Open Access Article
Open Access ArticleEvaluation of plasmid DNA stability against ultrasonic shear stress and its in vitro delivery efficiency using ionic liquid [Bmim][PF6]†
Satya Ranjan Sarkerab,
Andrew S. Ballc,
Suresh K. Bhargava *a and
Sarvesh K. Soni
*a and
Sarvesh K. Soni *ac
*ac
aCentre for Advanced Materials and Industrial Chemistry, RMIT University, Melbourne, Australia. E-mail: sarvesh.soni@rmit.edu.au; suresh.bhargava@rmit.edu.au
bDepartment of Biotechnology and Genetic Engineering, Jahangirnagar University, Savar, Dhaka 1342, Bangladesh
cSchool of Science, RMIT University, GPO Box 2476, Melbourne, VIC 3001, Australia
First published on 17th September 2019
Abstract
The hydrophobic ionic liquid (IL) 1-butyl-3-methylimidazolium hexafluorophosphate [Bmim][PF6] forms nanostructures with negatively charged plasmid DNA through electrostatic interactions. The formation of plasmid DNA/IL nanostructures was confirmed by measuring the zeta potential of plasmid DNA as well as plasmid DNA/IL nanostructures. The zeta potential of the nanostructures was positive, although plasmid DNA is negatively charged. The positive zeta potential is due to the complexation between plasmid DNA and positively charged ionic liquid [Bmim][PF6]. The ability of ionic liquid [Bmim][PF6] to protect plasmid DNA against ultrasonic shear stress was also investigated using an agarose gel electrophoretic assay and showed that ionic liquid stabilizes plasmid DNA against ultrasonication. The plasmid DNA and plasmid DNA/IL nanostructures were subjected to ultrasonic shear stress for different time periods and the biological functionality of pristine plasmid DNA (i.e., expression of the eGFP gene) as well as the self-assembled nanostructures was investigated in vitro using three different cell lines, COS7, HEK293 and HeLa. Ionic liquid [Bmim][PF6] protected the plasmid DNA against ultrasonic shear stress and also enhanced gene transfection efficiency in vitro. Furthermore, the cytotoxicity of ionic liquid [Bmim][PF6] was assayed in vitro using all three cell lines and the toxicity was very low. Therefore, the ionic liquid [Bmim][PF6] stabilizes plasmid DNA against ultrasonic shear stress and also enhances its in vitro delivery efficiency.
Introduction
Non-viral vectors are being extensively studied for their potential use in gene therapy to treat various complicated diseases. Non-viral vectors protect plasmid DNA against physical, chemical, and enzymatic degradation and deliver the DNA molecule to the target site. For example, cationic liposomes,1–3 chitosan4,5 and other positively charged nanoparticles6,7 form complexes with plasmid DNA through electrostatic interactions. However, the readily formed cationic liposomes/plasmid DNA complexes are relatively large (i.e., 300–400 nm) and heterogeneous in nature making them difficult to use in pharmaceutical applications.8,9 The large and heterogeneous plasmid DNA/liposomes,8 plasmid DNA/aerosols,10 and plasmid DNA/peptides11 complexes can be reduced to smaller, and homogeneous particles using ultrasonication. Moreover, the uncomplexed plasmid DNA gets degraded when exposed to high frequency ultrasonication, which is frequently used to reduce the hydrodynamic size of plasmid DNA/nonviral vector complexes.8,11 Because of the ultrasonic cavitation, the structure of plasmid DNA can be interrupted in many ways such as breakage of single strands, double strands, rupture of hydrogen bonds, base destruction, as well as cross linking.12 These breaks occur due to the exposure of DNA directly to the physical forces or the synthesis of highly toxic sonochemicals such as hydrogen peroxide12,13 during the sonication process. Ultrasonic shear stress also modifies the coiling structure of plasmid DNA and converts the supercoiled plasmid to open circular, linear, or even fragments them.10 Although ultrasonic shear stress degrades the plasmid DNA, many research groups have used high speed sonicative force to enhance the cellular uptake of DNA11 and to improve the encapsulation efficiency of DNA into liposomes.11,14Various polymers,15 peptides,11 and lipids8 protect the plasmid DNA against high frequency ultrasonication, and endonucleases have been reported. However, these polymers are often toxic, expensive, difficult to synthesize, and bring about immunogenicity.1,2
Ionic liquids (ILs) have been used as a green solvent to facilitate the polymerase chain reactions of DNA,16 and to extract double stranded (dsDNA) without using any other materials.17 They have also been used as a reaction solvent for water-insoluble macromolecules such as chitosan, cellulose, keratin, and silk fibroin.15,18 Furthermore, ionic liquids have been reported to enhance transcutaneous vaccination using proteins and peptide antigens19 and also help to preserve plasmid DNA for long-term storage at room temperature.20 The negatively charged plasmid DNA forms functional nanostructures with the positively charged hydrophobic ionic liquid [Bmim][PF6] through electrostatic interactions.21,22 ILs also interact with the hydrophobic bases of DNA through hydrophobic interactions23–25 and compact the coiling structure of DNA resulting in the reduced hydrodynamic diameter of the DNA/IL nanostructures.24 Recently, we have reported21 the application of hydrophobic ionic liquid as a non-viral plasmid DNA delivery vehicle to bacteria.
