Open Access Article
Open Access ArticleImpact of physical structure of granular sludge on methanogenesis and methanogenic community structure
Xiaofang
Pan
a,
Lina
Wang
b,
Nan
Lv
ac,
Jing
Ning
ac,
Mingdian
Zhou
ac,
Tao
Wang
ac,
Chunxing
Li
d and
Gefu
Zhu
 *a
*a
aKey Laboratory of Urban Pollutant Conversion, Institute of Urban Environment, Chinese Academy of Sciences, Xiamen 361021, China. E-mail: gfzhu@iue.ac.cn; Fax: +86-592-6190790; Tel: +86-592-6190790
bDepartment of Ophthalmology, China-Japan Union Hospital of Jilin University, 126 Xiantai Street, Changchun 130000, China
cUniversity of Chinese Academy of Sciences, Beijing 100049, China
dDepartment of Environmental Engineering, Technical University of Denmark, Kgs. Lyngby, DK-2800, Denmark
First published on 18th September 2019
Abstract
Physical structures of sludge are critical factors determining the performance of the anaerobic digestion process, especially for the rate-limiting step, methanogenesis. Thus, to evaluate the effect of granular physical structure on methanogenesis and methanogenic community variation, intact and disintegrated granules were applied as inocula with formate, hydrogen and acetate as sole substrates in batch reactors. Kinetics results revealed that the physical structure of sludge had little impact on methane yield potential from three substrates, while a significantly different impact on methanogenesis rates of formate, hydrogen and acetate. The methanogenesis rate of formate in disintegrated granules was higher than that in the intact granular system, the methanogenesis rate of H2/CO2 in the intact granular system was higher than that in the disintegrated granules and the methanogenesis rate of acetate was similar with the in intact and disintegrated granular systems. Besides, in both intact and disintegrated granular systems, methanogenesis rates of formate were the highest, then followed the H2/CO2 and acetate was the lowest, indicating formate consumption has an advantage over hydrogen in the studied system. A microbial assay indicated that Methanobacteriales, Methanosarcinales and Methanomicrobiales are dominant methanogens on the order level, and the physical structure of granular sludge has little influence on methanogenic communities on the order level but showed significant influence on the species level. It enlightens us that the physical structure of sludge could be considered for regulating the anaerobic digestion via influencing the methanogenesis rates.
Introduction
Anaerobic biological treatment technology is widely known as an important strategy for simultaneously achieving pollutant removal and energy generation.1,2 However, the traditional anaerobic digestion (AD) process still has the challenge of low treatment efficiency, which is limited by the slow syntrophic metabolism of fermentation intermediates (alcohol and volatile fatty acids (VFAs)).3–5 Many researchers have reported that syntrophic VFAs oxidization contributes to too much carbon flux in anaerobic methanogenic environments, which would directly lead to VFA accumulation.6 VFAs (mainly propionate and butyrate) are metabolized by syntrophic communities of fatty acid-oxidizing bacteria and methanogens in an anaerobic methanogenic environment. The thermodynamically unfavourable reactions of VFA degradation required the rapid consumption of intermediates as well as methanogenic precursors, such as formate, acetate and H2/CO2, to make VFA degradation feasible.7 Thus, in some cases, methanogenesis is considered as the rate-limiting pathway, since methanogens are relatively slow-growing microorganisms and have a limited range of available substrates,8 in addition, these substrates' removal determined the efficiency of syntrophic VFAs degradation. Therefore, the high efficiency of the methanogenesis process was essential for the performance of AD process.Methanogenesis process is dependent on several process factors of which the most important are nutrients, temperature, pH and inhibitors.2,9 These factors influence the microbial activities and change the methanogens community structure to affect the efficiency of methanogenesis. In addition to biological structure, the physical structure of sludge was also viewed as a significant factor affecting the process of methanogenesis, because the diffusion distance between the syntrophic bacteria and methanogens may have a large impact on the rate of VFAs conversion as well as the efficiency of methanogenesis. High conversion rates of syntrophic substrates in methanogenic bioreactors are due to the high cell densities and short interbacterial distance (<10 μm) between syntrophic–methanogenic associations in granular sludge system.10 Granular sludge, therefore, was supposed to a suitable system for anaerobic methanogenesis, since its structure could enhance the efficiency of interspecies transfer of formate, hydrogen and acetate,9 and further improve the efficiency of the anaerobic process. Previous studies about the physical structure of granules mainly focused on the diffusion distance and granular layer structure,10–12 while it is still unclear that whether the physical structure of granules can affect the microbial community structure shift.
