Open Access Article
Open Access ArticleNitrogen-containing three-dimensional biomass porous carbon materials as an efficient enzymatic biosensing platform for glucose sensing†
Baixi Shan‡
a,
Yanhua Ji‡a,
Youbao Zhonga,
Lai Chena,
Shanshan Lia,
Jie Zhanga,
Liling Chena,
Xuan Liua,
Yuan Chena,
Nan Yanb and
Yonggui Song *a
*a
aLaboratory Animal Science and Technology Center, College of Pharmacy, Jiangxi University of Traditional Chinese Medicine, 1688 Meiling Road, Nanchang 330006, PR China. E-mail: songyonggui1999@163.com; Fax: +86 791 87802135; Tel: +86 791 87802135
bNational Engineering Research Center for Carbohydrate Synthesis, Jiangxi Normal University, Nanchang 330022, PR China
First published on 15th August 2019
Abstract
A novel glucose biosensor was developed by immobilizing glucose oxidase (GOD) on a three-dimensional (3D) porous cane vine (wisteria) stem-derived carbon (3D-CVS), which was firstly proposed as novel support material for electrochemical biosensors using loaded biomolecules. Here, an integrated 3D-CVS electrode was fabricated by loading GOD molecule onto a whole piece of 3D-CVS electrode for a glucose biosensor. The morphologies of integrated 3D-CVS and 3D-CVS/GOD electrode were characterized by scanning electron microscopy (SEM) and transmission electron microscopy (TEM). SEM results show the 3D macroporous structure of the integrated 3D-CVS electrode. TEM results show that there are some micro-holes and defects in the 3D-CVS electrode. Electrochemical behaviors and electrocatalytic performance of integrated 3D-CVS/GOD electrode were evaluated by cyclic voltammetry and electrochemical impedance spectroscopy. The effects of pH and scanning rate on the electrochemical response of biosensors have been studied in detail. The glucose biosensor showed a wide linear range from 0.58 μM to 16 mM, with a high sensitivity of 86.17 μA mM−1 and a low detection limit of 0.19 μM. Furthermore, the glucose biosensor exhibited high selectivity, good repeatability and nice stability.
1. Introduction
The development of glucose detection techniques has attracted great interest in recent years due to their expanding practical applications in food and fermentation analysis, textile industry, environmental monitoring, medical diagnosis,1–5 etc. Many techniques have been used for monitoring glucose levels, such as surface plasmon resonance,6 near-infrared spectrometry,7 mass spectrometry,8–10 electrochemiluminescence,11 fluorescence,12 colorimetry,13 electrochemistry,14 etc. Compared with the large-scale instruments with complicated operation,15,16 electrochemical biosensors based on biomolecules immobilized on an electrode surface are promising tools owing to their high sensitivity, good selectivity, miniaturized compatibility, easy operation and low cost. Great efforts have been devoted to the immobilization of glucose oxidase (GOD) on an electrode surface, which is one of the main factors affecting the performance of glucose biosensors.17–19Over the past few years, nanomaterials with high electrical conductivity, large specific surface area and good catalytic activity have been introduced as support materials for the immobilization of GOD molecules.20–22 Among various nanomaterials, graphene has shown enormous potential for immobilization of GOD owing to its large specific surface area, two-dimensional honeycomb lattice and excellent electrical conductivity.23,24 For example, Liu et al. reported a novel highly efficient glucose biosensor by covalently attaching amines of GOD to carboxyl groups of graphene oxide sheets.25 Shan et al. also constructed a graphene-based electrochemical glucose biosensor with a low detection limit.26 However, graphene nanosheets were apt to aggregate after they were dropped on the electrode surface due to strong van der Waals' forces and π–π interactions between individual sheets.23,27 Accordingly, the performance of a graphene-based electrochemical glucose biosensor could only be improved to some extent. Carbon nanotubes (CNT), showing some better properties than graphene such as the closely-packed structures could be suppressed appropriately. Thus this material has also been extensively employed for immobilization of GOD.28–31 But this kind of material does not contain nitrogen element which may affect its biocompatibility32,33 and it is also impossible to fabricate an integrated electrode with three-dimensional structure to immobilize more enzymes. Therefore, it still can not meet the present demand for high-performance construction of glucose biosensors.