The plasmid delivery efficiency of non-viral vectors depends on their size, surface charge/zeta potential, biodegradability and biocompatibility, cargo loading ability and so on. To reduce the size of the non-viral vector/plasmid DNA complexes and maintain their size homogeneity, the complexes are subjected to sonication. The supercoiled structure of plasmid DNA shows the highest gene expression efficiency followed by circular DNA, with linear DNA showing the lowest gene expression efficiency.11
In this study, we focused on the potential application of hydrophobic ionic liquid [Bmim][PF6] to stabilize plasmid DNA against high frequency water bath ultrasonication. In vitro gene expression into various cell lines was also performed to confirm that DNA in the plasmid DNA/IL nanostructures subjected to ultrasonic shear stress was still biologically functional. At the same time, the potential role of IL in in vitro plasmid DNA delivery to various cell lines such as COS-7, HeLa and HEK293 was investigated. The cytotoxicity of ionic liquid [Bmim][PF6] was also investigated in all three cell lines.
Materials and methods
Materials
Ionic liquid (IL) 1-butyl-3-methylimidazolium hexafluorophosphate ([Bmim][PF6]) was purchased from Ionic Liquid Technologies (IoLiTec). Cervical Cancer Cells (HeLa cells), African Green Monkey Kidney Cells (COS7 cells), Human Embryonic Kidney Cells (HEK293 cells) were provided by the Micro Nano Research Facility (MNRF) at RMIT University. Dulbecco's Modified Eagles Medium (DMEM), fetal bovine serum (FBS), phosphate buffer saline (PBS) and penicillin–streptomycin were also obtained from the MNRF at RMIT University. 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide dye (MTT); pEGFP was obtained from the School of Science, RMIT University. Lipofectamine was purchased from Thermo Fisher Scientific, Australia.Preparation of plasmid DNA/ionic liquid (IL) complexes
Plasmid DNA, with a reporter ‘enhanced green fluorescent protein (pEGFP)’ tag formed nanostructures with ionic liquid [Bmim][PF6] through electrostatic interactions and the nanostructures were prepared according to the previously established protocol.21 The detailed protocol for the preparation of plasmid DNA/IL nanostructures has been described in the ESI Section.†Investigation of the stability of plasmid DNA, and plasmid DNA/IL nanostructures against ultrasonic shear stress
Plasmid DNA (200 ng) was subjected to high speed ultrasonic shear stress for different time periods (10, 20, 30, and 40 min) using water bath ultrasonicator. Similarly, pDNA/IL nanostructures were also subjected to high speed ultrasonic shear stress for different time periods (10, 20, 30, 40, 60, 90, and 120 min) by ultrasonication. The stability and functionality of plasmid DNA was investigated using agarose gel electrophoresis, and an in vitro GFP expression assay.Agarose gel electrophoresis
Agarose gel electrophoresis was performed to investigate the stability of plasmid DNA and plasmid DNA/IL nanostructures against ultrasonic shear stress. Plasmid DNA and plasmid DNA/IL nanostructures (after being subjected to ultrasonic shear stress for different time periods) were mixed with bromophenol blue and same amount of DNA (100 ng) was loaded onto the wells of 1% agarose gel containing SYBR safe dye. A DNA ladder was also loaded onto two different wells of both sides of the gel. The gel was then run for 30 min using Tris–EDTA buffer at 90 V ensuring that the dye migrated at least two-thirds of the gel. The subsequent gel was documented using a Bio-Rad Gel-doc system.Determination of the zeta potential