Besides, formate, hydrogen and acetate, as main methanogenic precursors, are intermediate products of syntrophic metabolism, and the methanogenesis rates from these substrates determined the efficiency of syntrophic VFAs degradation. Some researchers found that the physical structure of sludge influenced the relative importance of hydrogen/formate transfer, and it supported that hydrogen transfer dominates in granular system, and formate dominates in flocs sludge system.9 However, our previous study13 revealed that formate presented rapid consumption rate. And this result was not corresponded to the expectation that H2/CO2 consumption by methanogens in the granular system was faster than formate and acetate according to Angelidaki and Batstone (2010).9 Therefore, it is necessary to re-evaluate the effect of aggregated/flocculent structure of sludge on methanogenesis from the main intermediates during the AD process.
Comprehensive research is required to evaluate the impact of the physical structure of granular sludge on the shift of methanogens community structure and the methanogenesis rates from formate, acetate and H2/CO2. It could provide a better understanding of methanogenesis in anaerobic systems and further provide an efficient approach to improve the performance of AD. Therefore, batch reactors inoculating intact granules or disintegrated-granules with a different substrate (formate, acetate, or H2/CO2) were incubated for several days under mesophilic condition. Methane production from each reactor is measured along with the digestion time. Kinetic parameters of methanations from formate, acetate and H2 were simulated utilizing a modified Gompertz model. Microbial communities associated with various reactors were analyzed using Illumina 16S rRNA gene amplicon sequencing.
Results and discussion
Physical structure of granular sludge affects methanogenesis kinetics
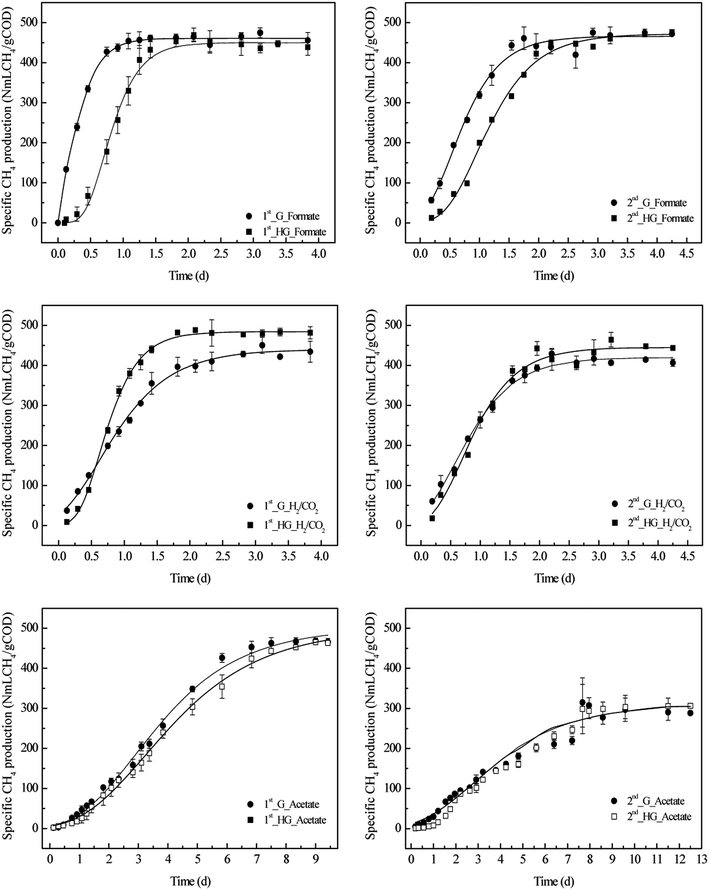
Under the assumption that methane production is paralleled with microbial growth in batch reactors, a modified Gompertz model was used to predict the kinetics parameters of methanogenesis from formate, acetate and H2/CO2 in both intact granular and disintegrated-granular systems. The predicted curves for specific CH4 yield along with digestion time fit well with observed values in each system, with R2 > 0.98 (Fig. 1, Table 1).| Generations | Parameters | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| A (N mLCH4 gCOD−1) | U (N mLCH4 gCOD−1 d−1) | λ (d−1) | R 2 | ||||||
| 1st | 2nd | 1st | 2nd | 1st | 2nd | 1st | 2nd | ||
| a H-Granule stands for disintegrated granules and n value is 3. | |||||||||
| Formate | Granule | 460.80 ± 5.62 | 465.76 ± 6.98 | 710.40 ± 11.35 | 381.70 ± 9.35 | 0.05 ± 0.01 | 0.08 ± 0.01 | 0.994 | 0.987 |
| H-Granule | 450.21 ± 7.83 | 471.66 ± 8.96 | 556.14 ± 10.63 | 319.05 ± 8.61 | 0.41 ± 0.03 | 0.41 ± 0.02 | 0.989 | 0.994 | |
| H2/CO2 | Granule | 439.69 ± 8.36 | 419.69 ± 10.36 | 271.55 ± 6.96 | 242.05 ± 11.32 | 0.02 ± 0.01 | 0.02 ± 0.01 | 0.995 | 0.992 |
| H-Granule | 484.31 ± 9.32 | 444.85 ± 5.63 | 540.33 ± 10.30 | 317.19 ± 10.23 | 0.29 ± 0.01 | 0.19 ± 0.03 | 0.998 | 0.986 | |
| Acetate | Granule | 499.79 ± 8.24 | 309.79 ± 7.96 | 94.38 ± 7.63 | 48.02 ± 2.67 | 0.95 ± 0.10 | 0.37 ± 0.01 | 0.996 | 0.972 |
| H-Granule | 497.97 ± 10.21 | 316.91 ± 10.36 | 84.90 ± 4.36 | 52.29 ± 3.26 | 1.09 ± 0.09 | 0.99 ± 0.04 | 0.998 | 0.985 | |