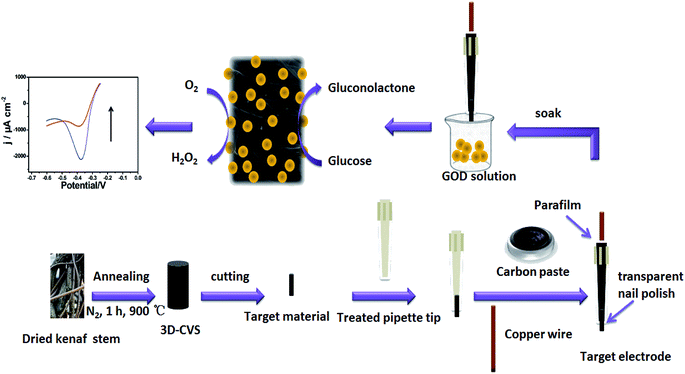
Herein, a novel electrochemical glucose biosensor is developed by using a low-cost and environmentally friendly 3D cane vine (wisteria) stem carbon materials (3D-CVS) as a new support material for loading GOD molecules effectively. As shown in Scheme 1, a whole 3D-CVS is used to fabricate integrated 3D-CVS electrodes, thus retaining the complete 3D macroporous structure, providing a large specific surface area, effectively fixing a large number of GOD molecules, greatly improving mass transfer. Some microporosities and defects in the 3D-CVS electrodes greatly enhance the immobilization of GOD molecules. Therefore, the 3D-CVS electrodes could be used to immobilize GOD molecules better and the resulting biosensor showed superior electrochemical performance with good stability and reproducibility. It is the first time that the 3D-CVS electrodes were used to immobilize biomolecules (e.g. GOD molecules) for electrochemical biosensing.
2. Experimental section
2.1. Materials and reagents
Cane vine (wisteria) stems (CVS) were obtained from Meiling Mountain (Nanchang, China) directly. Glucose oxidase (GOD, EC 1.1.3.4, 140 U mg−1). Graphite powder (99.95%, 325 meshes) was purchased from Aladdin Ltd (Shanghai, China). Potassium ferricyanide (K3Fe(CN)6), potassium chloride (KCl), ascorbic acid (AA), uric acid (UA), mannose, fructose, lactose, glucose, etc., were obtained from Beijing Chemical Reagent Factory (Beijing, China). All reagents were of analytical grade and used as received. Phosphate buffer solutions (PBS, 0.2 M, pH 7.0) were prepared with 0.2 M NaH2PO4 and 0.2 M Na2HPO4. GOD solution (65 μM) was prepared in 0.2 M PBS (pH 7.0). All solutions were prepared with ultra-pure water purified by a Millipore-Q System (18.2 MΩ cm).2.2. Apparatus
Cyclic voltammograms (CVs) and electrochemical impedance spectroscopy (EIS) were carried out with a CHI750D electrochemical workstation (CHI, China). A three-electrode configuration was used with a platinum wire as the auxiliary electrode, a saturated calomel electrode (SCE) as the reference electrode, and a 3D-CVS/GOD electrode as the working electrode. Effective surface areas (Aeff) of various 3D-CVS were estimated before use based on the CVs in 0.1 M KCl solution containing 5.0 mM Fe(CN)63−/4− at 50 mV s−1 according to the Randles–Sevcik equation as shown in Fig. S1 (ESI†). The currents had been normalized based on the Aeff. Other CVs were carried out in 10.0 mL of 0.2 M PBS (pH 7.0) at room temperature. EIS was characterized at an open circuit potential in the frequency range from 0.01 Hz to 105 Hz with amplitude 5 mV. Scanning electron microscopy (SEM) analysis was carried out using a XL30 ESEM-FEG SEM at an accelerating voltage of 20 kV equipped with a Phoenix energy dispersive X-ray analyzer (EDXA). Transmission electron microscopy (TEM) images were recorded on a JEOL JEM-2100 microscope operated at an acceleration voltage of 200 kV.2.3. Preparation of integrated 3D-CVS/GOD electrodes
The 3D-CVS was prepared by carbonization of dried cane vine stems in a high-temperature furnace (detailed steps are listed in the ESI, Fig. S2 and Table S1†). Then the 3D-CVS was cut into a cylindrical shape with the outside diameter almost equal to the inside diameter of the tip of the treated pipette and accordingly the cylindrical 3D-CVS could be firmly immobilized in the treated pipette tip. The processed 3D-CVS was washed with ethanol and ultra-pure water alternately, dried naturally, and inserted into the treated pipette tip leaving about a 0.2 mm long part outside. Then 1.0 g graphite powder and 0.25 g liquid paraffin were mixed and homogenized carefully in an agate mortar for 20 min. After that, the homogenized mixture was packed into the upper part of the pipette tip to contact the end of 3D-CVS. Here, about 0.2–0.3 mm of the bottom of the pipette tip had no homogenized mixture and accordingly the pasty mixture did not flow out. Then, a copper wire was inserted into the pipette tip to electrically connect it with the end of 3D-CVS via graphite powder. After the homogenized mixture paste was naturally dried at room temperature, the copper wire was further immobilized by using parafilm or epikote as shown in Scheme 1. Finally, a thin layer of transparent nail polish was coated on the gap between the treated pipette tip and the 3D-CVS to avoid the solution from entering the treated pipette tip. The schematic structure of the 3D-CVS electrode was illustrated in detail in Scheme 1. The 3D-CVS electrode was immersed into 65 μM GOD solution at 4 °C for 12 h. Finally, the modified electrode was rinsed with ultra-pure water to remove weakly bound molecules and stored at 4 °C for further use. The resulting electrode was referred to as the 3D-CVS/GOD electrode. The preparation procedure is also illustrated in Scheme 1.3. Results and discussion