The zeta potential of both plasmid DNA and plasmid DNA/IL nanostructures was determined using a Zetasizer (Malvern, Nano Range, UK).Cell cultures
COS7 cells (African Green Monkey Kidney Cells), HeLa cells (Cervical Cancer Cells), and HEK293 (Human Embryonic Kidney Cells 293) were obtained from the PC2 mammalian cell laboratory's cell repository, MicroNano Research facility at RMIT University and cultured in DMEM supplemented with heat inactivated 10% FBS, 100 IU mL−1 penicillin, and 100 μg mL−1 penicillin/streptomycin. All cell cultures were maintained in an atmosphere of 5% CO2 at 37 °C.In vitro gene expression assay
COS7, HEK293, and HeLa cells were used to evaluate the stability, biological functionality as well as the gene delivery efficiency of pDNA/IL nanostructures. COS7 cells (1 × 105 cells per well), HEK293 cells (1 × 105 cells per well), and HeLa cells (1 × 105 cells per well) were seeded into 24 well plates and cultured with DMEM containing 10% FBS and 1% penicillin/streptomycin and incubated in an atmosphere of 5% CO2 and 37 °C until the cells reached ∼80% confluency. The medium in each well of the cell culture plates was then exchanged with pDNA/lipofectamine nanostructures sonicated for different time points (10, 20, 30 and 40 min) in the presence of 10% FBS and incubated for 24 h at 37 °C and 5% CO2. Furthermore, pDNA/IL nanostructures, sonicated for different time points (10, 20, 30, 40, 60, 90 and 120 min) were mixed with either 1 μL or 2 μL lipofectamine (LA) and incubated for 20 min at room temperature to allow the formation of pDNA/IL/LA nanostructures. An aliquot (1 μL) of pDNA/IL/LA nanostructures, prepared with LA were incubated with HeLa cells and the nanostructures prepared with LA (2 μL) were incubated with the COS7 and HEK293 cells. All the cells lines were incubated for 24 h in the presence of 10% FBS in a 5% CO2 incubator at 37 °C. The medium was then replaced with fresh complete DMEM containing 10% FBS and incubated for another 24 h under the same condition. Finally, the green fluorescent protein (GFP) positive cells were observed using a fluorescent microscope (ZOE, Biorad) and GFP positive cells were counted.MTT assay
Cytotoxicity was evaluated using the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay method.26 Before the cell viability assay, COS7, HeLa and, HEK293 cells were seeded in a 96-well plate at a density of 5 × 103 cells per well and incubated overnight in a 5% CO2 incubator at 37 °C. After incubation (1 × 105 cells per well), the old media was replaced with 100 μL complete DMEM (DMEM containing 10% FBS, and 1% penicillin/streptomycin) containing different volumes of ionic liquid (0.015 μL to 4.43 μg, 0.03 μL to 8.87 μg, 0.06 μL to 17.75 μg, 0.125 μL to 35.5 μg, 0.25 μL to 71 μg, and 0.5 μL to 142 μg) and incubated for 24 h in a 5% CO2 incubator at 37 °C. After 24 h, the medium containing different volumes of ionic liquid was aspirated and 100 μL of culture media (i.e., DMEM) containing 5 mg mL−1 MTT (thiazolyl blue tetrazolium bromide) was added to each well and cells were further incubated for 4 h in dark at 37 °C. After 4 h incubation, the media containing MTT was removed and DMSO (100 μL) was added to each well to solubilize the crystallized formazan product. The plates were read on a micro-plate reader at 570 nm and a reference wavelength of 630 nm.The wells treated with only DMEM medium were used as control. Cell viability was calculated as follows: cell viability = OD570 (sample)/OD570 (control) × 100%. All the experiments were carried out in triplicate to ensure the reproducibility.