In the first generation, all of the batch reactors were inoculated with the same amount of sludge, the only difference was the physical structure of the sludge, consisting of intact granular sludge and disintegrated-granular sludge. Assuming that the inoculating error was within an acceptable range, the difference in methanogenesis from a specific substrate in a granular system and a homogeneous-granular system could completely be attributed to the difference in the physical structure of the sludge.
In general, the cumulative production of CH4 increased along with digestion time in both granular and homogeneous-granular systems. CH4 reached a stable value when the substrate was completely consumed (Fig. 1). Initially, formate was metabolized immediately in the granular system with a short lag time, while a little longer lag time (0.41 d) occurred in the disintegrated-granular system. This could be resulted from the high microbial densities in the granules, which would minimize formate transfer resistance.9 The maximum CH4 production rate from formate in the granular system was higher than that in the disintegrated-granular system (UG_formate > UHG_formate). The methane yield potential in the two systems seemed to have no significant differences, and the values were about 450–471 N mLCH4 gCOD−1. This indicated that the physical structure of granular sludge had an impact on methanation rate rather than the methane production potential. In addition, these values of methane production potential were significantly larger than that in digested manure (85–105 NmLCH4 gCOD−1) and sewage sludge (57.74 N mLCH4 gCOD−1) systems examined in a previous study.13 This might due to the varying quantities and activities of methanogens in these systems.
The observed trend of specific methane production from H2/CO2 along with incubation time was quite similar to it in the formate added system. In addition, methane production also required a longer lag time in granular system than that in disintegrated-granular system (Table 1). However, the maximum CH4 production rate from H2/CO2 in disintegrated-granular system was higher than that in granular system during the two-generation incubation (UHG_H2/CO2 > UG_H2/CO2), which is contrary to the results obtained from formate added system. The physical structure of granule had a significantly different impact on methane production rates from formate and H2/CO2. This could be explained by the previous finding that more hydrogen-consuming methanogens would be expected to be freely suspended in the medium after the disintegration of granules.14
Generally, acetate metabolism by methanogens required a longer adaption time than formate and H2/CO2, and the lag time in the disintegrated-granular system can reach approximate one day. The values of methane production potential were quite close to both the granular and disintegrated-granular systems. Moreover, the maximum CH4 production rate in the granular system was also similar to the rate in the disintegrated-granular system (UHG_acetate ≈ UG_acetate), which means the physical structure of a granule had little influence on methanogenesis from acetate (Table 1).
In conclusion, the consumption of three substrates in disintegrated-granular systems required longer lag time. The physical structure of granular sludge has little impact on methane production potential in different systems, while influenced the maximum methane production rates.