3.1. Characterization of 3D-CVS/GOD electrodes
SEM was firstly employed to investigate the morphology of 3D-CVS and 3D-CVS/GOD electrodes. As shown in Fig. 1A–C and S3,† The 3D-CVS was cylindrical and many macropores appeared in it, revealing the hollow structure inside. Their diameters were estimated to be about 2.5 μm which is much smaller than previously reported.5,34 At the same time, it could be seen from Fig. 1B and C that the material also had ultra-thin lamellar structure. Moreover, the TEM image clearly revealed many black and white spots on the 3D-CVS electrode (Fig. 1D), which indicated microporous structures and some defects in the graphite nanosheets. These above special structures may useful for proton transport and catalytic performance of electrodes. Energy disperse spectroscopy (EDS) mapping images were also used to characterize the 3D-CVS (Fig. 1E and F), and the C, N elements uniformly appeared on it, which was consistent with the results of EDS (Fig. S5†), indicating the electrode had good biocompatibility. Magnified SEM images (Fig. 1G and H) showed that when GOD molecules were assembled on the 3D-CVS electrode, the lamellar structure becomes smooth (Fig. 1G), and the diameters of holes in the 3D-CVS also decreases (Fig. 1H). At the same time, S elements also could be seen on the surface of the electrode by EDS mapping images (Fig. 1I). These above results clearly demonstrated the successful immobilization of GOD molecules on the 3D-CVS electrodes.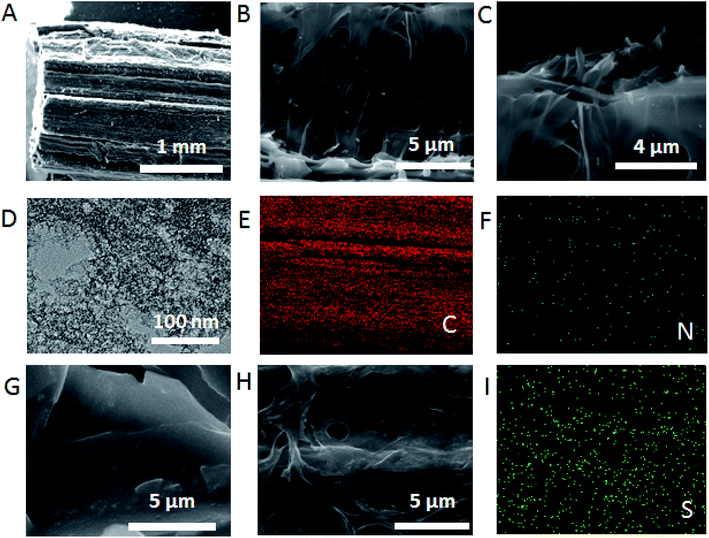 | ||
| Fig. 1 (A–C) SEM images of 3D-CVS and (D) TEM image of 3D-CVS. (E and F) EDS mapping images of the 3D-CVS. (G and H) SEM images of 3D-CVS/GOD. (I) EDS mapping images of the 3D-CVS/GOD. | ||
3.2. Electrochemical behaviors of 3D-CVS/GOD electrodes
The EIS had been a popular tool to monitor the electrode assembly process due to its immediate and sensitive response to the electrode surface changes.35 Fig. 2 shows the EIS of the 3D-CVS/GOD electrode (curve a) and the 3D-CVS electrode (curve b) in 5 mM Fe(CN)63−/4− containing 0.1 KCl as the supporting electrolyte. The inset shows a Randles equivalence circuit model which was chosen to fit the impedance data.36 In the Randles circuit, the resistance to charge transfer (Rct) and the diffusion impedance (W) were both assumed to be in parallel to the interfacial capacity (Cdl). The parallel structure of Rct and Cdl gave rise to a semicircle in the complex plane plot of Zim against Zre. The Rct of the 3D-CVS electrode (50 Ω, curve b) was extremely small. After the GOD was coated on the 3D-CVS electrode surface, an obvious increase of Rct (168 Ω, curve a) was observed (fitting results of the impedance spectra are listed in Table S2, ESI†). It might be ascribed not only to hindrance of the GOD molecule layer to Fe(CN)63−/4− but also to electrostatic repulsion between Fe(CN)63−/4− and negatively charged GOD molecules.37 Therefore, the increase of Rct also indicated that the successful immobilization of GOD molecules on the surface of the 3D-CVS electrode.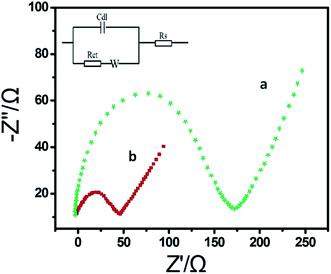 | ||
| Fig. 2 EIS of the 3D-CVS/GOD electrode (curve a) and the 3D-CVS electrode (curve b) in 5.0 mM Fe(CN)63−/4− containing 0.1 KCl. Inset is the equivalent circuit. Frequency range is 0.01–10 kHz. | ||
Fig. S4A† shows the CVs of the 3D-CVS/GOD electrode in 0.2 M N2-saturated PBS with pH from 6.0 to 8.0 at a scan rate of 100 mV s−1. The formal potential (E0 = (Epa + Epc)/2) displayed a linear relativity to pH with a slope of −60.5 mV per pH (R = −0.998) (Fig. S4B†). This value was very close to the theoretical value of −58.6 mV per pH for the two-electron and two-proton transportation process of FAD/FADH2 redox couples in the GOD.38 The effect of the potential scan rate on the electrochemical performance of the 3D-CVS/GOD electrode was shown in Fig. S4C.† The redox peak current increased linearly as the potential scan rate increased from 50 to 1000 mV s−1 (Fig. S4D†), indicating a quasi-reversible surface-controlled electrochemical process. The surface coverage (Γ, in mol cm−2) of GOD on the 3D-CVS electrode could be determined based on Faraday's law:39
