycopatterns among clinical samples. The lectin microarrays were produced according to the protocol.232.4 Lectin blotting
To normalize the differences between subjects and to tolerate individual variation, samples in each group were pooled, respectively. The expression level of specific glycans was further analyzed by lectin blotting according to the protocol.24 The pooled tissue proteins of each group were subjected to 10% SDS-PAGE electrophoresis, and transferred to PVDF membranes incubated with Cy5-labeled PSA and LCA.2.5 Real-time quantitative PCR
RNA isolation, cDNA synthesis and quantitative real-time polymerase chain reaction (qRT-PCR) were described in the previous manuscript.25 The gene specific primers used in this study including FUT8 (F: 5′-AACTGGTTCAGCGGAGAATAACR-3′; R: 5′-TGAGATTCCAAGATGAGTGTTCG-3′); and GAPDH (F: 5′-GGAGCGAGATCCCTCCAAAAT-3′; R: 5′-GGCTGTTGTCATACTTCTCATGG -3′). The mRNA levels of the genes were normalized to the amount of GAPDH in each sample, and quantization of gene expression level was determined using the 2−ΔΔCT comparative method.2.6 Fluorescence-based lectin histochemistry
Cy3-labeled lectins were applied to detect the specific glycan structures present on the lung tissue sections as described.24 Briefly, formalin-fixed paraffin-embedded tissue microarrays were dewaxed and hydrated in a series of different concentrations of dimethylbenzene and ethanol. The tissue microarrays were rinsed 3 times with PBS for 5 min and blocked with blocking buffer (PBS supplemented with 5% BSA) at 25 °C for 30 min. Thereafter, the sections were incubated with a solution containing a final concentration of 100 μg mL−1 Cy3-labeled lectins and 5% (w/v) BSA for 3 h at room temperature, in the dark. Finally, sections were stained with DAPI (1 μg mL−1 in PBS; Roche) for 10 min before the final rinse. A laser scanning confocal microscope FV 1000 (Olympus, Tokyo, Japan) was used to collect the images using the merge channels of Cy3 and DAPI.2.7 Isolation and purification of N-linked glycans