Comparison of methanogenesis rates from different substrates
In an intact granular system during second-generation incubation, the order of maximum CH4 production rates from three substrates was as follows: UG_formate > UG_H2/CO2 > UG_acetate. This finding is supported by previous research that formate was consumed faster than H2/CO2 in a granular system derived from an IC reactor.13 Although part of the reason for the lower H2/CO2 degradation rate in the granular system was that H2/CO2 was supplemented as an external carbon source in a gas phase. The hydrogen has to transfer from the gas phase to the bulk liquid phase. This process could increase the mass transfer resistance compared to a real anaerobic granular system in which H2/CO2 is formed by syntrophic fatty acid bacteria closed to the methanogens.9,13 Although there is no study to show the direct relation between methanation rate from formate/H2 and the relative importance of interspecies formate/H2 transfer, the rate of CH4 conversion from methanogenic substrates has a positive correlation with the efficiency of interspecies electron transfer.15 Since interspecies electron transfer during the anaerobic methanogenic process is generated using an electron-donating partner and carried by formate, acetate and hydrogen, and eventually sinks into methane. Dong and Stams (1995)15 revealed that only formate-lux could result in a fast methane production rate. It concluded that formate was the main interspecies electron carrier in mesophilic suspended co-cultures, which indicated that formate consumption and formate transfer has close relations. Therefore, the high consumption of formate in the studied granular system revealed that formate might play an important role in the studied granular systems as an intermediate product. The lowest methanation rate from acetate was reasonable, since acetate is more complex than the other two substrates, which had been proven in a previous study performed by Pan et al. (2016).13 In addition, research focusing on layered structure and juxtaposition of syntrophs and methanogens in the consortia revealed that the inner layer is mostly comprised of acetoclastic methanogens and the outer layer consists of fermentative bacteria.11,16Similarly, the order in a homogeneous-granular system during second-generation incubation was also UHG_formate > UHG_H2/CO2 > UHG_acetate. But actually, the variation in methanation rates between formate and H2 was quite small. As mentioned above, the physical structure of sludge had opposite effect on formate and H2/CO2 methanation, with the disrupted granules decreasing the methanation rate from formate and increasing rate from H2/CO2. A higher methanation rate than observed may be obtained by deducting the increased mass transfer resistance of H2/CO2. Thus, it is difficult to define whether formate or H2/CO2 consumed faster in the studied homogeneous-granular system.
Therefore, the results revealed that either in the granular system or disintegrated-granular system, formate had a rapid conversion. Although there was no direct evidence to prove that formate was the dominant interspecies electron carrier in the studied granular system, it can be deduced that the role of formate played in a granular according to the result of rapid formate consumption. Thus, more attention should be paid to formate conversion when regulating and controlling of anaerobic digesters.
Microbial community variation
| Levels | G0 | G1 | HG1 | G2 | HG2 | G3 | HG3 |
|---|---|---|---|---|---|---|---|
| a G1 and HG2 were the samples from formate added granular and homogeneous-granular system; G2 and HG2 were the samples from acetate supplemented granular and homogeneous-granular system; G3 and HG3 stand for the samples from H2/CO2 injected granular and disintegrated-granular system. | |||||||
| Orders | 5 | 5 | 5 | 5 | 5 | 5 | 5 |
| Families | 15 | 14 | 15 | 15 | 15 | 13 | 14 |
| Species | 28 | 21 | 24 | 23 | 26 | 23 | 26 |
| Sequence reads | 46![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 342 342 |
46![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 961 961 |
46![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 736 736 |
41![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 544 544 |
42![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 648 648 |
48![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 109 109 |
56![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 489 489 |
| Chao | 37.00 | 26.00 | 25.33 | 27.60 | 33.25 | 24.00 | 33.00 |
| Ace | 37.43 | 29.66 | 26.38 | 29.38 | 34.41 | 24.45 | 36.28 |
| Shannon | 1.51 | 1.56 | 1.22 | 1.24 | 1.72 | 1.47 | 1.47 |
| Simpson | 0.68 | 0.70 | 0.54 | 0.59 | 0.73 | 0.71 | 0.61 |
| Shannoneven | 0.42 | 0.50 | 0.38 | 0.38 | 0.49 | 0.46 | 0.43 |
For experimental systems, the relative abundance of Methanosarcinales increased compared to the abundance in the original granular sludge, and Methanosarcinales dominated in both intact granular and disintegrated-granular systems after two generations of incubation. Methanomicrobiales also showed a significant increase in both systems as well, with a relative abundance of 22.06% in the G1 reactor (formate added granular system) and 19.86% in the HG1 reactor (formate added disintegrated-granular system). However, the relative abundance of Methanobacteriales presented an obvious reduction, and the values dwindled to 8.01% and 4.17% in the G1 and HG1 reactors, respectively (Fig. 2a). Therefore, the G1 reactor had more hydrogenotrophic methanogens (30.07%) than the HG1 reactor (24.03%).