Results and discussion
Analysis of DNA stability through electrophoretic assay
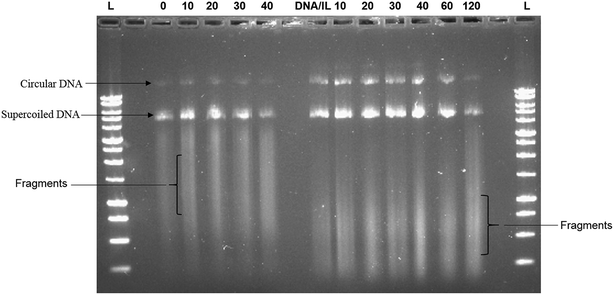
The ability of the hydrophobic ionic liquid [Bmim][PF6] to protect plasmid DNA against high frequency ultrasonic shear stress was investigated through agarose gel electrophoresis assay. Fig. 1 shows the presence of two distinct bands (i.e., one at the bottom and the other one at the middle) and a smear band in the gel. The intensity of the bands for DNA/IL nanostructures was greater at all time points of sonication when compared to that of the native DNA. Furthermore, the ultrasonic shear stress converts supercoiled plasmid to open circular, linear or even fragmented DNA.10 The positively charged hydrophobic IL forms electrostatic interactions with the negatively charged plasmid DNA21 protecting the plasmid DNA against high frequency ultrasonic shear stress.Furthermore, Fig. 1 also showed that the smear bands of the IL treated samples migrate consistently farther through the gel than those of the non-IL treated samples. The fact that the unsonicated IL-treated sample (0 hour) showed the same smear band shift as the sonicated samples (10–90 min) suggested that this increase in fragment migration was not due to sonication. In our previous report,21 for the CD spectra of DNA-IL, we observed an intense long wave positive band for DNA-IL that restrict various degree of freedom of DNA fragments and it led for farther and faster migration of DNA-IL fragments in agarose gel.
Determination of the zeta potential of plasmid DNA and plasmid DNA/IL nanostructures
The zeta potentials of native plasmid DNA and plasmid DNA/IL nanostructures were −0.259 ± 0.03 mV, and 0.360 ± 0.04 mV, respectively (Table S1†). Zeta potential of the nanostructures plays important role in their stability as well as their cellular uptake efficiency.27 As the zeta potential increases in both the positive and negative scale, the stability of the nanostructures increases. Usually, particles with positive zeta potential interact more with the negatively charged cell membrane and their cellular uptake efficiency is also very high.28Analysis of the biological functionality of the pDNA/IL nanostructures
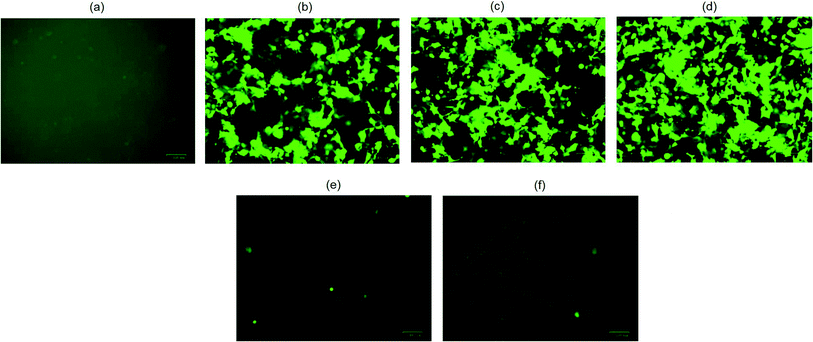
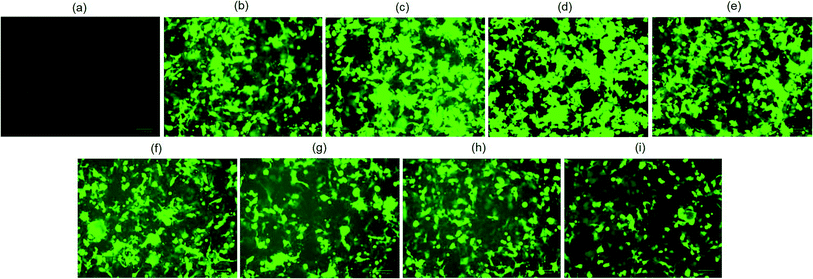
The biological functionality of pristine plasmid DNA and plasmid DNA/IL nanostructures were investigated using three different cell lines, COS7, HEK293 and HeLa cells in terms of their transfection efficiency. All transfection assays were performed in the presence of 10% fetal bovine serum (FBS). When native plasmid DNA was incubated with COS7 cells, there was no fluorescent positive cells (Fig. 2a) observed due to electrostatic repulsion among the negatively charged pDNA and COS7 cells. In contrast, when pDNA (200 ng) was subjected to ultrasonic shear stress for 0, 10, 20, 30 and 40 min, and thereafter complexed with lipofectamine, no fluorescent positive COS7 cells was observed for pDNA subjected to ultrasonication for 30, and 40 min (Fig. 2e and f). This is likely due to extensive fragmentation of DNA11 through rupturing of the DNA double helical structure, damage to the phosphodiester bonds as well as the ribose sugars due to the high speed ultrasonic shear stress.8 The