The N-glycans released by PNGase F glycosidase (New England Biolabs, Beverly, MA) according to previous protocols.26 Briefly, 200 μg glycoproteins were concentrated and desalted by adding to a 3K centrifugal ultrafiltration. Then, the obtained glycoproteins were denatured with 8 M urea, 10 mM DTT and 10 mM IAM, and exchanged buffer into 40 mM NH4HCO3 by using 3K centrifugal ultrafiltration. After that, 2 μg trypsin was added to digest the glycoproteins overnight at 37 °C. The mixture was heated at 80 °C for 5 min to deactivate the activity of trypsin. After the temperature of mixture restored to room temperature, 2 μL PNGase F was added and incubated at 37 °C overnight to release the N-linked glycans from the glycopeptides. Finally, the filtrates that contained peptides and N-linked glycans were collected by centrifugation, and the released glycans were purified with HyperSep Hypercarb SPE cartridges (25 mg, 1 mL; Thermo Scientific) according to the manufacturer's recommendation. The glycans were eluted by 0.5 mL of elution solution (50% (v/v) acetonitrile with 0.1% (v/v) TFA). The purified glycans were collected and lyophilized.2.8 Characterization of the N-linked glycans by MALDI-TOF/TOF-MS
The purified glycans were characterized by MALDI-TOF/TOF-MS (UltrafleXtreme, Bruker Daltonics). Glycans were resuspended in 10 μL of methanol, and 1 μL was spotted directly on the MTP AnchorChip sample target and dried. Then 1 μL of 10 mg mL−1 DHB in 50% (v/v) methanol solution was spotted to the recrystallize glycans. Mass calibration was performed using peptide calibration standards (250 calibration points; Bruker). Measurements were taken in positive-ion mode, m/z data were generated and analyzed using FlexAnalysis and GlycoWorkbench software, respectively. Relative intensity was calculated by dividing the intensity of a given type of glycan by the highest glycans intensity of its MS spectra.3. Results
3.1 The alterations of glycopatterns in LUSC tissues versus PCTs