A small variation occurred between H2/CO2 that served as substrate reactors (G3 and HG3) at the order level (Fig. 2a). This means the physical structure of sludge has little effect on the microbial community shift on order level. In the G3 reactor (H2/CO2 added granular system), Methanosarcinales (54.60%), Methanomicrobiales (36.69%) and Methanobacteriales (4.04%) were detected, and Methanosarcinales (60.12%) was also the predominant methanogenic group in HG3 (H2/CO2 added the disintegrated-granular system), followed by Methanomicrobiales (29.83%) and Methanobacteriales (7.50%). At the species level, Methanothermobacter thermautotrophicus accounted for large percentages (43.85%) in the HG3 reactor. In addition, Methanobacterium beijingense (3.32%) and Methanobacterium formicicum were also detected in HG3. While the dominant specie of Methanobacteriales in the G3 system was Methanothermobacter thermautotrophicus (accounting for 3.53%). As for Methanomicrobiales, the HG3 reactor had a higher diversity than the G3, in which five species (relative abundance > 0.5%) were detected. While the G3 reactor contained fewer species, it had a higher relative abundance of unclassified_Methanolinea (20.72%) and Methanospirillum (4.52%).
Compared to the original granules, the Methanosaeta concilii outcompeted Methanosaeta thermophila_PT in both G2 (acetate added granular system) and HG2 (acetate added disintegrated-granular system) reactors. Surprisingly, no Methanosarcina spp. was observed in the studied system. However, comparing Fig. 2a and b, it was obvious that approximately 15% and 20% unclassified Methanosarcinales existed in the G2 and HG2 reactors, respectively. The uncultured methanogens might mostly consist of Methanosarcina spp. It has been reported that both Methanosaeta spp. (formerly Methanothrix) and Methanosarcina spp. were identified as important acetoclastic methanogens in granular sludge.20 The similar microbial community structure and a similar amount of abundance of acetoclastic methanogens in the G2 and HG2 reactors could explain the similar values of Umax in the acetate added anaerobic batch systems. Theoretically, the structure of the sludge would have an impact on the rate of substrate consumption by microorganisms and thus influence the activity of functional microbes. This will finally cause a variation in microbial diversity and abundance, which would influence substrate degradation in turn. Therefore, it can be concluded that the physical structure of sludge had no significant effect on acetate methanation.
In the disintegrated-granular system, the microbial communities structure in HG1, HG2 and HG3 showed a difference (Fig. 3), which indicated that different substrates had a different effect on methanogens community variation with inoculating disrupted granular sludge. These results agree with the previous statement that microbial community structures were related to substrates.24 In addition, the similar amount of relative abundance of hydrogenotrophic methanogens in HG3 and HG1 supports the kinetics results that UH2/CO2 was quite close to Uformate in the second generation.
Considering the small change in the methanogens community structure in the two systems and the kinetics results in the second generation, it can be concluded that the physical structure affects the methane production rate, and substrate instead of the physical structure of granular sludge contributed to the methanogens variation in different systems. Substrate conversion rate reflect the types and numbers of methanogens involved in substrate degradation.22,25
Experimental
Batch reactor experiments
The substrates of formate and acetate were supplied by HCOONa and CH3COONa (Sigma Chemicals). Stock solutions of 0.8 M HCOONa and 0.2 M CH3COONa were prepared with distilled water. Thus, a 5 mL stock solution complementation could theoretically result in a 1 mmol CH4 product (Table 3). A mixture gas of hydrogen and carbon dioxide with a ratio (v/v) of 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 was prepared using pure H2 and CO2 with a purity of 99.999%. The determination of the ratio of 4
1 was prepared using pure H2 and CO2 with a purity of 99.999%. The determination of the ratio of 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 was based on the stoichiometric coefficient of methanogenesis of H2/CO2 (Table 3).
1 was based on the stoichiometric coefficient of methanogenesis of H2/CO2 (Table 3).
| Reactions | Stoichiometric coefficient of methanogenesis | ΔG0 (kJ molCH4−1) | Microorganisms |
|---|---|---|---|
| 4H2 + CO2 → CH4 + 2H2O | 4/1 | −135 | Hydrogenotrophic methanogens and some Methanosarcina |
| 4HCOOH → CH4 + 3CO2 + 2H2O | 4/1 | −130 | Many hydrogenotrophic methanogens |
| CH3COOH → CH4 + CO2 | 1/1 | −33 | Methanosarcina and Methanosaeta |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v) mixture of gas was added to the systems, and 5 mmol of mixture gas was injected into serum bottles using a syringe (maximum span of 50 mL). The actual injected volume was calculated and modified using laboratory temperature and pressure methods according to the ideal gas law (PV = nRT), and 5 mL distilled water, instead of a substrate, was supplemented to make the same working volume of 50 mL. In addition, 5 mL of distilled water, instead of a substrate, was added in the control experiments, following the same procedures as found in experimental batch reactors. To reduce heterogeneity from the inoculum, all the performances were conducted in triplicate.
