Open Access Article
Open Access ArticleThe mutual influence between corrosion and the surrounding soil microbial communities of buried petroleum pipelines†
Hong Su ab,
Shuofu Mia,
Xiaowei Penga and
Yejun Han*a
ab,
Shuofu Mia,
Xiaowei Penga and
Yejun Han*a
aNational Key Laboratory of Biochemical Engineering, Institute of Process Engineering, Chinese Academy of Sciences, Beijing 100190, China. E-mail: yjhan@ipe.ac.cn; Tel: +86 18810182857
bSchool of Chemical Engineering, University of Chinese Academy of Sciences, Beijing 100049, China
First published on 17th June 2019
Abstract
Buried petroleum pipeline corrosion and leaks cause inevitable changes in the microbial communities of the surrounding soils. In addition, soils with different microbial communities can make different contributions to buried pipeline corrosion. Three kinds of soil samples of buried petroleum pipelines under different corrosion and petroleum contamination conditions were collected from the Shengli Oilfield of China to investigate the mutual influence between corrosion and the microbial communities of the surrounding soil. The 16S rRNA gene high-throughput Illumina MiSeq sequencing was used to analyze the microbial communities of different surrounding soils. Electrochemical tests were performed for steel corrosion investigation. The results showed that the microbial diversity of the surrounding soils of corroded pipelines with/without petroleum contamination (O-soil and C-soil, respectively) decreased significantly as compared with that of the non-corroded and non-contaminated ones (NC-soil). The C-soil contained more abundant Balneolaceae (Balneola, KSA1), Flavobacteriaceae (Muricauda, Gramella) and Desulfuromonadaceae (Pelobacter, Geoalkalibacter). The O-soil possessed a greater abundance of Halomonas, Pseudoalteromonas, Psychrobacter and Dietzia, which were reported to have a capacity for hydrocarbon degradation. Moreover, electrochemical measurements indicated that the microcosm of the C-soil and NC-soil promoted steel corrosion, while the C-soil community showed a slightly higher corrosion rate. However, the O-soil community mitigated the steel corrosion. These observations suggested that pipeline corrosion increased proportions of microorganisms, which are likely related to fermentation, sulfur respiration, iron respiration and manganese respiration in surrounding soils and enhanced the soil corrosivity, while petroleum contamination weakened the corrosion ability and promoted the growth of hydrocarbon-degrading organisms in the microbial community.
1. Introduction
The corrosion of iron-based infrastructures causes serious societal and economic consequences since iron-based materials are widely used in various industries and services, such as water, oil and gas distribution systems.1,2 It is estimated that the corrosion of oil and gas transmission pipelines costs over US$7 billion a year in the United States.3 Buried steel pipelines in the petroleum industry are susceptible to corrosion and oil leakage due to corrosion perforation occurs occasionally. The corrosion of buried pipes in soils is a complicated process, involving multiscale interactions and multiple abiotic and biotic factors.4–6 Corrosion driven by the microbial characteristics of soil, namely microbiologically influenced corrosion (MIC), was estimated to be responsible for more than 50% of buried pipeline and cable failures.7 MIC was found to cause severe localized pitting corrosion that originated from the exterior of the buried pipelines in field.8It is reported that diverse physiological groups, including sulfate-reducing bacteria (SRB), nitrate-reducing bacteria (NRB), thiosulfate-reducing bacteria, acetogens, methanogens, Fe(II) oxidizers, Fe(III) reducers, and fermenting bacteria, are related to MIC.9–17 MIC is a complex and integral process influenced by various microorganisms, which display distinct electrochemical reactions and extracellular active metabolites, rather than a consequence of a specific microbial species or group.6,18,19 The main corroding microorganisms usually work as biofilms on the metal surface in MIC, and microbes in surrounding soils are generally the origin.6,20 Therefore, the composition and characteristics of soil microbial communities have a marked impact on the corrosion process of buried pipelines. Besides, microbial community composition, structure and function are closely linked with environmental factors. For instance, anaerobic microorganisms, with the ability to directly oxidize metallic iron and consume the electrons from iron and thus have higher corrosion rates, are usually found in organic carbon-poor environments.9,12,21–23 Hence, buried pipeline corrosion and petroleum contamination caused by corrosion perforation would inevitably modify the surrounding soil microbial communities. However, the interaction between corrosion and the surrounding soil microbial community is still unclear and needs further investigation.
In the present study, three kinds of soil samples of buried petroleum pipelines under different corrosion and petroleum contamination conditions were collected from Shengli Oilfield in China. The samples were used to investigate the mutual influence between corrosion and the surrounding soil microbial communities of buried petroleum pipelines. The 16S rRNA gene high-throughput Illumina MiSeq sequencing, along with LEfSe (Linear discriminant analysis Effect Size) and FAPROTAX (Function Annotation of Prokaryotic Taxa) were used to analyze the surrounding soil microbiota, which was influenced by pipeline corrosion and/or petroleum contamination. The corrosion behaviors of Q235 carbon steel in the collected soil samples were investigated by electrochemical tests under laboratory conditions.