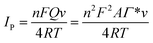 | (1) |
This equation could be transferred as the following expression:
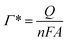 | (2) |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 493 C mol−1 (ref. 39)) and Aeff was the effective surface area of the electrode (Fig. S1†). The calculated Γ value of the 3D-CVS/GOD electrode was 3.55 × 10−10 mol cm−2, which was much higher than the theoretical molecular monolayer surface coverage (2.86 × 10−12 mol cm−2).38 The principle of calculating the surface coverage of monolayer40–43 is mainly based on the maximum cross-sectional area of the modified protease, and the molar number of protease molecules arranged on the surface of 1 cm2 electrode (excluding any stacking). The corresponding formulas are as follows:
493 C mol−1 (ref. 39)) and Aeff was the effective surface area of the electrode (Fig. S1†). The calculated Γ value of the 3D-CVS/GOD electrode was 3.55 × 10−10 mol cm−2, which was much higher than the theoretical molecular monolayer surface coverage (2.86 × 10−12 mol cm−2).38 The principle of calculating the surface coverage of monolayer40–43 is mainly based on the maximum cross-sectional area of the modified protease, and the molar number of protease molecules arranged on the surface of 1 cm2 electrode (excluding any stacking). The corresponding formulas are as follows:
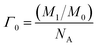 | (3) |
The large Γ might be ascribed to the 3D porous structure integrated with the 3D-CVS electrode, which provides a large specific surface area to effectively immobilize a large number of GOD molecules. Moreover, some microporosities and defects in the integrated 3D-CVS electrode might also enhance the immobilization of GOD molecules.
At a scan rate of 100 mV s−1, ΔEp = 45 mV and nΔEp = 90 mV < 200 mV, the electron transfer coefficient (αs) was determined to be 0.5. The electron-transfer rate constant (ks) could be estimated based on Laviron theory:44
| Ks = mnFυ/RT | (4) |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 493 C mol−1), υ was the scan rate, R was the gas constant (R = 8.314 J mol−1 K−1), T was the temperature in kelvin (T = 298 K) and m was a constant which relates to ΔEp. The Ks was calculated to be 2.30 s−1 for the 3D-CVS/GOD electrode, larger than that of the previously reported gelatin-multiwalled carbon nanotube/GOD/glutaraldehyde/GCE of 1.08 s−1 (ref. 28) and GOD/PDDA/multi-walled carbon nanotube (MWCNT)-AuNP/GCE of 1.01 s−1.55 The experimental results indicated that the 3D-CVS electrode could effectively promote the electron transfer of GOD molecule. Fast electron transfer might be attributed to the special 3D porous and layered structure of the 3D-CVS electrode, which effectively improves the electrolyte diffusion between GOD and the electrode surface to facilitate rapid electron transfer. The good electrical conductivity of the 3D-CVS electrode36 and the uniform distribution of GOD molecules on the 3D-CVS electrode further enhance the electron transfer.
493 C mol−1), υ was the scan rate, R was the gas constant (R = 8.314 J mol−1 K−1), T was the temperature in kelvin (T = 298 K) and m was a constant which relates to ΔEp. The Ks was calculated to be 2.30 s−1 for the 3D-CVS/GOD electrode, larger than that of the previously reported gelatin-multiwalled carbon nanotube/GOD/glutaraldehyde/GCE of 1.08 s−1 (ref. 28) and GOD/PDDA/multi-walled carbon nanotube (MWCNT)-AuNP/GCE of 1.01 s−1.55 The experimental results indicated that the 3D-CVS electrode could effectively promote the electron transfer of GOD molecule. Fast electron transfer might be attributed to the special 3D porous and layered structure of the 3D-CVS electrode, which effectively improves the electrolyte diffusion between GOD and the electrode surface to facilitate rapid electron transfer. The good electrical conductivity of the 3D-CVS electrode36 and the uniform distribution of GOD molecules on the 3D-CVS electrode further enhance the electron transfer.