1, v/v) mixture of gas was added to the systems, and 5 mmol of mixture gas was injected into serum bottles using a syringe (maximum span of 50 mL). The actual injected volume was calculated and modified using laboratory temperature and pressure methods according to the ideal gas law (PV = nRT), and 5 mL distilled water, instead of a substrate, was supplemented to make the same working volume of 50 mL. In addition, 5 mL of distilled water, instead of a substrate, was added in the control experiments, following the same procedures as found in experimental batch reactors. To reduce heterogeneity from the inoculum, all the performances were conducted in triplicate.
The first-generation incubation was finished when the substrates were consumed completely by methanogens in the batch reactors. To evaluate the variation of methanogens communities and the methanogenesis rates in the cultivated sludge using a sole substrate, a second-generation incubation was conducted. 5 mL of supernatant liquid was extracted from each batch reactor by a syringe. The rest of the slurry mixture was used for second-generation incubation. Then, substrates of formate, acetate, and H2/CO2 were added separately, and the subsequent procedures were the same as the first generation.
Methane production monitoring
Methane production was assayed in closed serum bottles with a headspace of 260 mL. Blank experiments were conducted where 5 mL distilled water was used for replacing the substrate solution as described above. The methane production from the three substrates was calculated by deducting the methane produced from the blank reactors.The CH4 production determination was conducted using gas chromatography FULI GC9790II with a column of TDX-01 (2 m long and a 3 mm inner diameter) and thermal conductivity detector (TCD). Helium served as the carrier gas. The temperatures of the packed column, detector, and injection port were set to 120 °C, 160 °C, and 160 °C, respectively.
The cumulative volume of CH4 generated in a serum bottle was calculated by multiplying the headspace volume (260 mL) by the CH4 percentage (mL of CH4 per mL) in the headspace as determined by GC analysis. It must be noted that gas samples taken from batch reactors should equilibrate in batch reactors, which means the pressure in batch reactors have to be taken into consideration. In addition, the obtained value of cumulative CH4 production was normalized in standard temperature and pressure (STP) conditions (0 °C and 1 atm) according to the ideal gas law (PV = nRT). The methane production assay was referenced to sample mass or chemical oxygen demand (N mLCH4 gVS−1 or N mLCH4 gCOD−1, N: normalized in STP). In this study, to ensure the unity of units, the mass of the substrate was converted to g COD, and the conversion factors used (COD/VS, g g−1) for the formate, acetate, and H2/CO2 were 0.35, 1.07, and 8.00, respectively.
The modified Gompertz model
Recently, many studies have used the modified Gompertz model to predict biogas (CH4 and CO2) production potential by assuming that the rate of gas production is proportional to the microbial activity in anaerobic digesters.29–31 The modified Gompertz equation is shown as follows:where P – cumulative methane production, N mL gVS−1 at any digestion time t; A – methane yield potential, N mL gVS−1; U – maximum rate of methane production, N mL (gVS d)−1; λ – lag phase period to produce methane, days; t – digestion time at which cumulative methane production P is measured, days; e – mathematical constant (2.718282).
The kinetic parameters of A, U, and λ were simulated for each batch reactor using non-linear regression with the help of SPSS software. These parameters were determined for best fit with high R2 (>0.95).
DNA extraction and Illumina 16S rRNA gene amplicon sequencing
At the end of second generation incubation, the DNA from 500 mg of the sludge samples from each reactor was extracted after centrifuging 5 mL sludge for 20 minutes at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm. The DNA extraction protocol followed the manufacturer's instructions from the Fast DNA Spin Kit for Soil (MP bio, USA). The extracted DNA was subjected to electrophoresis in agarose gels (1.0% w/v) and visualized under a blue-ray. The extracted DNA was stored at −20 °C until further diversity and abundance analyses. The archaea 16S rRNA gene was amplified using the barcoded primer pair ARC-787f (5′-ATTAGATACCCSBGTAGTCC-3′) and ARC-1059r (5′-GCCATGCACCWCCTCT-3′).32 Sequencing of archaeal amplicons was performed on Illumina MiSeq at Genenergy, Shanghai, China. The Illumina sequencing raw data were deposited in the NCBI Sequence Read Archive database (accession no. PRJNA491806).