2. Materials and methods
2.1 Experimental design and sampling
All of the soil samples were collected during excavation maintenance of a petroleum pipeline of Shengli Oilfield in Dongying, Shandong Province, China. Three kinds of soils were sampled and named NC-soil, C-soil, and O-soil (Table 1). NC-soil was collected from where the external coating of the pipeline was intact and the pipeline was not corroded. C-soil and O-soil were collected from where the external coating of the pipeline was damaged and the pipeline was corroded but not yet perforated. O-soil was contaminated by petroleum, while C-soil and NC-soil were not. Five sites for each type of soil were chosen and the soil from 0 to 5 cm of the pipe surface was sampled. All of the soil samples were stored at −80 °C before use. NC-soil samples were used as the control group in this study.| Sample description | Physicochemical properties | |||||||
|---|---|---|---|---|---|---|---|---|
| Group (abbreviation) (sample code) | Status of soil | Status of pipeline surrounded by sampling soil | Moistureb (%) | pH | Water-soluble sulphate (g kg−1) | Acid-soluble sulphate (g kg−1) | Organic carbon (g kg−1) | Total Fe (g kg−1) |
| a All data are shown as means ± standard deviations (n = 5). Superscript letters of values in a row represent significant differences at P < 0.05 as calculated by ANOVA.b Moisture for air-dried soil. | ||||||||
| NC-soil (NC) (NC1, NC2, NC3, NC4, NC5) | Not contaminated by petroleum | With intact external coating and not corroded | 0.89 ± 0.09a | 6.98 ± 0.20a | 2.50 ± 0.23b | 2.26 ± 0.21b | 20.93 ± 0.91b | 29.06 ± 1.07a |
| C-soil (C) (C1, C2, C3, C4, C5) | With damaged external coating and corroded but not yet perforated | 1.00 ± 0.10a | 7.03 ± 0.15a | 0.49 ± 0.03a | 0.45 ± 0.03a | 38.17 ± 0.94a | 30.02 ± 2.18a | |
| O-soil (O) (O1, O2, O3, O4, O5) | Contaminated by petroleum | 2.92 ± 0.12b | 7.35 ± 0.21a | 2.06 ± 0.15c | 2.98 ± 0.56b | 56.33 ± 0.57c | 28.64 ± 1.56a | |
2.2 Soil physicochemical factor determination
Air-dried soils were prepared as described by Bao.24 Moisture was determined using a gravimetric method by drying air-dried soils at 105 °C to a constant weight. The pH was measured in the slurry (with a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2.5, w/v ratio of soil to deionized water) using a digital pH meter (PB-10 Basic Meter, Sartorius). Water-soluble sulfate was extracted by vibrating the mixture of soil and deionized water with a ratio of 1
2.5, w/v ratio of soil to deionized water) using a digital pH meter (PB-10 Basic Meter, Sartorius). Water-soluble sulfate was extracted by vibrating the mixture of soil and deionized water with a ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 (w/v) at 25 °C, 150 rpm overnight. Besides, acid-soluble sulfate was prepared according to the standard of state environmental protection of HJ635-2012. Subsequently, the samples were centrifuged and then, the supernatant was used for sulfate measurement using an ICS-900 ion chromatographic system (Dionex, Sunnyvale, CA, USA).25 The organic carbon was determined with the potassium dichromate titration method.24 The total iron element was detected using inductively coupled plasma atomic emission spectroscopy (ICPE-9000, Shimadzu), and the soil solution was prepared by the melting method using sodium carbonate as flux.24
5 (w/v) at 25 °C, 150 rpm overnight. Besides, acid-soluble sulfate was prepared according to the standard of state environmental protection of HJ635-2012. Subsequently, the samples were centrifuged and then, the supernatant was used for sulfate measurement using an ICS-900 ion chromatographic system (Dionex, Sunnyvale, CA, USA).25 The organic carbon was determined with the potassium dichromate titration method.24 The total iron element was detected using inductively coupled plasma atomic emission spectroscopy (ICPE-9000, Shimadzu), and the soil solution was prepared by the melting method using sodium carbonate as flux.24
2.3 DNA extraction and 16S rRNA gene amplicon sequencing
Total DNA was extracted using a PowerSoil DNA Isolation Kit (MO BIO Laboratories, Inc. Cat# 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 888-50). The concentration and quality of the extracted DNA were measured using the Genova Nano of JENWAY. The DNA concentration was adjusted to 10 ng μL−1 before PCR amplification.
888-50). The concentration and quality of the extracted DNA were measured using the Genova Nano of JENWAY. The DNA concentration was adjusted to 10 ng μL−1 before PCR amplification.
The V4–V5 hypervariable region of the prokaryotic 16S rRNA gene (∼400 bp) was amplified using the universal primer 515F (5′-GTGCCAGCMGCCGCGGTAA-3′) with 12 nt unique barcode at its 5′-end and 909R (5′-CCCCGYCAATTCMTTTRAGT-3′).26 Amplicon sequencing was performed on an Illumina Miseq system at the Environmental Genome Platform of Chengdu Institute of Biology. The details of amplification and sequencing samples preparation are described elsewhere.27
2.4 Data analysis
The raw sequences were sorted, quality filtered and deionised using QIIME Pipeline.28 Uchime algorithm29 was used to remove chimera sequences. The resulting sequences were clustered into operational taxonomic units (OTUs) at 97% similarity.30 The OTUs containing only one read were filtered out. The Ribosomal Database Project (RDP) classifier was used to annotate taxonomy at a confidence level of 80%.31Each sample was rarefied to 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 reads to deal with the difference of sequencing depths among samples. Alpha diversity indices, including chao1 estimator of richness, observed OTUs, Shannon index and Good's coverage, were calculated based on the randomly-resampled 12
000 reads to deal with the difference of sequencing depths among samples. Alpha diversity indices, including chao1 estimator of richness, observed OTUs, Shannon index and Good's coverage, were calculated based on the randomly-resampled 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 reads through the QIIME pipeline.