number of fluorescent positive COS7 cells were similar when pDNA was subjected to ultrasonication for 0, 10, and 20 min (Fig. 2b–d and S1†). These results confirm that the structure and functionality of the plasmid DNA is retained for up to 20 minutes of ultrasonication.When plasmid DNA was complexed with hydrophobic ionic liquid [Bmim][PF6] to make pDNA/IL nanostructures and incubated with African green monkey kidney cells (i.e., COS7 cells), there was no fluorescent positive cells (Fig. 3a). This may be due to the almost neutral zeta potential of the nanostructures (Table S1†) which results in their aggregation and therefore no cellular uptake. However, when the pDNA/IL nanostructures were subjected to ultrasonic shear stress for 0, 10, 20, 30, 40, 60, 90, and 120 min and complexed with the commercially available cationic gene delivery agent lipofectamine, the percent of fluorescent positive cells were 80%, 98%, 97%, 85%, 78%, 65%, 65%, and 50%, respectively (Fig. 3b–i, and S2†). The percentage of fluorescent positive cells increased when the nanostructures were subjected to ultrasonic shear stress for 10, and 20 min, and thereafter decreased. The observed increased transfection efficiency may be due to the ultrasonic shear stress that reduces the nanostructures size and increases the stability against serum endonuclease.11,29 When the small sized pDNA/IL nanostructures interact with the positively charged lipofectamine, they form positively charged stable pDNA/IL/LA complexes that are readily taken up by the cells and bring about higher transfection efficiency. The size of the particles play an important role in the cellular uptake efficiency.27 The larger the particles size, the lower the cellular uptake efficiency and vice versa. The percentage of fluorescent positive cells reduced as the time of exposure of the pDNA/IL nanostructures to ultrasonic shear stress increased from 40 min to 120 min (Fig. 3e–i, and S2†). This reduction of the fluorescent positive cells is due to the extensive fragmentation and disruption of the DNA structure.11
Plasmid DNA/IL nanostructures were also delivered to human embryonic kidney 293 (HEK293) cells in the presence of 10% FBS. In HEK293 cells, plasmid DNA and pDNA/LA complexes were taken as negative control, and positive control, respectively. There were no fluorescent positive cells when only pDNA was incubated with HEK293 cells (Fig. 4a). However, the number of fluorescent positive cells in case of pDNA/IL/LA nanostructures transfected cells almost doubled compared to that of the pDNA/LA transfected cells (Fig. 4b and c, and S3†). Hydrophobic ionic liquid [Bmim][PF6] also enhances the transfection efficiency of pDNA/IL/LA nanostructures to cervical cancer cells, HeLa (Fig. S4†). In the presence of IL, the gene delivery efficiency of pDNA/LA to HEK293 cells, and HeLa cells increased almost 2-fold. This increase in the gene delivery efficiency in the presence of the ionic liquid [Bmim][PF6] is due to the protection of DNA against serum endonucleases and the hydrophobic interactions with the endosomal membranes.30,31 This results in the rupture of endosomes and the release of plasmid DNA which then enter into the nucleus through the nuclear pore complex present on the membrane of the nucleus.