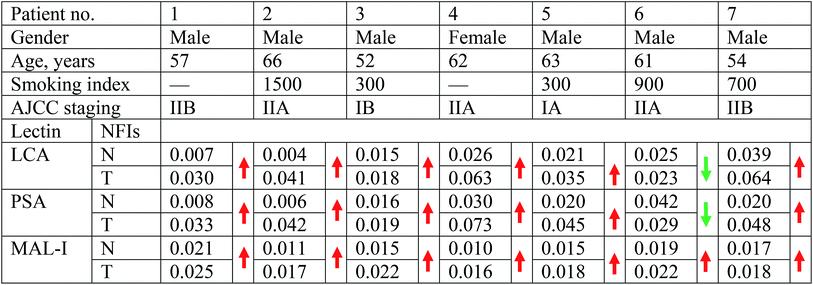
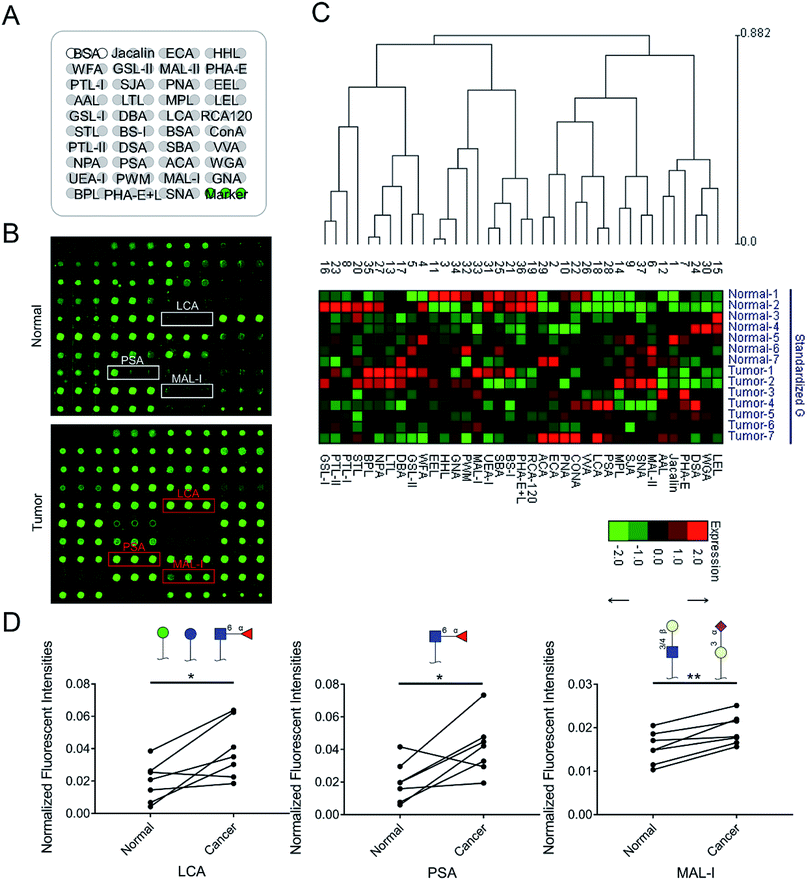
To identify the abnormal glycopatterns associated with LUSC, the lectin microarrays were utilized to compare the glycopatterns between the paired PCTs and LUSC tissues. The layout of the lectin microarrays and one pair of Cy3-labeled proteins from PCTs and LUSC tissues bound to the lectin microarrays were shown in Fig. 1A and B. The generated data from each sample was imported into EXPANDER software and furtherly analyzed using a hierarchical clustering method to exhibit the overview of the glycopatterns among all the paired tissue samples (Fig. 1C). By the paired t-test analysis, it was found that the Galβ-1,3/4 GlcNAc, Siaα2-3Gal binder MAL-I exhibited significantly increased the normalized fluorescent intensities (NFIs) in the LUSC tissues against PCTs (P < 0.05) among all the paired tissue samples, and the α-D-Man, Fucα-1,6GlcNAc (core fucose), α-D-Glc binder LCA and Fucα-1,6GlcNAc binder PSA showed significantly increased the NFIs among six paired tissue samples (Table 1). However, the NFIs of other lectins were not significantly different between LUSC tissues and PCTs. Therefore, the abnormal expression of Fucα-1,6GlcNAc structure was perceived as the most important changes between LUSC tissues and PCTs.3.2 Validation of the differences glycopatterns between LUSC tissues and PCTs
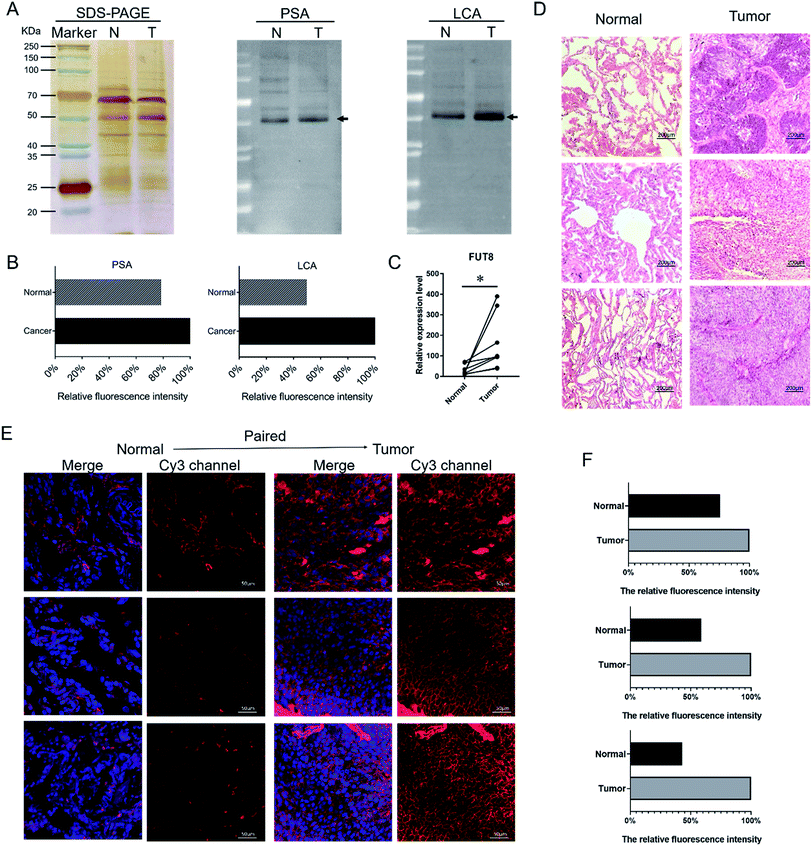
According to the results of the lectin microarrays, there was an abnormal expression of Fucα-1,6GlcNAc structure recognized specifically by PSA and LCA in LUSC tissues. SDS-PAGE and lectin blotting analysis were performed with silver staining, Cy5-labeled PSA and LCA staining to rapidly validate the target glycan structures, respectively (Fig. 2A). The results of SDS-PAGE demonstrated that the protein bands from each paired tissue sample were similar. The results of the lectin blotting analysis showed that there was only one major band with molecular weight of approximately 53 kDa that