000 reads through the QIIME pipeline.
The overall structural changes of different prokaryotic communities were evaluated by the principal coordinate analysis (PCoA) based on the unweighted and weighted UniFrac distance matrices in Fast UniFrac (http://bmf.colorado.edu/fastunifrac/). Permutational Multivariate Analysis of Variance (PerMANOVA), based on the weighted PCoA scores, was applied to assess the statistical significance among different groups. PerMANOVA was conducted in PAST (http://folk.uio.no/ohammer/past/).
Linear discriminant analysis Effect Size (LEfSe)32 was used to determine the specific prokaryotic taxa with significant difference for each group. LEfSe was conducted on the website (http://huttenhower.sph.harvard.edu/galaxy/) with a LDA threshold score of 3.5.
Function Annotation of Prokaryotic Taxa (FAPROTAX)33 was used to predict the functions of the prokaryotic clades detected in each sample.
2.5 Nucleotide sequence accession number
The original sequence data are available in the National Center for Biotechnology Information (BioProject ID: PRJNA414370, https://www.ncbi.nlm.nih.gov/bioproject/PRJNA414370).2.6 Steel corrosion test for different soil samples under laboratory conditions
Electrochemical corrosion tests under laboratory conditions were performed to investigate the influence of differently modified microbial communities on steel corrosion. A three-electrode setup was used to perform the electrochemical tests. The working electrode (WE) was prepared by the following procedure: Q235 carbon steel cube (composition in wt%: 99.231% Fe; 0.3% C; 0.01% Si; 0.42% Mn; 0.029% S; 0.01% P) with a side length of 10 mm was contacted with a copper wire by soldering. Only one surface of the carbon steel cube was exposed and the rest was embedded in epoxy resin. The exposed surface was polished with 600, 1000, 1500 and 3000-grit silicon carbide metallurgical papers, then ultrasonically cleaned with ethanol, dried with nitrogen gas, and finally stored in a desiccator until use. A platinum plate was used as the counter electrode (CE), and a silver–silver chloride (Ag–AgCl) electrode was used as the reference electrode (RE). A Luggin capillary (LC) filled with a mixture of 2% agar in 3 M KCl was introduced into the cell to eliminate the IR-drop. WE, CE and LC were all pierced through the rubber stopper to make contact with the culture. Considering the saline-alkaline soil condition of sampling sites and the facultative anaerobic condition of new buried pipes, the culture media included 12 g (counted by dry weight) of collected soils (C-soil, NC-soil and O-soil) and 60 mL of artificial seawater medium (ASW), sealed in a 100 mL glass bottle with a sterile air-filled headspace. Bottles with equivalent sterilized soil and ASW were set as abiotic controls. All the bottles were incubated at 35 °C, 100 rpm. ASW was prepared as described by ASTM standard D1141-98,34 composed of (g L−1): 24.5NaCl; 5.20MgCl2; 4.09Na2SO4; 1.16CaCl2; 0.695KCl; 0.201NaHCO3; 0.101KBr; 0.027H3BO3; 0.025SrCl2; 0.003NaF.The open circuit potential (OCP), electrochemical impedance spectroscopy (EIS) and potentiodynamic polarization tests were performed using a CS310 electrochemical workstation (Corrtest, CS310, China). EIS was obtained using a sinusoidal signal of 10 mV in frequencies ranging from 10 mHz to 1 MHz. The collected data were analysed using ZSimpWin (Version 3.10) software from Princeton Applied Research. All EIS spectra in NC-soil, O-soil samples, abiotic controls and 1st day of C-soil group were simulated using the equivalent electrical circuit of the one-time constant model (Fig. 5g). A two-time constant circuit (Fig. 5h) was used to analyze the rest spectra of C-soil samples. Herein, Rs refers to the solution resistance, Q or Qb and Rb represent the capacitance and the resistance of the biofilm or the corrosion product film, Rp stands for the polarization resistance and Qdl and Rct the double layer capacitance and the charge transfer resistance. The chi-square (χ2) values for fitting results were all less than 0.01. Potentiodynamic polarization curves were conducted with a scan rate of 0.2 mV s−1 and in the range from −200 mV to + 200 mV vs. the OCP. Tafel analysis based on the polarization data was performed to obtain electrochemical parameters related to corrosion.
3. Results
3.1 Soil physiochemical properties
The comparison of the physiochemical characteristics of three kinds of soil samples is displayed in Table 1. Compared with NC-soil, the water-soluble sulfate (Sw) and acid-soluble sulfate (Sa) contents of C-soil decreased significantly (P < 0.05). The total sulfate content of C-soil declined by 80.3% as compared to the control group. Levels of soil moisture, pH, and total iron element content were not significantly different between the C-soil and NC-soil. The moisture and organic carbon contents in O-soil were significantly higher than those of the other two groups (P < 0.05). Nevertheless, Sw in C-soil (0.49 ± 0.03) was 80.4% and 76.2% lower than NC-soil (2.50 ± 0.23) and O-soil (2.06 ± 0.15), respectively.3.2 Microbial community richness, diversity, and structure of surrounding soils
The alpha diversity indices of prokaryotic microbes in different samples are displayed in Table 2. Rarefaction curves (Fig. S1†) tended to be saturated under 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 reads per sample and the Good's coverage values ranged from 0.93 to 0.95 (data not shown), suggesting that the sequencing depth was sufficient. The rarefaction curves also indicated that the NC-soil community had a greater OTU-level richness than C-soil and O-soil communities. Chao1 richness and Shannon index of diversity showed consistent results. Furthermore, the microbial diversity of NC-soil microbiota was significantly higher than that of C-soil and O-soil communities (P < 0.05). All these observations indicated that pipeline corrosion and oil contamination reduced the microbial community richness and diversity of surrounding soils.