3.3. Electrocatalytic properties of 3D-CVS/GOD electrodes
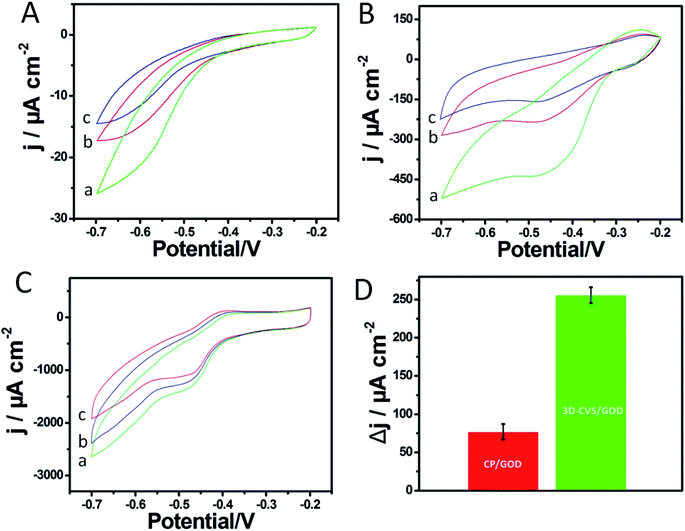
In order to explore the electrochemical properties of the 3D-CVS/GOD electrode, the CVs of various electrodes including glass carbon/GOD (GC/GOD) electrode, carbon paste/GOD (CP/GOD) electrode and 3D-CVS/GOD electrode were investigated in 0.2 M PBS (pH = 7.0) containing 0.0, 5.0 and 10.0 mM glucose, respectively (Fig. 3A–C). As shown by curve a in Fig. 3C, the CVs of 3D-CVS/GOD exhibited an obvious cathodic peak and a disappeared anodic peak in the presence of O2, indicating a typical electrocatalytic reduction of O2.45 With the increase of glucose concentration, the cathodic current of the electrode decreases gradually, indicating that the oxygen in the solution is consumed. Therefore, the glucose could be catalytically oxidized into gluconolactone by GOD on the 3D-CVS/GOD and the generated GOD (FADH2) could be oxidized into GOD (FAD) by O2. The possible mechanism could be shown by the following equations.46,47| Glucose + GOD (FAD) → gluconolactone + GOD (FADH2) | (5) |
| GOD (FADH2) + O2 → GOD (FAD) + H2O2 | (6) |
In total:
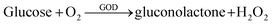 | (7) |
Simultaneously, it could be seen that the cathodic current at the 3D-CVS/GOD electrode (Fig. 3C) exhibited the regular decrease with the addition of glucose, while the CP/GOD electrode (Fig. 3B) showed an irregular change at the investigated potential range, and the peak current response toward the 5.0 to 10.0 mM of glucose was only 1/3 that of the 3D-CVS/GOD electrode (Fig. 3D). The ability of 3D-CVS/GOD electrode to enhance electrocatalytic activity might have originated from the nitrogen-doped porous architecture, which enables a large number of GOD molecules to be firmly immobilized on 3D-CVS and effectively maintain their biological activity. As shown in Fig. 3A, no obvious cathodic and anodic current appeared at the GC/GOD electrode due to the lack of defects and active sites to catalyze the reduction of O2. Furthermore, it was difficult to immobilize GOD on the smooth surface of the GC electrode, which resulted in a poor response toward glucose accordingly.21
3.4. Voltammetric detection of glucose
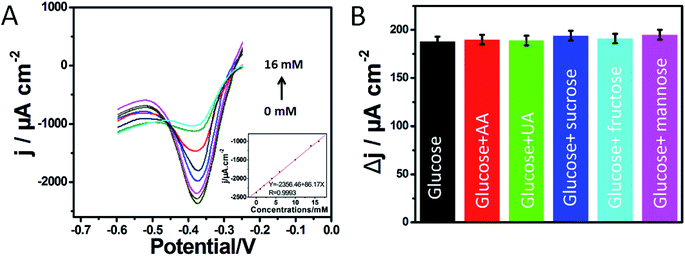
To further illustrate the relationship between the electrocatalytic current and the concentration of glucose, DPV measurements were performed in 0.2 M PBS (pH 7.0) in the voltage range from −0.6 to −0.2 V (Fig. 4A). The sharp and well-defined reduction peak currents decreased with the successive addition of glucose. At the same time, the reduction potential remained steady. The calibration curve (inset of Fig. 4A) fitted to determine the relationship between the peak current and the concentration of glucose was described by Ipa (μA) = −2356.46 + 86.17C (mM) in the concentration range of 0.58 μM to 16 mM with R = 0.9993, and the sensitivity was estimated to be 86.17 μA cm−2 mM−1. The detection limit was calculated to be 0.19 μM (S/N = 3). To exhibit the advantages of proposed biosensor, the analytical performances of developed biosensor were compared with other glucose sensors in Table 1. The results showed that the designed biosensor of the carbon material exhibited a wide linear range and a high sensitivity toward glucose detection. As mentioned above, some defects and microporosities in the 3D-CVS electrode could adsorb GOD molecules well. The 3D porous structure of 3D-CVS might improve the mass transfer and provide a large specific surface area, thus effectively loading a large number of GOD molecules. The good electrical conductivity of 3D-CVS electrode and uniform distribution of GOD molecules further enhanced the electron transfer. Therefore, the biosensor we studied has good conductivity. The synthesis of carbon materials is always very complex, compared with 3D-CVS, which is a natural and simple material. As could be seen, the comparison of analytical performance of our proposed glucose biosensors with selected ones that have been proposed by others in earlier publications in Table 1 perhaps don't have a very clearly significant advancement. However, it is possible that further treatment by N-doping or fabricating a lot of porosities or defects in the 3D-CVS electrode should improve the performance of our developed biosensor.| Glucose sensors | Linear range (mM) | Detection limit (μM) | Sensitivity (μA cm−2 mM−1) | Reference |