000 rpm. The DNA extraction protocol followed the manufacturer's instructions from the Fast DNA Spin Kit for Soil (MP bio, USA). The extracted DNA was subjected to electrophoresis in agarose gels (1.0% w/v) and visualized under a blue-ray. The extracted DNA was stored at −20 °C until further diversity and abundance analyses. The archaea 16S rRNA gene was amplified using the barcoded primer pair ARC-787f (5′-ATTAGATACCCSBGTAGTCC-3′) and ARC-1059r (5′-GCCATGCACCWCCTCT-3′).32 Sequencing of archaeal amplicons was performed on Illumina MiSeq at Genenergy, Shanghai, China. The Illumina sequencing raw data were deposited in the NCBI Sequence Read Archive database (accession no. PRJNA491806).
Statistical analysis
Principal components analysis (PCA) was used to characterize the structure and relative abundance of microbial communities in the experimental batch reactors. The dominant microorganisms at the family level were displayed. The software, CANOCO 5.0, was used for data analysis.33 Besides, Origin 8.0 was used for the production of graphics.Conclusions
The physical structure of granular sludge had significantly different effects on formate and H2/CO2 conversion, whereas it had little impact on acetate conversion. And the physical structure of granules showed no significant effect on the methanogen composition shift in three systems on order level, while the substrate affects the succession of methanogens composition. Formate had a significantly rapid consumption rate in the granular system, which meant the role of formate in the anaerobic system might be undervalued. This study enlightened that enhancing formate transfer might be an effective approach for improving the performance of the AD process in the studied granular systems. Applying this approach to industrial methane production process could reach higher waste (water) treatment efficiency and energy product (methane) production.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors would like to thank the National Key Research and Development Program of China (Contract No. 2018YFD0500202-4), the National Natural Science Foundation of China (Contract No. 51678553, 21876167, 21477122, 51808525), the Project of the Natural Science Foundation of Fujian Province (Contract No. 2017J05092), the IUE CAS Young Talents Frontier Project (Contract No. IUEQN201501), the Xiamen Science and Technology Project (Contract No. 3502Z20182003), the FY2015 Japanese-China Research Cooperative Program (Contract No. 2016YFE0118000), the Strategic Priority Research Program of the Chinese Academy of Sciences (Grant No. XDA23030301) and Innovation Fund Project (2019000011) for their supports for this study. We would like to thank LetPub (http://www.letpub.com) for providing linguistic assistance during the preparation of this manuscript. We are grateful to Genenergy for help with Illumina sequencing raw data processing.References
- D. J. Batstone and B. Virdis, Curr. Opin. Biotechnol., 2014, 27, 142–149, DOI:10.1016/j.copbio.2014.01.013.
- H. Li, D. Ji and L. Zang, IOP Conf. Ser. Earth Environ. Sci., 2018, 112, 012006, DOI:10.1088/1755-1315/112/1/012006.
- M. J. McInerney and M. P. Bryant, in Biomass Conversion Processes for Energy and Fuels, ed. S. S. Sofer and O. R. Zaborsky, New York, 1981, pp. 277–296 Search PubMed.
- B. Schink, Microbiol. Mol. Biol. Rev., 1997, 61, 262–280, DOI:10.1016/j.ijpharm.2004.07.010.
- G. Capson-Tojo, R. Moscoviz, D. Ruiz, G. Santa-Catalina, E. Trably, M. Rouez, M. Crest, J. P. Steyer, N. Bernet, J. P. Delgenès and R. Escudié, Bioresour. Technol., 2018, 260, 157–168, DOI:10.1016/j.biortech.2018.03.097.
- B. Schink and A. J. M. Stams, in The Prokaryotes: An Evolving Electronic Resource for the Microbiological Community, ed. O. R. Dworkin, S. Falkow, E. Rosenberg, K. H. Schleifer and E. Stackebrandt, New York, 4th edn, 2006, pp. 309–335 Search PubMed.
- A. J. Stams and C. M. Plugge, Nat. Rev. Microbiol., 2009, 7, 568–577, DOI:10.1038/nrmicro2166.
- Y. Liu and W. B. Whitman, Ann. N. Y. Acad. Sci., 2008, 1125, 171–189, DOI:10.1196/annals.1419.019.
- I. Angelidaki and D. J. Batstone, in Solid Waste Technology & Management, ed. T. H. Christensen, Wiley, West Sussex, UK, 2010, pp. 583–600 Search PubMed.
- F. A. de Bok, C. M. Plugge and A. J. Stams, Water Res., 2004, 38, 1368–1375, DOI:10.1016/j.watres.2003.11.028.