000 reads per sample and the Good's coverage values ranged from 0.93 to 0.95 (data not shown), suggesting that the sequencing depth was sufficient. The rarefaction curves also indicated that the NC-soil community had a greater OTU-level richness than C-soil and O-soil communities. Chao1 richness and Shannon index of diversity showed consistent results. Furthermore, the microbial diversity of NC-soil microbiota was significantly higher than that of C-soil and O-soil communities (P < 0.05). All these observations indicated that pipeline corrosion and oil contamination reduced the microbial community richness and diversity of surrounding soils.
| Sample | Chao1 | Observed | Shannon |
|---|---|---|---|
| OTU | Index | ||
a All data are calculated based on a cutoff of 97% similarity of 16S rRNA sequences of 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 reads per sample and shown as means ± standard deviations (n = 5). Superscript letters represent significant differences at P < 0.05 as calculated by ANOVA. 000 reads per sample and shown as means ± standard deviations (n = 5). Superscript letters represent significant differences at P < 0.05 as calculated by ANOVA. |
|||
| C-soil | 2157.0 ± 110.5a | 1203.8 ± 43.2a | 6.70 ± 0.14a |
| NC-soil | 2747.2 ± 186.3b | 1553.2 ± 102.1b | 7.34 ± 0.22b |
| O-soil | 2555.5 ± 277.2ab | 1329.1 ± 119.8a | 6.67 ± 0.43a |
PCoA was performed with unweighted as well as weighted UniFrac distances to analyse the beta diversity among 15 soil samples. Unweighted UniFrac PCoA (Fig. 1a) and weighted UniFrac PCoA (Fig. 1b) showed the consistent results that five samples in each type of soil tended to cluster and these three types of soils were obviously distinct from each other in view of the microbial community structure. Furthermore, PerMANOVA analysis confirmed that the difference between any two of them was significant (P < 0.01).
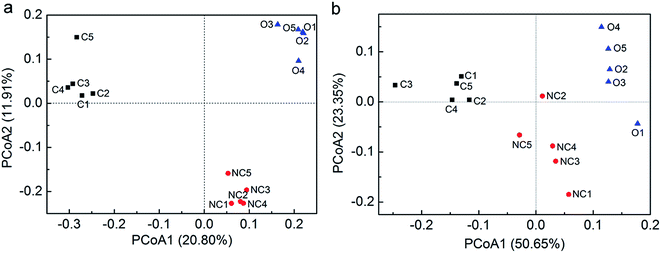 | ||
| Fig. 1 The principal coordinate analysis (PCoA) using unweighted UniFrac distance matrices (a) and weighted UniFrac distance matrices (b). PCoA was conducted in fast UniFrac (http://bmf.colorado.edu/fastunifrac/). Sample abbreviations refer to Table 1. | ||
3.3 Microbial community composition and comparison of surrounding soils
On the whole, 177![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 764 qualified reads were categorized into 4571 OTUs, in which singleton OTUs were excluded. A total of 4420 OTUs (97.9% of total reads) were annotated as bacterial phyla and 150 OTUs (2.05% of total reads) were affiliated with archaeal phyla. Two reads were assigned as unclassified.
764 qualified reads were categorized into 4571 OTUs, in which singleton OTUs were excluded. A total of 4420 OTUs (97.9% of total reads) were annotated as bacterial phyla and 150 OTUs (2.05% of total reads) were affiliated with archaeal phyla. Two reads were assigned as unclassified.
Fig. 2a shows the microbial community composition of each sample at the phylum level (relative abundance > 0.3%). Across all samples, the dominant bacteria phyla were Proteobacteria (mean ± SEM = 54.04% ± 3.67% in C-soil samples, 50.81% ± 8.34% in NC-soil samples, and 58.75% ± 6.84% in O-soil samples) and Bacteroidetes (41.79% ± 3.44% in C-soil samples, 36.20% ± 6.76% in NC-soil samples and 32.17% ± 7.94% in O-soil samples). Actinobacteria and Firmicutes were significantly decreased in the C-soil group (0.36% ± 0.18% and 0.28% ± 0.27%, respectively) when compared with the NC-soil (3.01% ± 0.50% and 0.95% ± 0.14%, respectively) and O-soil groups (3.81% ± 0.74% and 1.58% ± 0.57%, respectively). Besides, Euryarchaeota in NC-soil samples and Chloroflexi in O-soil samples reached the highest average abundance of 5.03% and 0.84%, respectively. The significance testing was performed using one-way ANOVA at P < 0.05 unless otherwise stated.