showed an apparently increased fluorescence intensities in LUSC tissues compared with PCTs (Fig. 2B). Besides, the increased expression level of Fucα-1,6GlcNAc structure in LUSC tissues was also verified at the genetic level. FUT8 as the only enzyme responsible for α1,6-linked core fucosylation by adding fucose to the innermost GlcNAc residue of an N-linked glycan was detected using qRT-PCR. The data showed that the gene expression level of FUT8 was significantly increased in LUSC tissues compared to PCTs (Fig. 2C). These results were basically consistent with the results of lectin microarrays.3.3 Expression and distribution of the specific glycan in the tissue sections
To further validate and assess the expression and distribution of the Fucα-1,6GlcNAc in PCTs and LUSC tissues, fluorescence-based lectin histochemistry was performed with PSA lectins. The hematoxylin–eosin (H&E) pathological staining were performed to verify the LUSC tissue (Fig. 2D). The result of H&E illustrated that nuclear atypia of LUSC tissues was more clear than that of PCTs. Lectin histochemistry indicated that PSA strongly bind to the cytoplasm and the membrane of lung cells (Fig. 2E). As a result, it is obvious that the fluorescence signal intensity of PSA was significantly increased in LUSC tissues compared to PCTs (Fig. 2E and F). The result demonstrated that the core fucosylated glycan increased on the surface and cytoplasm of lung cells when LUSC occurred. And the result was in accord with the above experiments and verifications.3.4 Alterations of fucosylated N-linked glycan profiles in LUSC tissues versus PCTs
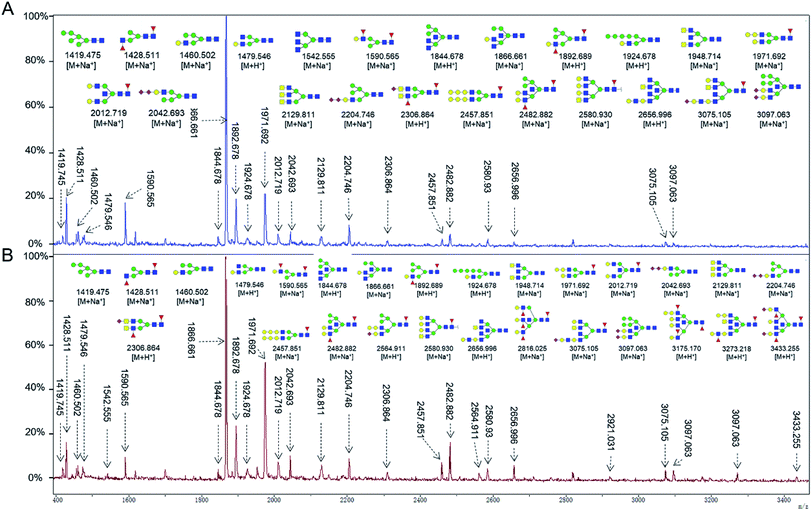
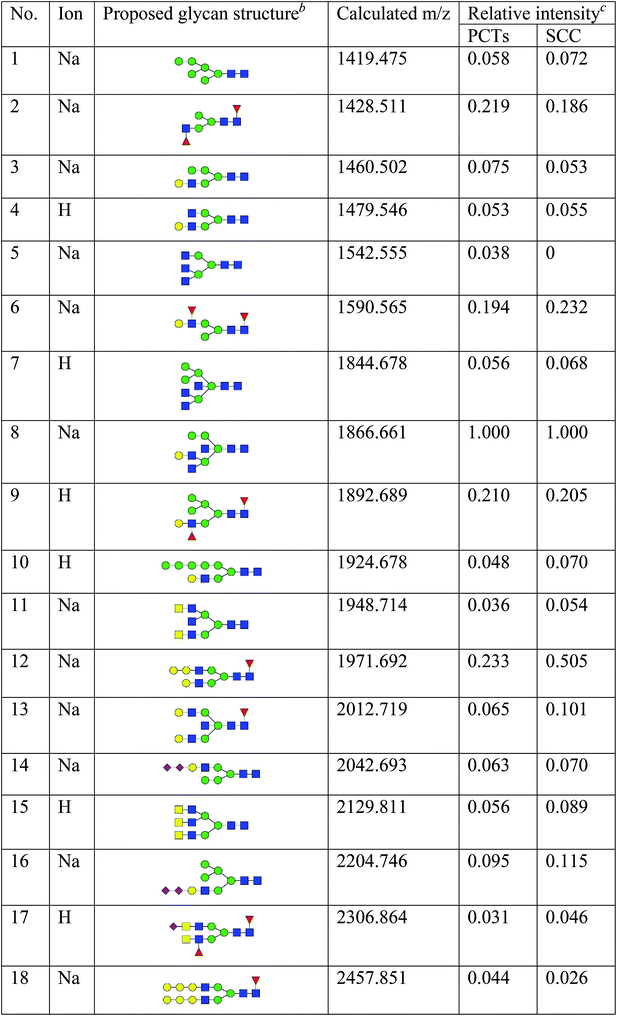
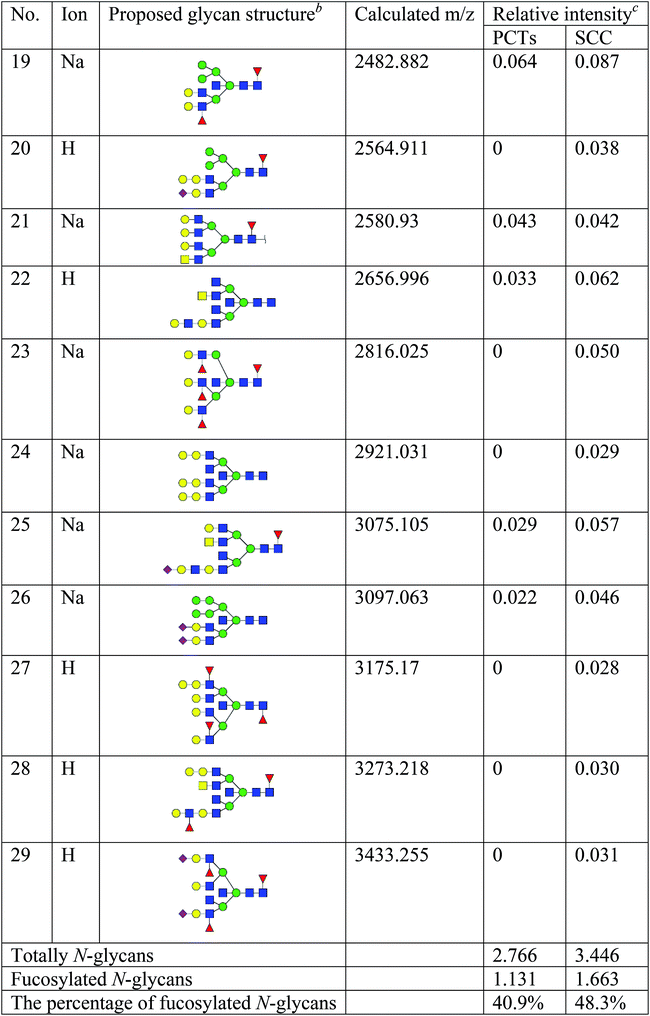