|---|---|---|---|---|
| a Iron phthalocyanine.b Quantum dots.c Electrochemically reduced graphene oxide.d Multiwalled carbon nanotubes.e Polyvinyl alcohol.f Partially prehydrolyzed tetraethyl orthosilicate.g 11-Mercaptoundecanoic acid.h 6-Mercapto-1-hexanol.i Polydopamine. | ||||
| GOD@TiO2/FePca-CNTs | 0.05–4.0 | 30.0 | 65.3 | 48 |
| GOD/Mn-doped ZnS QDsb | 0.01–0.1 and 0.1–1.0 | 3.0 | — | 49 |
| GOD/ERGOc-MWCNTsd | 0.01–6.5 | 4.7 | 7.95 | 50 |
| GOD/PVAe-Au-pphTEOSf | 1.0–8.0 | 0.7 | 43.22 | 51 |
| 4-Amino thiophenol/AuNP/GOD-HRP/MUAg-MCHh/Au | 0.0165–10.0 | 5.4 | 41.78 | 52 |
| GOD/CNTs | 0.04–1.0 | 20.0 | 2.4 | 53 |
| GOD/Ag-Pdopi@CNTs | 0.05–1.1 | 17.0 | 3.1 | 54 |
| GOD/graphene–chitosan | 0.08–12.0 | 20.0 | 37.93 | 55 |
| GOD/CVS electrode | 0.00058–16 | 0.19 | 86.17 | In this work |
3.5. Repeatability and stability of the biosensor
The repeatability of biosensor was investigated by successively detecting 5.0 mM glucose for 6 times, and the RSD was about 3.52%, indicating a good repeatability. The current responses were checked daily up to 14 days, and the current response was still retained 96.9% (Fig. S6†) value of the initial response. The results indicated that the 3D-CVS/GOD electrode showed good repeatability and stability.3.6. Interference and glucose detection in human blood serum
Some possibly coexisting materials such as some carbohydrates, AA and UA were used to test the selectivity of the glucose biosensor. The 3D-CVS/GOD electrode was first kept in a stirred 0.2 M O2-saturated PBS (pH 7.0) at scan rate of 100 mV s−1 and then a 2.0 mM glucose solution was added. After that, 6.0 mM AA, 6.0 UA mM, 6.0 mM sucrose, 6.0 fructose and 6.0 mM mannose were added sequentially. The results indicated that the possibly coexisted materials have no obvious interference for the detection of glucose as shown in Fig. 4B. Hence, a highly selective response to glucose was obtained without the use of a permselective membrane or enzymatic peroxidation.In order to evaluate the practical application of the developed biosensor in clinical analysis, the 3D-CVS/GOD was utilized to detect glucose in human serum samples. As shown in Table S3,† different amounts of glucose were added into human serum samples for recovery tests. The test results from five repeated experiments showed acceptable recoveries between 98.95% and 101.19% and the RSD was lower than 3.21%. The results indicated that the developed method was reliable and applicable in real human sample analysis.
4. Conclusion
In summary, a promising glucose biosensor based on GOD molecules immobilized on the 3D-CVS electrode was successfully achieved. The results proved that the integrated 3D-CVS electrode could be used to effectively load a large number of biomolecules for electrochemical biosensing. The 3D porous structure obviously improved the mass transfer. The excellent conductivity was favorable for electron transfer between biomolecules and electrode surface. Some defects and microporosities formed in the 3D-CVS electrode resulted in a good adsorption of GOD molecules. The CVs results showed a pair of well-defined redox peaks corresponding to the electron transfer of GOD, which indicated that the direct electron transfer between GOD and the 3D-CVS electrode could be achieved. With wide linear range, high sensitivity and good selectivity, the new designed biosensor might be used for the detection of glucose concentrations in practical samples. Moreover, it is possible to fabricate other analogous enzyme electrode based on the integrated 3D-CVS electrode.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This research is financially supported by Special Research Project of Jiangxi University of Traditional Chinese Medicine (2016ZR002), Double First-class Discipline (Chinese materia medica) Construction Project Fund of Jiangxi Province (JXSYLXK-ZHYAO069, JXSYLXK-ZHYAO106), the Natural Science Foundation of Jiangxi Province (20171BAB213007, 2018BBE50004), the Education Department of Jiangxi Province (GJJ160326).References
- J. Wang, Chem. Rev., 2008, 108, 814–825 CrossRef CAS PubMed.