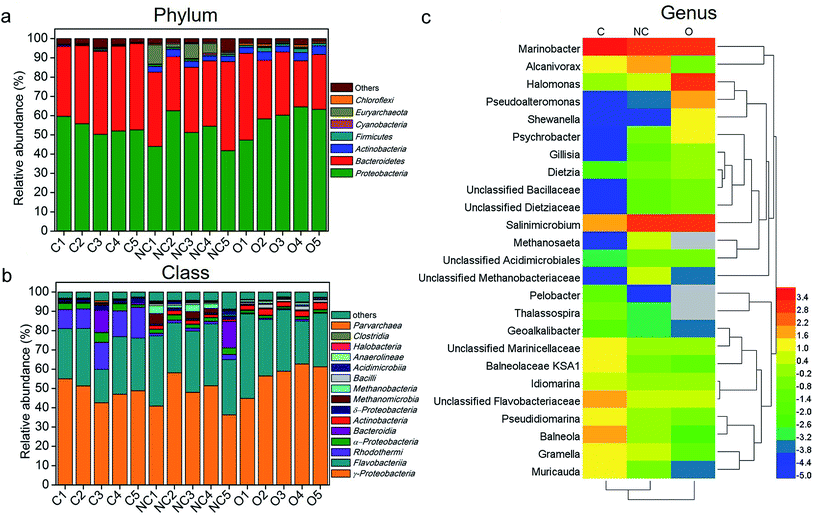 | ||
| Fig. 2 The composition of different soil communities at the (a) phylum (relative abundance >0.3%), (b) class (relative abundance >0.3%), and (c) genus (top 25) levels. Taxonomic classification was based on RDP classifier with a confidence threshold of 80%. The average relative abundance values of top 25 genera in each group were processed with logarithmic normalization with the base of the euler number. Heat plots were compiled using a heatmap illustrator 1.0.3.3. Hierarchical clustering analysis using average linkage based on the Bray–Curtis similarity index was applied for the row and column. Empty values were set as grey. Sample abbreviations refer to Table 1. | ||
Further comparison of the microbial communities was conducted at the class level and depicted in Fig. 2b. γ-Proteobacteria and Flavobacteria were the first two abundant taxa of all samples, accounting for an average of 75.07% in the C-soil group, 77.98% for the NC-soil group, and 87.9% for the O-soil group. In the C-soil group, Rhodothermi showed significantly higher abundance (12.63% ± 2.65%) when compared with the other two groups. Similarly, Methanomicrobia (2.46% ± 2.30%) and Methanobacteria (2.31% ± 1.46%) showed significantly higher abundance in the NC-soil group. Actinobacteria (2.87% ± 0.55%), Bacilli (1.33% ± 0.54%) and Anaerolineae (0.65% ± 0.22%) showed significantly higher abundance in the O-soil group.
More specifically, the representative genera (top 15) of each sample and their comparison with each other are shown in Fig. 2c. Across all the samples, 8 of the top 15 genera were shared, including Marinobacter, Salinimicrobium, unclassified Flavobacteriaceae, unclassified Marinicellaceae, Alcanivorax, Pseudidiomarina, Idiomarina, and Halomonas. The unique dominant genera of the C-soil community included Muricauda, Thalassospira, Pelobacter and Geoalkalibacter. It was found that the relative abundances of Balneola, Balneolaceae KSA1 and Gramella were significantly higher in the C-soil group. Halomonas, Pseudoalteromonas, Shewanella, Psychrobacter, Dietzia, Gillisia and unclassified Dietziaceae were the specific dominant genera of the O-soil community. In addition, the relative abundances of Methanosaeta and unclassified Methanobacteriaceae in the NC-soil group were significantly higher than that in the other two groups.
3.4 Differential taxa for different soil microbial communities
LEfSe was performed to identify the specific prokaryotic taxa with significantly higher abundance in each soil microbial community group. As shown in Fig. 3, a total of 56 prokaryotic clades were screened out with a LDA threshold score of 3.5. Taxa with significantly higher abundance in the C-soil group mainly belonged to class Rhodothermi (including genus Balneola and KSA1), class Alphaproteobacteria (including genus Thalassospira), and class Deltaproteobacteria (including genus Pelobacter and Geoalkalibacter). Genus Muricauda and Gramella, affiliated with the family Flavobacteriaceae were also enriched in the C-soil group. Taxa with abundant advantages in the NC-soil group were mainly archaeal taxa related to methane formation, including the genus Methanosaeta and family Methanobacteriaceae. As for the O-soil community, Halomonas, Pseudoalteromonas, Shewanella, Psychrobacter belonging to Gammaproteobacteria and Dietzia, which belongs to Actinobacteria, had a significantly higher relative abundance.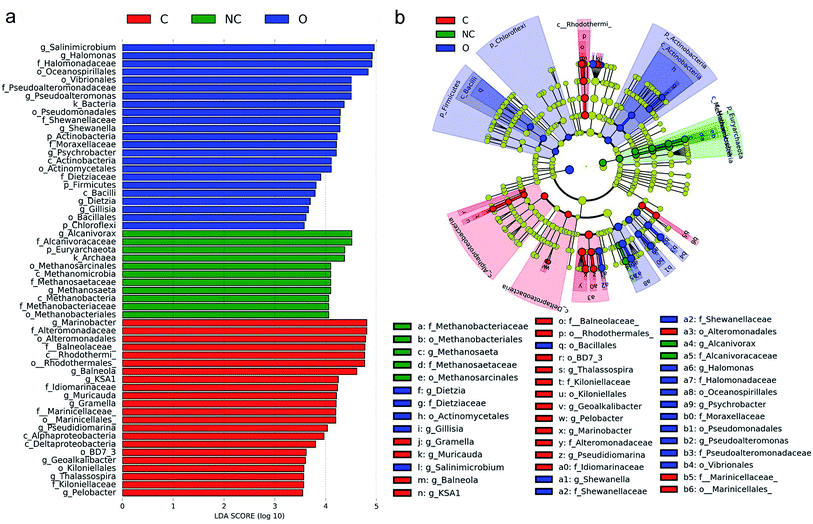 | ||
| Fig. 3 The linear discriminant analysis effect size (LEfSe) analysis of microbial abundance among different soil samples. (a) Taxa with significantly difference in different soil groups were detected by LEfSe analysis with a LDA threshold score of 3.5 and a significant α of 0.05. (b) The cladogram of detected prokaryotic taxa for each soil community. LEfSe analysis was performed on the website http://huttenhower.sph.harvard.edu/galaxy/. Sample abbreviations refer to Table 1. | ||