To obtain detail structural information of glycoproteins from the tumor and PCTs. The N-linked glycans were released from glycoproteins by PNGase F and identified by MALDI-TOF/TOF-MS. The identified N-linked glycans and their proposed structures from the pooled tissue samples of LUSC and PCTs were shown in Fig. 3 and Table 2. A total of 23 and 28 N-linked glycans were identified and annotated, respectively. Of these, 22 N-linked glycans (e.g., m/z 1419.475, 1428.511 and 1460.502) were observed both in PCTs and LUSC tissues, while one N-linked glycans (m/z 1542.555) detected only in PCTs and six N-linked glycans (e.g., m/z 2564.911, 2816.025, and 2921.031) detected only in LUSC tissues. The MS/MS spectra of the precursor ions m/z 1866.661, 2204.746, and 2580.930 were illustrated in Fig. 4A–C.| a Monosaccharides are represented according to MS-tools from the GlycoWorkbench software (GlcNAc, blue square; Man, green circle; Gal, yellow circle; Fuc, red triangle; NeuAc, purple diamond).b The proposed glycan structure.c The relative intensity was calculated by dividing the intensity of a given type of glycan by the highest glycans intensity of its MS spectra. |
|---|
 |
 |
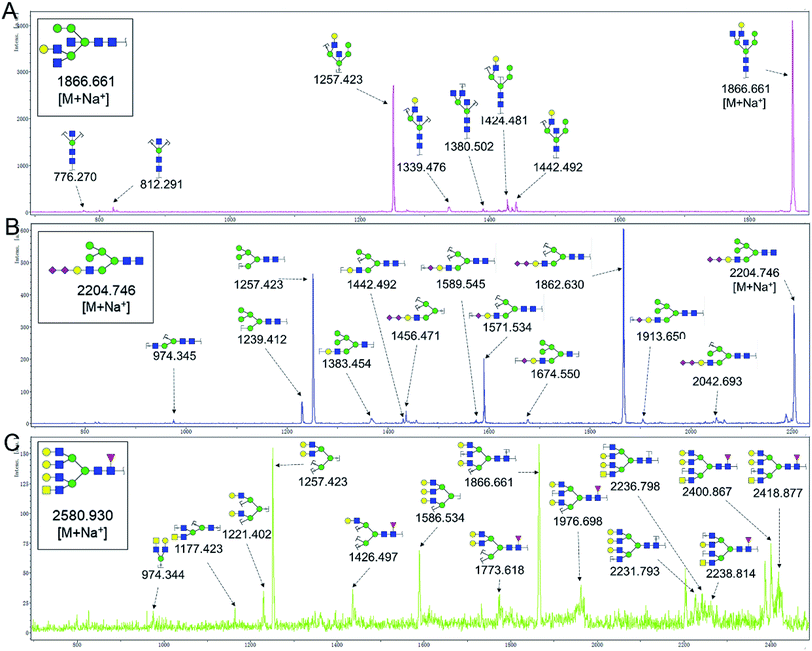 | ||
| Fig. 4 MALDI-TOF/TOF MS/MS analyzing the N-glycan precursor ion from MS spectra. With example, the three N-glycan peaks (A) m/z 1866.661, (B) 2204.746, and (C) 2580.930, subjected to MS/MS analysis. | ||
Focusing on the fucosylated N-glycans, it is noticeable that there were 10 and 15 fucosylated N-linked glycans were detected in PCTs and LUSC tissues, respectively. Of these fucosylated N-glycans, there was an overlap of 10 fucosylated N-glycan peaks (e.g., m/z 1428.51,1 1590.565 and 1892.689) between PCTs and LUSC tissues. Besides, five fucosylated N-glycans (including m/z 2564.911, 2816.025, 3175.17, 3273.218, and 3433.255) were unique to LUSC tissues. In fact, not only the numbers but also the relative intensity of fucosylated glycans were shown significant increasing in the LUSC tissues compared with that in PCTs. The sum relative intensity of fucosylated glycans were increased from 1.131 in PCTs to 1.663 in LUSC tissues. And the proportion of the fucosylated N-linked glycans was significantly increased from 40.9% (PCTs) to 48.3% (LUSC).