Analysis of in vitro cytotoxicity of ionic liquids
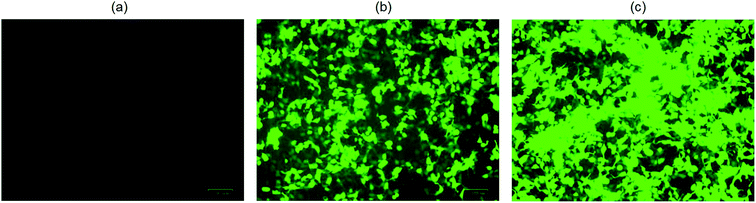
The cytotoxicity of the ionic liquid [Bmim][PF6] was assayed in vitro in three different cell lines viz. COS7 cells, HEK293 cells, and HeLa cells. The ionic liquid is not toxic to all the cell lines in the presence of [Bmim][PF6] (0.015 μL). However, it shows toxicity to all the cell lines as the volume of IL increased from 0.03 μL to 0.5 μL (Fig. 5a–c). HEK 293 cells were more sensitive to IL, showing toxicity when the volume of IL was increased from 0.015 μL to 0.03 μL. The degree of cytotoxicity varies for different cell lines due variation in their membrane molecular structure and composition32 which may bring about different degrees of interaction with the IL. The low cytotoxicity of the IL is due to the smaller alkyl chain length33 and ILs with C8 are more toxic when compared to those with C6 and C4, regardless of the type of cation they contain.34 It has been reported that the longer alkyl chain brings about cytotoxicity upon inhibiting the acetylcholinesterase enzyme.34 The elongated alkyl chain length also increases the lipophilicity of the ILs which is responsible for the incorporation of IL with the phospholipid bilayers of biological membranes through hydrophobic interactions.34 The toxicity of ILs can also be attributed to their membrane permeability altering the physical properties of the lipid bilayer.29,35,36 Furthermore, the structure of [Bmim][PF6] contains a imidazolium ring which is similar to the structure of detergents, pesticides and antibiotics that disturbs the cell membrane lipid structure or cationic surfactants that cause membrane-bound protein disruption37 and cause the cell death.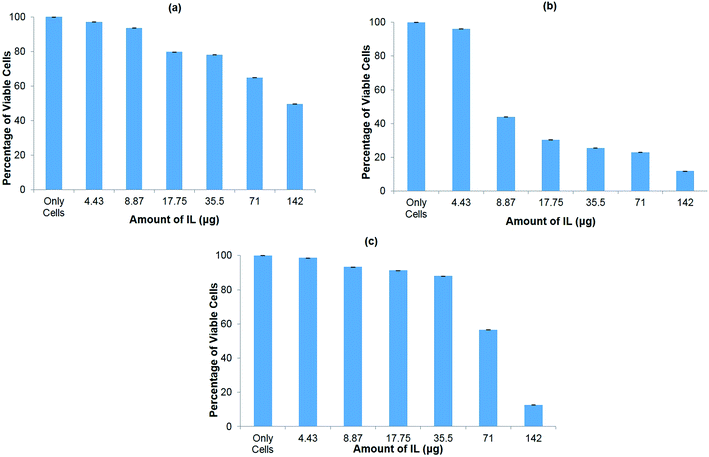 | ||
| Fig. 5 In vitro cytotoxicity assay for hydrophobic ionic liquid (IL) [Bmim][PF6] to different cell lines. (a) COS7 cells; (b) HEK293 cells; (c) HeLa cells. | ||
Conclusions
The hydrophobic ionic liquid [Bmim][PF6] protects plasmid DNA against high frequency ultrasonic shear stress for up to 120 min. Plasmid DNA usually gets degraded after 30 min of ultrasonication. Thus, IL could be used as a potential agent to protect plasmid DNA against mechanical forces as well as various serum endonucleases. Ionic liquid also enhances DNA transfection efficiency to a variety of cell lines such as COS7 cells, HEK293 cells as well as HeLa cells in the presence of serum proteins. Therefore, ionic liquid [Bmim][PF6] stabilizes plasmid DNA against ultrasonic shear stress and enhances their in vitro delivery efficiency. Due to its low cytotoxicity and ability to increase the gene transfection efficiency, IL can be used as a co-delivery agent to deliver nucleic acids, oligonucleotides (e.g., siRNA, and shRNA), and other biological macromolecules such as antibodies.Conflicts of interest
There is no conflict to declare.Acknowledgements
The authors acknowledge Micro Nano Research Facility of RMIT University. S. R. S. thanks the Department of Education and Training, Govt. of Australia, for the award of an Endeavour Executive Fellowship.References
- S. R. Sarker, S. Arai, M. Murate, H. Takahashi, M. Takata, T. Kobayashi and S. Takeoka, Int. J. Pharm., 2012, 422, 364–373 CrossRef CAS PubMed.