3.5 Function prediction and comparison of different soil microbial communities
In order to interpret the metabolic and functional profiles of the detected communities, FAPROTAX was used to map the prokaryotic taxa to putative diverse functional groups according to identified functions of cultured strains in current literature.33 A total of 18 functional groups probably associated with steel corrosion and petroleum contamination were screened out based on the FAPROTAX database and current knowledge. Fig. 4 shows the relative abundance (OTU proportion) comparison of these functional groups in each type of soil community. For the C-soil community, prokaryotic clades associated with fermentation, sulfur respiration, iron respiration and manganese respiration were significantly richer as compared to the other two groups (P < 0.05). The NC-soil community was predicted to contain a significantly higher abundance of methanogenic archaea, catalyzing acetoclastic or hydrogenotrophic methanogenesis, and taxa related to methanol oxidation and methylotrophy (P < 0.05). Taxa predicted by hydrocarbon degradation in the O-soil community had a slight advantage in terms of abundance over others. In addition, the relative abundances of predicted nitrite respiration and denitrification microorganisms in the NC-soil and O-soil communities were significantly higher than the C-soil community (P < 0.05).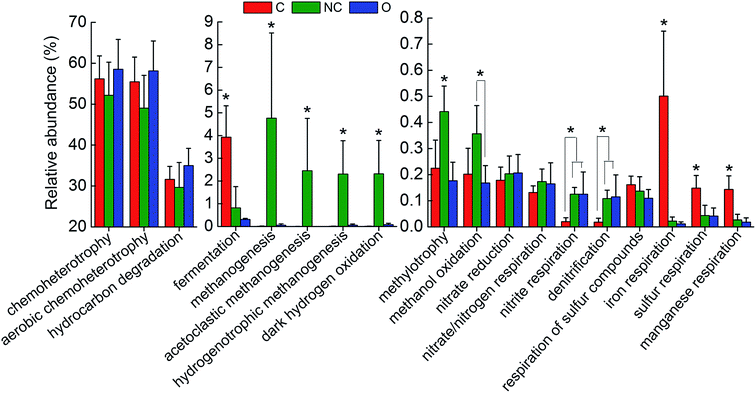 | ||
| Fig. 4 The comparison of predicted functions of different soil communities based on FAPROTAX database. A total of 18 functional groups were screened out based on functional annotation of prokaryotic taxa (FAPROTAX) database. The average relative abundance (OTU proportion) of these functions in each soil community was depicted as histogram. The error bars denote the standard deviation of 5 samples in each group. The asterisks indicated the significant differences at P < 0.05 as calculated by ANOVA. Sample abbreviations referred to Table 1. | ||
3.6 Comparison and analysis of corrosion electrochemical measurements
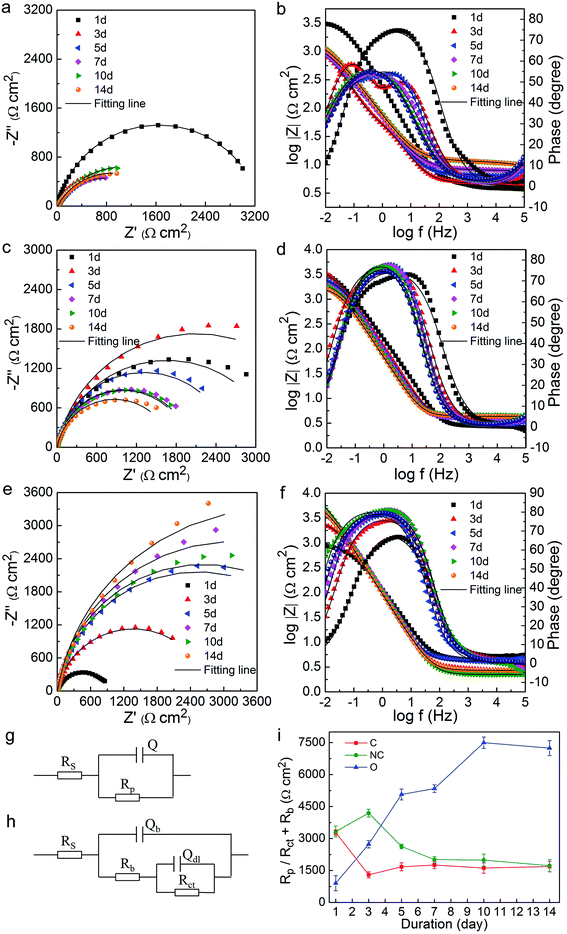
The variation of OCP vs. Ag/AgCl with time during the 14 day incubation is presented in Fig. S2.† The OCP values in three soil groups at the very beginning (day 0) were all around −0.675 V (data not shown). On the whole, the OCP values of the C-soil group shifted in the positive direction, while the NC-soil and O-soil groups showed a slight negative shift. OCP values of all samples changed little over time after the 6th or 7th day.The EIS data were collected under stable OCP for different soil groups on the 1st, 3rd, 5th, 7th, 10th, and 14th days. Equivalent electrical circuit (Fig. 5g and h) was used to simulate the impedance spectrum. The Nyquist (Fig. 5a, c and e) and Bode (Fig. 5b, d and f) plots and Rp or Rct + Rb values (Fig. 5i) obtained from fitting results are shown in Fig. 5. Like OCP, the impedance on day 0 for all samples was averaging 1362 Ω cm2 (data not shown). It was found that the impedance in the C-soil group decreased, but it increased in the O-soil group over time. On the other hand, the impedance in the NC-soil group initially increased, and then subsequently decreased. In the final stage of incubation, the C-soil and NC-soil groups attained very close impedance values, which were significantly lower than that of the O-soil group. The fitting values of Rp or Rct + Rb showed a consistent conclusion. The impedance of three abiotic control groups was around 2500 Ω cm2 and with no significant difference after a 14 day incubation (data not shown). Since a higher value of impedance and Rp or Rct + Rb means a lower corrosion rate,23,35 more severe corrosion occurred in C-soil and NC-soil samples when compared to O-soil samples. In comparison with the sterile control, the C-soil and NC-soil communities slightly facilitated steel corrosion, while the O-soil community mitigated steel corrosion.