4. Discussion
Glycosylation involve in a wide range of biological processes including cellular communication, adhesion, interaction, malignant transformation and metastasis.27,28 The synthesis of glycan is a complex process with the coordination of multiple organelles and strictly regulated by a set of enzymes, which result innumerous possibilities for branching and anomeric linkage.13,29 Given the complexity of protein glycosylation and its fundamental effects on a variety of biological processes, it is no doubt that aberrant glycosylation not only represents a characteristic of cancer, but also significantly impact the biology of cells, thus analysis of the abnormal glycopatterns in malignant tissues are contribute to understand the mechanisms of cancerization.13,30–33Our previous study investigated the glycopatterns of serum proteins in non-small cell lung cancer (NSCLC) patients at different stages using a lectin microarray, and systematically compare the alterations of serum glycopatterns between NSCLC patients and healthy controls. There were 18 lectins (e.g., AAL, Jacalin, GSL-I and DBA) and 16 lectins (e.g., Jacalin, HHL, and PHA-E + L) exhibited significantly alterations of serum glycopatterns in lung adenocarcinoma and LUSC patients compared to healthy group, respectively.34 Meanwhile, the levels of core fucosylation was elevated and showed positive correlated with the development of lung adenocarcinoma, but not in LUSC patients.34 In the present study, the different glycopatterns between LUSC tissues and PCTs were detected by the lectin microarrays. The MAL-I, LCA and PSA showed significantly increased NFIs in the LUSC tissues. And the level of the core fucosylation identified by PSA and LCA and FUT8 was significantly elevated in LUSC tissues in this study. Previous studies also showed that the expression level of FUT8 was elevated in NSCLC tissues and further indicated that up-regulation of FUT8 contributes to tumor progression through multiple mechanisms.22,35,36 The elevated core fucosylation and transcription of FUT8 have been also observed in sera of hepatocellular carcinoma, prostate and ovarian cancer patients.37–39
In conclusion, the present study systematically investigated the glycopatterns between LUSC tissues and PCTs. The glycopattern of core fucosylation was significantly increased in LUSC tissues compared with PCTs. Furthermore, the numbers of N-glycans with fucosylation were increased remarkably in LUSC tissues, five fucosylated N-glycans were detected only in LUSC tissues, and the abundance of fucosylated N-glycans was increased from 40.9% (PCTs) to 48.3% (LUSC). Our data provided useful information in order to elucidate the molecular mechanisms of LUSC and develop new treatment strategies. However, the association among abnormal glycosylation and malignant transformation, infiltration, and migration are still needed to further explore.
Funding
This work is supported by National Natural Science Foundation of China (Grant No. 81871955) and the Northwest University Doctorate Dissertation of Excellence Funds (Grant No. YYB17018).Conflicts of interest
The authors declare that there are no conflicts of interest.Abbreviations
| FUT8 | Fucosyltransferase 8 |
| HCA | Hierarchical cluster analysis |
| H&E | Hematoxylin–eosin |
| LCA | Lens culinaris agglutinin |
| LUSC | Lung squamous cell carcinoma |
| MAL-I | Maackia amurensis lectin I |
| NSCLC | Non-small cell lung cancer |
| PCTs | Pericarcinomatous tissues |
| PSA | Pisum sativum agglutinin |
| qRT-PCR | Quantitative real-time polymerase chain reaction |
References
- F. Bray, J. Ferlay, I. Soerjomataram, R. L. Siegel, L. A. Torre and A. Jemal, Ca-Cancer J. Clin., 2018, 68, 394–424 CrossRef.
- W. D. Travis, E. Brambilla, A. G. Nicholson, Y. Yatabe, J. H. M. Austin, M. B. Beasley, L. R. Chirieac, S. Dacic, E. Duhig, D. B. Flieder, K. Geisinger, F. R. Hirsch, Y. Ishikawa, K. M. Kerr, M. Noguchi, G. Pelosi, C. A. Powell, M. S. Tsao and I. Wistuba, J. Thorac. Oncol., 2015, 10, 1243–1260 CrossRef.
- W. D. Travis, Clin. Chest. Med., 2011, 32, 669–692 CrossRef.