- F. A. MacLeod, S. R. Guiot and J. W. Costerton, Appl. Environ. Microbiol., 1990, 56, 1598–1607, DOI:10.1002/bit.260360114.
- J. Wu, R. U. Z. Afrifi and Z. Cao, Bioresour. Technol., 2016, 202, 165–171, DOI:10.1016/j.biortech.2015.12.006.
- X. Pan, I. Angelidaki, M. Alvarado-Morales, H. Liu, Y. Liu, X. Huang and G. F. Zhu, Bioresour. Technol., 2016, 218, 796–806, DOI:10.1016/j.biortech.2016.07.032.
- J. E. Schmidt and B. K. Ahring, Appl. Environ. Microbiol., 1991, 35, 681–685, DOI:10.1007/bf00169637.
- X. Z. Dong and A. J. M. Stams, Anaerobe, 1995, 1, 35–39, DOI:10.1016/S1075-9964(95)80405-6.
- S. R. Guiot, A. Pauss and J. W. Costerton, Water Sci. Technol., 1992, 25, 1–10, DOI:10.2166/wst.1992.0133.
- K. Ma, X. Liu and X. Dong, Int. J. Syst. Evol. Microbiol., 2005, 55, 325–329, DOI:10.1099/ijs.0.63254-0.
- N. L. Schauer and J. G. Ferry, J. Bacteriol., 1980, 142, 800–807 CrossRef CAS PubMed.
- L. W. Hulshoff Pol, S. I. de Castro Lopes, G. Lettinga and P. N. Lens, Water Res., 2004, 38, 1376–1389, DOI:10.1016/j.watres.2003.12.002.
- J. E. Schmidt and B. K. Ahring, Biotechnol. Bioeng., 2015, 49, 229–246, DOI:10.1002/(SICI)1097-0290(19960205)49:3<229::AID-BIT1>3.0.CO;2-M.
- J. T. Grotenhuis, M. Smit, C. M. Plugge, Y. S. Xu, A. A. van Lammeren and A. J. Stams, Appl. Environ. Microbiol., 1991, 57, 1942–1949, DOI:10.1002/yea.320070614.
- C. Lee, J. Kim, K. Hwang, V. O'Flaherty and S. Hwang, Water Res., 2009, 43, 157–165, DOI:10.1016/j.watres.2008.09.032.
- A. Wasserfallen, J. Nölling, P. Pfister, J. Reeve and D. M. E. Conway, Int. J. Syst. Evol. Microbiol., 2000, 50, 43–53, DOI:10.1099/00207713-50-1-43.
- D. J. Batstone, J. Keller and L. L. Blackall, Water Res., 2004, 38, 1390–1404, DOI:10.1016/j.watres.2003.12.003.
- J. Yuan, Y. Yuan, Y. Zhu and L. Cao, Sci. Total Environ., 2018, 627, 770–781, DOI:10.1016/j.scitotenv.2018.01.233.
- M. Soto, R. Méndez and J. M. Lema, Water Res., 1993, 27, 1361–1376, DOI:10.1016/0043-1354(93)90224-6.
- W. E. Federation and A. P. H. Association, in Standard Methods for the Examination of Water and Wastewater, American Public Health Association (APHA), Washington, DC, USA, 2005 Search PubMed.
- I. Angelidaki and W. Sanders, Rev. Environ. Sci. Bio/Technol., 2004, 3, 117–129, DOI:10.1007/s11157-004-2502-3.
- N. I. Budiyono Widiasa, S. Johari and S. Sunarso, Int. J. Chem. Biol. Eng., 2010, 3, 39–44 Search PubMed.
- S. Adiga, R. Ramya, B. Shankar, J. H. Patil and C. Geetha, Biol. Environ. Eng., 2012, 42, 73–78, DOI:10.7763/IPCBEE.2012.V42.15.
- S. Weiss, A. Zankel, M. Lebuhn, S. Petrak, W. Somitsch and G. M. Guebitz, Bioresour. Technol., 2011, 102, 4353–4359, DOI:10.1016/j.biortech.2010.12.076.
- R. Rathaur, S. H. Dhawane, A. Ganguly, M. K. Mandal and G. Halder, Process Saf. Environ. Prot., 2018, 113, 413–423, DOI:10.1016/j.psep.2017.11.014.
- V. N. Roth, T. Dittmar, R. Gaupp and G. Gleixner, PLoS One, 2015, 10, e0119188, DOI:10.1371/journal.pone.0119188.
| This journal is © The Royal Society of Chemistry 2019 |