Potentiodynamic polarization measurements were conducted after 14 day incubation and plotted in Fig. 6. The electrochemical parameters from the Tafel analysis, corrosion current density (icorr), corrosion potential (Ecorr), anodic and cathodic slopes (Ba and Bc), are shown in Table 3. The C-soil and NC-soil samples had higher corrosion current densities (5.36 and 4.72 μA cm−2, respectively) than the O-soil group (0.98 μA cm−2). The corrosion current densities of sterile controls were averaging 3.27 μA cm−2 and with no significant difference (data not shown). The results of potentiodynamic polarization measurements were accordant with EIS results.
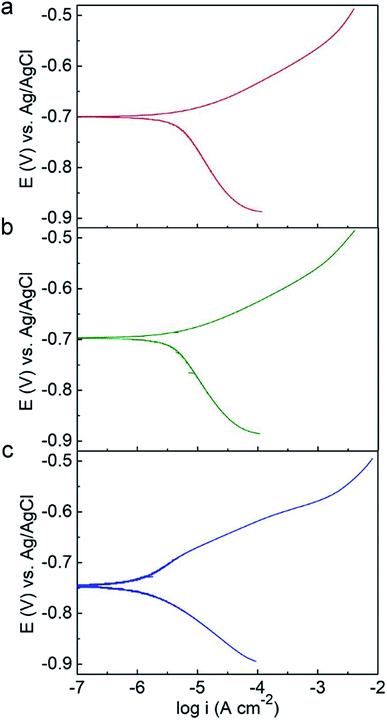 | ||
| Fig. 6 Potentiodynamic polarization curves of Q235 steel in (a) C-soil, (b) NC-soil and (c) O-soil samples on the 14th day. | ||
| Group | icorr (μA cm−2) | Ecorr (V vs. Ag/AgCl) | Ba (mV dec−1) | Bc (mV dec−1) |
|---|---|---|---|---|
| C-soil | 5.36 ± 0.52 | −0.71 ± 0.05 | 66.4 ± 4.7 | 168.3 ± 5.5 |
| NC-soil | 4.72 ± 0.43 | −0.71 ± 0.04 | 61.4 ± 5.8 | 165.8 ± 7.6 |
| O-soil | 0.98 ± 0.24 | −0.73 ± 0.03 | 52.0 ± 3.4 | 90.4 ± 4.6 |
4. Discussion
In the present study, three kinds of surrounding soils of buried petroleum pipelines were collected to study the mutual influence between the soil microbial community and pipeline corrosion or petroleum contamination using 16S rRNA analysis and electrochemical tests. The results revealed different soil microbial communities influenced by pipeline corrosion or petroleum contamination and the discrepancy in their influence on steel corrosion. Herein, we mainly focus on differential microorganisms in these communities and their potential relations to corrosion and petroleum contamination and hope to provide more detailed information for buried pipeline corrosion and control.It is generally acknowledged that sulfate-reducing bacteria have major responsibility for MIC in anoxic environments (e.g., buried petroleum pipelines), and carbon steel or iron corrosion was observed to positively correlate with sulfate loss in incubation media.36,37 Based on this, significantly lower sulfate contents in C-soil and O-soil samples could be a sign of pipeline corrosion and the pipeline surrounded by C-soil endured more severe corrosion, as their sulfate content was the lowest (Table 1). Taken together, from the microbial community composition and LEfSe analysis, Balneolaceae genera (Balneola and KSA1), Flavobacteriaceae genera (Muricauda and Gramella), and Desulfuromonadaceae genera (Pelobacter, Geoalkalibacter) were significantly more abundant in the C-soil population (Fig. 2c and 3a). Muricauda species was reported to produce acid from various simple sugars and Muricauda ruestringenesis could grow in facultative anaerobic conditions.38 The genus Pelobacter was originally found to anaerobically ferment acetoin, acetylene and other infrequent substrates. As a relative of genus Desulfuromonas and Geobacter, Pelobacter species was found to have respiratory metabolisms with Fe(III) and S0, which serve as terminal electron acceptors.39 Individual species were reported to indirectly reduce Fe(III) via an elemental sulfur/sulfide cycle and sulfide formation.40 It was also reported that Geoalkalibacter bacteria could grow by reducing Fe(III), Mn(IV) or elemental sulfur.41 All of these findings may support the functional prediction results that the functional groups associated with fermentation, sulfur respiration, iron respiration and manganese respiration were increased in the C-soil community (Fig. 4). Moreover, since the external corrosion products of buried steel pipelines are usually composed of iron oxides and iron sulfide, it is reasonable that iron/manganese/sulfur reducers were higher in the soil surrounding corroded pipelines. Electrochemical tests indicated that the C-soil community slightly promoted steel corrosion as compared with its sterile control (Table 3, Fig. 5, and 6). Pelobacter and Geoalkalibacter were detected in the corrosive biofilms of oil production facilities and were speculated to promote corrosion by eliminating Fe(III) oxide passivation layers and re-exposing iron to corroded products.42,43 In addition, the ability of Muricauda and Pelobacter to produce acetate or H2 may sustain the growth of other corrosive microbes and therefore contribute to the steel corrosion.38,43 In the present study, the prokaryotic communities of the surrounding soil of buried petroleum pipelines influenced by corrosion were analyzed; few studies are available concerning the direct effect of the identified microorganisms on corrosion. The actual contributions of these microorganisms to corrosion need to be further investigated in future research.