- J. Brahmer, K. L. Reckamp, P. Baas, L. Crino, W. E. Eberhardt, E. Poddubskaya, S. Antonia, A. Pluzanski, E. E. Vokes, E. Holgado, D. Waterhouse, N. Ready, J. Gainor, O. Aren Frontera, L. Havel, M. Steins, M. C. Garassino, J. G. Aerts, M. Domine, L. Paz-Ares, M. Reck, C. Baudelet, C. T. Harbison, B. Lestini and D. R. Spigel, N. Engl. J. Med., 2015, 373, 123–135 CrossRef CAS.
- S. Senoo, K. Ninomiya, K. Hotta and K. Kiura, Int. J. Clin. Oncol., 2019, 24, 461–467 CrossRef.
- T. Tamura, Y. Kato, K. Ohashi, K. Ninomiya, G. Makimoto, H. Gotoda, T. Kubo, E. Ichihara, T. Tanaka, K. Ichimura, Y. Maeda, K. Hotta and K. Kiura, Biochem. Biophys. Res. Commun., 2018, 495, 360–367 CrossRef CAS.
- H. Isozaki, K. Hotta, E. Ichihara, N. Takigawa, K. Ohashi, T. Kubo, T. Ninomiya, K. Ninomiya, N. Oda, H. Yoshioka, H. Ichikawa, M. Inoue, I. Takata, T. Shibayama, S. Kuyama, K. Sugimoto, D. Harada, S. Harita, T. Sendo, M. Tanimoto and K. Kiura, Clin. Lung Cancer, 2016, 17, 602–605 CrossRef CAS PubMed.
- K. T. Bui, W. A. Cooper, S. Kao and M. Boyer, J. Clin. Med., 2018, 7, E192 CrossRef.
- T. Shukuya, T. Takahashi, R. Kaira, A. Ono, Y. Nakamura, A. Tsuya, H. Kenmotsu, T. Naito, K. Kaira, H. Murakami, M. Endo, K. Takahashi and N. Yamamoto, Cancer Sci., 2011, 102, 1032–1037 CrossRef CAS.
- X. Song, X. Han, F. Yu, X. Zhang, L. Chen and C. Lv, Theranostics, 2018, 8, 2217–2228 CrossRef CAS.
- X. Zhang, N. He, Y. Huang, F. Yu, B. Li, C. Lv and L. Chen, Sens. Actuators, B, 2019, 282, 69–77 CrossRef CAS.
- M. Cumberbatch, X. Tang, G. Beran, S. Eckersley, X. Wang, R. P. Ellston, S. Dearden, S. Cosulich, P. D. Smith, C. Behrens, E. S. Kim, X. Su, S. Fan, N. Gray, D. P. Blowers, I. I. Wistuba and C. Womack, Clin. Cancer Res., 2014, 20, 595–603 CrossRef CAS PubMed.
- S. R. Stowell, T. Ju and R. D. Cummings, Annu. Rev. Pathol.: Mech. Dis., 2015, 10, 473–510 CrossRef CAS PubMed.
- C. Reily, T. J. Stewart, M. B. Renfrow and J. Novak, Nat. Rev. Nephrol., 2019, 15, 346–366 CrossRef.
- Q. Yang, R. Zhang, H. Cai and L. X. Wang, J. Biol. Chem., 2017, 292, 14796–14803 CrossRef CAS.
- Y. Wang, T. Fukuda, T. Isaji, J. Lu, W. Gu, H. H. Lee, Y. Ohkubo, Y. Kamada, N. Taniguchi, E. Miyoshi and J. Gu, Sci. Rep., 2015, 5, 8264 CrossRef CAS.
- M. Ueda, Y. Kamada, S. Takamatsu, M. Shimomura, T. Maekawa, T. Sobajima, H. Fujii, K. Nakayama, K. Nishino, M. Yamada, Y. Kobayashi, T. Kumada, T. Ito, H. Eguchi, H. Nagano and E. Miyoshi, Pancreatology, 2016, 16, 238–243 CrossRef CAS.
- J. Ma, M. Sanda, R. Wei, L. Zhang and R. Goldman, J. Proteomics, 2018, 189, 67–74 CrossRef CAS PubMed.
- H. Fujii, S. Shinzaki, H. Iijima, K. Wakamatsu, C. Iwamoto, T. Sobajima, R. Kuwahara, S. Hiyama, Y. Hayashi, S. Takamatsu, N. Uozumi, Y. Kamada, M. Tsujii, N. Taniguchi, T. Takehara and E. Miyoshi, Gastroenterology, 2016, 150, 1620–1632 CrossRef CAS.