- C. Guo, H. Huo, X. Han, C. Xu and H. Li, Anal. Chem., 2014, 86, 876–883 CrossRef CAS PubMed.
- W. Lu, X. Qin, A. M. Asiri, A. O. Al-Youbi and X. Sun, Analyst, 2013, 138, 417–420 RSC.
- D. Su, B. Feng, P. Xu, Q. Zeng, B. Shan and Y. Song, Anal. Methods, 2018, 10, 4320–4328 RSC.
- Y. Song, D. Su, Y. Shen, H. Liu and L. Wang, Anal. Bioanal. Chem., 2017, 409, 161–168 CrossRef CAS PubMed.
- H. He, X. Xu, H. Wu, Y. Zhai and Y. Jin, Anal. Chem., 2013, 85, 4546–4553 CrossRef CAS PubMed.
- M. Mesch, C. Zhang, P. V. Braun and H. Giessen, ACS Photonics, 2015, 2, 475–480 CrossRef CAS.
- R. Chen, W. Xu, C. Xiong, X. Zhou, S. Xiong, Z. Nie, L. Mao, Y. Chen and H. C. Chang, Anal. Chem., 2011, 84, 465–469 CrossRef PubMed.
- K. Liu, Y. Song, Y. Liu, M. Peng, H. Li, X. Li, B. Feng, P. Xu and D. Su, J. Pharm. Biomed. Anal., 2017, 139, 165–178 CrossRef CAS PubMed.
- D. Su, W. Li, Q. Xu, Y. Liu, Y. Song and Y. Feng, Fitoterapia, 2016, 112, 45–55 CrossRef CAS PubMed.
- J. Kremeskotter, R. Wilson, D. Schiffrin, B. J. Luff and J. S. Wilkinson, Meas. Sci. Technol., 1995, 6, 1325 CrossRef.
- T. Wang, X. Hu, X. Zhang, H. Cao, Y. Huang and P. Feng, Anal. Methods, 2019, 11, 415–420 RSC.
- Y. Bao, X. Hua, H. Ran, J. Zeng and F. Wu, J. Mater. Chem. B, 2019, 7, 296–304 RSC.
- N. A. B. Ismail, A. W. Firdaus, N. I. Ramli, M. M. Bader and W. W. A. W. Salim, Nanotechnology: Applications in Energy, Drug and Food, 2019, pp. 423–439 Search PubMed.
- Y. Song, B. Shan, H. Li, B. Feng, H. Peng, P. Xu, Q. Zeng, Z. Liao, K. Liu and D. Su, J. Ethnopharmacol., 2019, 235, 435–445 CrossRef CAS PubMed.
- Q. Zeng, Y. Liu, Y. Song, B. Feng, P. Xu, B. Shan, Z. Liao, K. Liu, Y. Zhong, L. Chen and D. Su, J. Pharm. Biomed. Anal., 2019, 169, 215–224 CrossRef CAS PubMed.
- W. Ma, Q. Jiang, P. Yu, L. Yang and L. Mao, Anal. Chem., 2013, 85, 7550–7557 CrossRef CAS PubMed.
- Y. Song, H. Liu, Y. Wang and L. Wang, Electrochim. Acta, 2013, 93, 17–24 CrossRef CAS.
- P. Yang, L. Wang, Q. Wu, Z. Chen and X. Lin, Sens. Actuators, B, 2014, 194, 71–78 CrossRef CAS.
- D. Xiang, L. Yin, J. Ma, E. Guo, Q. Li, Z. Li and K. Liu, Analyst, 2015, 140, 644–653 RSC.
- Y. Song, J. Chen, H. Liu, Y. Song, F. Xu, H. Tan and L. Wang, Electrochim. Acta, 2015, 158, 56–63 CrossRef CAS.
- S. Palanisamy, C. Karuppiah and S.-M. Chen, Colloids Surf., B, 2014, 114, 164–169 CrossRef CAS PubMed.
- Q. Xu, S.-X. Gu, L. Jin, Y.-E. Zhou, Z. Yang, W. Wang and X. Hu, Sens. Actuators, B, 2014, 190, 562–569 CrossRef CAS.
- D. Li, M. B. Mueller, S. Gilje, R. B. Kaner and G. G. Wallace, Nat. Nanotechnol., 2008, 3, 101–105 CrossRef CAS PubMed.
- Y. Liu, D. Yu, C. Zeng, Z. Miao and L. Dai, Langmuir, 2010, 26, 6158–6160 CrossRef CAS PubMed.
- C. Shan, H. Yang, D. Han, Q. Zhang, A. Ivaska and L. Niu, Langmuir, 2009, 25, 12030–12033 CrossRef CAS PubMed.