- S. R. Sarker, Y. Aoshima, R. Hokama, T. Inoue, K. Sou and S. Takeoka, Int. J. Nanomed., 2013, 8, 1361–1375 CrossRef PubMed.
- Y. Aoshima, R. Hokama, K. Sou, S. R. Sarker, K. Iida, H. Nakamura, T. Inoue and S. Takeoka, ACS Chem. Neurosci., 2013, 4, 1514–1519 CrossRef CAS PubMed.
- B. Shi, Z. Shen, H. Zhang, J. Bi and S. Dai, Biomacromolecules, 2012, 13, 146–153 CrossRef CAS PubMed.
- J. Deng, Y. Zhou, B. Xu, K. Mai, Y. Deng and L. M. Zhang, Biomacromolecules, 2011, 12, 642–649 CrossRef CAS PubMed.
- J. J. Green, E. Chiu, E. S. Leshchiner, J. Shi, R. Langer and D. G. Anderson, Nano Lett., 2007, 7, 874–879 CrossRef CAS PubMed.
- C. H. Jones, C. K. Chen, M. Jiang, L. Fang, C. Cheng and B. A. Pfeifer, Mol. Pharmaceutics, 2013, 10, 1138–1145 CrossRef CAS PubMed.
- E. K. Wasan, D. L. Reimer and M. B. Bally, J. Pharm. Sci., 1996, 85, 427–433 CrossRef CAS PubMed.
- H. Hofland and L. Huang, Biochem. Biophys. Res. Commun., 1995, 207, 492–496 CrossRef CAS PubMed.
- S. J. Eastman, J. D. Tousignant, M. J. Lukason, H. Murray, C. S. Siegel, P. Constantino, D. J. Harris, S. H. Cheng and R. K. Scheule, Hum. Gene Ther., 1997, 8, 313–322 CrossRef CAS PubMed.
- R. C. Adami, W. T. Collard, S. A. Gupta, K. Y. Kwok, J. Bonadio and K. G. Rice, J. Pharm. Sci., 1998, 87, 678–683 CrossRef CAS PubMed.
- T. Kondo, S. Arai, M. Kuwabara, G. Yoshii and E. Kano, Radiat. Res., 1985, 104, 284–292 CrossRef CAS PubMed.
- D. L. Miller, R. M. Thomas and M. E. Frazier, Ultrasound Med. Biol., 1991, 17, 729–735 CrossRef CAS PubMed.
- I. Leibiger, B. Leibiger, D. Sarrach, R. Walther and H. Zuhlke, Biomed. Biochim. Acta, 1990, 49, 1193–1200 CAS.
- H. Chen, S. Cui, Y. Zhao, C. Zhang, S. Zhang and X. Peng, PLoS One, 2015, 10, e0121817 CrossRef PubMed.
- Y. Shi, Y. L. Liu, P. Y. Lai, M. C. Tseng, M. J. Tseng, Y. Li and Y. H. Chu, Chem. Commun., 2012, 48, 5325–5327 RSC.
- J. H. Wang, D. H. Cheng, X. W. Chen, Z. Du and Z. L. Fang, Anal. Chem., 2007, 79, 620–625 CrossRef CAS PubMed.