- M. Noda, H. Okayama, Y. Kofunato, S. Chida, K. Saito, T. Tada, M. Ashizawa, T. Nakajima, K. Aoto, T. Kikuchi, W. Sakamoto, H. Endo, S. Fujita, M. Saito, T. Momma, S. Ohki and K. Kono, PLoS One, 2018, 13, e0200315 CrossRef.
- L. Veillon, C. Fakih, H. Abou-El-Hassan, F. Kobeissy and Y. Mechref, ACS Chem. Neurosci., 2018, 9, 51–72 CrossRef CAS.
- C. Y. Chen, Y. H. Jan, Y. H. Juan, C. J. Yang, M. S. Huang, C. J. Yu, P. C. Yang, M. Hsiao, T. L. Hsu and C. H. Wong, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 630–635 CrossRef CAS.
- J. Shu, H. Yu, X. Li, D. Zhang, X. Liu, H. Du, J. Zhang, Z. Yang, H. Xie and Z. Li, Oncotarget, 2017, 8, 35718–35727 Search PubMed.
- Y. Qin, Y. Zhong, M. Zhu, L. Dang, H. Yu, Z. Chen, W. Chen, X. Wang, H. Zhang and Z. Li, J. Proteome Res., 2013, 12, 2742–2754 CrossRef CAS.
- H. Yu, M. Zhu, Y. Qin, Y. Zhong, H. Yan, Q. Wang, H. Bian and Z. Li, J. Proteome Res., 2012, 11, 5277–5285 CrossRef CAS.
- J. Shu, H. Yu, H. Du, J. Zhang, K. Zhang, X. Li, H. Xie and Z. Li, Cancer Biomarkers, 2018, 22, 669–681 CAS.
- J. Shu, L. Dang, D. Zhang, P. Shah, L. Chen, H. Zhang and S. Sun, FEBS J., 2019, 286, 1594–1605 CrossRef CAS.
- M. Dalziel, M. Crispin, C. N. Scanlan, N. Zitzmann and R. A. Dwek, Science, 2014, 343, 1235681 CrossRef.
- K. W. Moremen, M. Tiemeyer and A. V. Nairn, Nat. Rev. Mol. Cell Biol., 2012, 13, 448–462 CrossRef CAS PubMed.
- L. Oliveira-Ferrer, K. Legler and K. Milde-Langosch, Semin. Cancer Biol., 2017, 44, 141–152 CrossRef CAS.
- J. H. Rho, J. J. Ladd, C. I. Li, J. D. Potter, Y. Zhang, D. Shelley, D. Shibata, D. Coppola, H. Yamada, H. Toyoda, T. Tada, T. Kumada, D. E. Brenner, S. M. Hanash and P. D. Lampe, Gut, 2018, 67, 473–484 CrossRef CAS.
- X. Liu, H. Yu, Y. Qiao, J. Yang, J. Shu, J. Zhang, Z. Zhang, J. He and Z. Li, EBioMedicine, 2018, 28, 70–79 CrossRef.
- A. Silsirivanit, Adv. Clin. Chem., 2019, 89, 189–213 Search PubMed.
- Y. Liang, P. Han, T. Wang, H. Ren, L. Gao, P. Shi, S. Zhang, A. Yang, Z. Li and M. Chen, Clin. Proteomics, 2019, 16, 20 CrossRef.
- R. Honma, I. Kinoshita, E. Miyoshi, U. Tomaru, Y. Matsuno, Y. Shimizu, S. Takeuchi, Y. Kobayashi, K. Kaga, N. Taniguchi and H. Dosaka-Akita, Oncology, 2015, 88, 298–308 CrossRef CAS.
- C. F. Tu, M. Y. Wu, Y. C. Lin, R. Kannagi and R. B. Yang, Breast Cancer Res., 2017, 19, 111 CrossRef.
- W. R. Alley Jr, J. A. Vasseur, J. A. Goetz, M. Svoboda, B. F. Mann, D. E. Matei, N. Menning, A. Hussein, Y. Mechref and M. V. Novotny, J. Proteome Res., 2012, 11, 2282–2300 CrossRef.
- X. Wang, J. Chen, Q. K. Li, S. B. Peskoe, B. Zhang, C. Choi, E. A. Platz and H. Zhang, Glycobiology, 2014, 24, 935–944 CrossRef CAS.
- A. Mehta, P. Norton, H. Liang, M. A. Comunale, M. Wang, L. Rodemich-Betesh, A. Koszycki, K. Noda, E. Miyoshi and T. Block, Cancer Epidemiol., Biomarkers Prev., 2012, 21, 925–933 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2019 |