- J. C. Meyer, A. K. Geim and M. I. Katsnelson, et al., Nature, 2007, 446(7131), 60 CrossRef CAS PubMed.
- H. Wang, Q. Lang, L. Li, B. Liang, X. Tang, L. Kong, M. Mascini and A. Liu, Anal. Chem., 2013, 85, 6107–6112 CrossRef CAS PubMed.
- M. Zhao, Y. Gao, J. Sun and F. Gao, Anal. Chem., 2015, 87, 2615–2622 CrossRef CAS PubMed.
- X. Lu, H. Cheng, P. Huang, L. Yang, P. Yu and L. Mao, Anal. Chem., 2013, 85, 4007–4013 CrossRef CAS PubMed.
- J. M. Goran, S. M. Mantilla and K. J. Stevenson, Anal. Chem., 2013, 85, 1571–1581 CrossRef CAS PubMed.
- S. S. Karajanagi, A. A. Vertegel and R. S. Kane, et al., Langmuir, 2004, 20(26), 11594–11599 CrossRef CAS PubMed.
- K. Matsuura, T. Saito and T. Okazaki, et al., Chem. Phys. Lett., 2006, 429(4–6), 497–502 CrossRef CAS.
- Y. Song, B. Shan, B. Feng, P. Xu, Q. Zeng and D. Su, RSC Adv., 2018, 8, 27008 RSC.
- W. R. Yang, J. J. Gooding and D. B. Hibbert, J. Electroanal. Chem., 2001, 516, 10–16 CrossRef CAS.
- H. O. Finklea, D. A. Snider, J. Fedyk, E. Sabatani, Y. Gafni and I. Rubinstein, Langmuir, 1993, 9, 3660–3667 CrossRef CAS.
- X. Kang, J. Wang, H. Wu, I. A. Aksay, J. Liu and Y. Lin, Biosens. Bioelectron., 2009, 25, 901–905 CrossRef CAS PubMed.
- C. Deng, J. Chen, Z. Nie and S. Si, Biosens. Bioelectron., 2010, 26, 213–219 CrossRef CAS PubMed.
- R. Freeman, B. Willner and I. Willner, J. Phys. Chem. Lett., 2011, 2, 2667–2677 CrossRef CAS.
- J. Hodak, R. Etchenique and E. J. Calvo, et al., Langmuir, 1997, 13(10), 2708–2716 CrossRef CAS.
- C. Shan, H. Yang, J. Song, D. Han, A. Ivaska and L. Niu, Anal. Chem., 2009, 81(6), 2378–2382 CrossRef CAS PubMed.
- Y. Huang, W. Zhang and H. Xiao, et al., Biosens. Bioelectron., 2005, 21(5), 817–821 CrossRef CAS PubMed.
- H. J. Hecht, H. M. Kalisz and J. Hendle, et al., J. Mol. Biol., 1993, 229(1), 153–172 CrossRef CAS PubMed.
- E. Laviron, J. Electroanal. Chem., 1979, 101, 19–28 CrossRef CAS.
- Q. Zeng, J.-S. Cheng, X.-F. Liu, H.-T. Bai and J.-H. Jiang, Biosens. Bioelectron., 2011, 26, 3456–3463 CrossRef CAS PubMed.
- M. Wooten, S. Karra, M. Zhang and W. Gorski, Anal. Chem., 2014, 86, 752–757 CrossRef CAS PubMed.
- F. Qu, Y. Zhang, A. Rasooly and M. Yang, Anal. Chem., 2014, 86, 973–976 CrossRef CAS PubMed.
- H. Cui, K. Zhang, Y. Zhang, Y. Sun, J. Wang, W. Zhang and J. H. T. Luong, Biosens. Bioelectron., 2013, 46, 113–118 CrossRef CAS PubMed.
- P. Wu, Y. He, He. Wang and X. Yan, Anal. Chem., 2010, 82, 1427–1433 CrossRef CAS PubMed.
- V. Mani, B. Devadas and S. Chen, Biosens. Bioelectron., 2013, 41, 309–315 CrossRef CAS PubMed.
- U. Lad, G. M. Kale and R. Bryaskova, Anal. Chem., 2013, 85, 6349–6355 CrossRef CAS PubMed.
- C. Gong, J. Chen, Y. Song, M. Sun, Y. Song, Q. Guo and L. Wang, Anal. Methods, 2016, 8, 1513–1519 RSC.
- X. Luo, A. J. Killard and M. R. Smyth, Electroanalysis, 2006, 18, 1131–1134 CrossRef CAS.
- Y. Wang, L. Liu, M. Li, S. Xu and F. Gao, Biosens. Bioelectron., 2011, 30, 107–111 CrossRef CAS PubMed.
- X. Kang, J. Wang, H. Wu, I. A. Aksay, J. Liu and Y. Lin, Biosens. Bioelectron., 2009, 25, 901–905 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9ra04008k |
| ‡ These authors contribute equally to this work. |
| This journal is © The Royal Society of Chemistry 2019 |