- R. P. Swatloski, S. K. Spear, J. D. Holbrey and R. D. Rogers, J. Am. Chem. Soc., 2002, 124, 4974–4975 CrossRef CAS PubMed.
- S. Araki, R. Wakabayashi, M. Moniruzzaman, N. Kamiya and M. Goto, Med. Chem. Commun., 2015, 6, 2124–2128 RSC.
- R. Vijayaraghavan, A. Izgorodin, V. Ganesh, M. Surianarayanan and D. R. MacFarlane, Angew. Chem., 2010, 49, 1631–1633 CrossRef CAS PubMed.
- S. K. Soni, S. Sarkar, N. Mirzadeh, P. R. Selvakannan and S. K. Bhargava, Langmuir, 2015, 31, 4722–4732 CrossRef CAS PubMed.
- D. H. Cheng, X. W. Chen, J. H. Wang and Z. L. Fang, Chemistry, 2007, 13, 4833–4839 CrossRef CAS PubMed.
- I. Khimji, K. Doan, K. Bruggeman, P.-J. J. Huang, P. Vajha and J. Liu, Chem. Commun., 2013, 49, 4537 RSC.
- S. Satpathi, A. Sengupta, V. M. Hridya, K. Gavvala, R. K. Koninti, B. Roy and P. Hazra, Sci. Rep., 2015, 5, 9137 CrossRef.
- A. Chandran, D. Ghoshdastidar and S. Senapati, J. Am. Chem. Soc., 2012, 134, 20330–20339 CrossRef CAS PubMed.
- T. S. Reddy, H. Kulhari, V. G. Reddy, V. Bansal, A. Kamal and R. Shukla, Eur. J. Med. Chem., 2015, 101, 790–805 CrossRef CAS PubMed.
- H. K. Ha, J. W. Kim, M. R. Lee, W. Jun and W. J. Lee, Asian-Australas. J. Anim. Sci., 2015, 28, 420–427 CrossRef CAS PubMed.
- P. Yingchoncharoen, D. S. Kalinowski and D. R. Richardson, Pharmacol. Rev., 2016, 68, 701–787 CrossRef CAS PubMed.
- A. Vidiš, C. A. Ohlin, G. Laurenczy, E. Küsters, G. Sedelmeier and P. J. Dyson, Adv. Synth. Catal., 2005, 347, 266–274 CrossRef.
- Y. Zhang, X. Chen, J. Lan, J. You and L. Chen, Chem. Biol. Drug Des., 2009, 74, 282–288 CrossRef CAS PubMed.
- R. R. Mazid, A. Cooper, Y. Zhang, R. Vijayaraghavan, D. R. MacFarlane, C. Cortez-Jugo and W. Cheng, RSC Adv., 2015, 5, 43839–43844 RSC.
- K. Romoren, B. J. Thu, N. C. Bols and O. Evensen, Biochim. Biophys. Acta, 2004, 1663, 127–134 CrossRef CAS PubMed.
- X. Liu, C. Ji, Q. Yang, Z. Bao, X. Fan, Y. Yang and H. Xing, ACS Sustainable Chem. Eng., 2017, 5, 1974–1981 CrossRef CAS.
- X. Wang, C. A. Ohlin, Q. Lu, Z. Fei, J. Hu and P. J. Dyson, Green Chem., 2007, 9, 1191 RSC.
- M. T. Garcia, N. Gathergood and P. J. Scammells, Green Chem., 2005, 7, 9 RSC.
- B. Jastorff, K. Mölter, P. Behrend, U. Bottin-Weber, J. Filser, A. Heimers, B. Ondruschka, J. Ranke, M. Schaefer, H. Schröder, A. Stark, P. Stepnowski, F. Stock, R. Störmann, S. Stolte, U. Welz-Biermann, S. Ziegert and J. Thöming, Green Chem., 2005, 7, 362 RSC.
- R. J. Bernot, E. E. Kennedy and G. A. Lamberti, Environ. Toxicol. Chem., 2005, 24, 1759 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9ra03414e |
| This journal is © The Royal Society of Chemistry 2019 |