Prokaryotic clades with significantly higher relative abundances in the O-soil community include Halomonas, Pseudoalteromonas, Psychrobacter, Shewanella and Dietzia (Fig. 2c and 3a). Species of the genus Halomonas, Psychrobacter and Dietzia, were all reported to be capable of degrading hydrocarbons and have potential applications in bioremediation.44–46 This is consistent with our results. The organic carbon content in O-soil samples was markedly higher and the hydrocarbon degraders in them were predicted to be richer (Table 1, Fig. 4). In addition, species of the genus Pseudoalteromonas are generally associated with higher organisms and produce various biologically active extracellular agents, such as extracellular enzymes, polysaccharides, and toxins.47 Pseudoalteromonas and Shewanella species could also reduce Fe(III) oxides.48–51 Considering that O-soil samples were influenced by the pipeline corrosion, Pseudoalteromonas and Shewanella might be also upgraded by iron oxide corrosion products in soil. Notably, the total Fe content in soils around corroded pipelines (C-soil and O-soil) are supposed to be higher than that of non-corroded ones (NC-soil). However, in the present study, the average value of the total Fe content in C-soil was slightly higher, and that in O-soil was slightly lower as compared to NC-soil, but there was no significant difference among the three soil groups (Table 1). There may have been differences in the original total Fe of three soil groups before being influenced by pipeline corrosion, and the corrosion products (such as iron oxides and iron sulfide) are probably not detached from the pipeline and dispersed into the surrounding soils, which may be the reasons for the observed non-significant difference in total Fe contents. Contrary to C-soil and NC-soil communities, the O-soil community showed an inhibiting effect on steel corrosion (Table 3, Fig. 5, and 6). Abiotic steel corrosion usually takes place in the presence of oxygen and moisture, and the corrosion rate would markedly decrease when oxygen was depleted.36 In the O-soil community, aerobic hydrocarbon degraders, such as Halomonas, would consume most of the oxygen in the sealed incubation bottle. Moreover, Halomonas and Pseudoalteromonas were reported to produce surfactants and exopolysaccharides and they would drastically change the interfacial properties of the solid surface and hence be used in corrosion inhibition.52 Steel corrosion inhibition from Pseudoalteromonas was investigated and it was believed to be closely associated with oxygen depletion and compact biofilm formation.51 Therefore, oxygen consumption, as well as the production of surfactants and exopolysaccharides, might be a factor in the corrosion inhibition of the O-soil community.
5. Conclusions
The mutual influence between corrosion or petroleum contamination and the surrounding soil microbial communities of buried petroleum pipelines were investigated. Both pipeline corrosion and petroleum contamination significantly decreased the surrounding soil microbial diversity and altered the microbial composition. The corrosion-modified microbial community showed the promotion of steel corrosion, while the microbial community influenced by petroleum contamination exhibited the reverse effects. Several unique microbes were identified in differently modified microbial populations. Although the actual concrete effects of these observed differential microorganisms on corrosion remains uncertain, the results will enlighten future research on MIC analysis and control.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was financially support by the Foundation of National Key Laboratory of Biochemical Engineering, the Chinese Academy of Sciences Strategic Biological Resources Service Network (ZSYS-015), and National Natural Science Foundation of China (No. 21676279). We thank Guangshan Wei from Key Laboratory of Marine Genetic Resources, Third Institute of Oceanography, SOA (Xiamen, China) for assistance of bioinformatics analysis.Notes and references
- G. Schmitt, M. Schütze, G. F. Hays, W. Burns, E.-H. Han, A. Pourbaix and G. Jacobson, Global needs for knowledge dissemination, research, and development in materials deterioration and corrosion control, Word Corrosion Organization, New York, NY, 2009 Search PubMed.
- X. Li, D. Zhang, Z. Liu, Z. Li, C. Du and C. Dong, Nature, 2015, 527, 441–442 CrossRef CAS PubMed.
- N. Muthukumar, Role of Colloidal Systems in Environmental Protection, 2014, pp. 527–571 Search PubMed.
- I. S. Cole and D. Marney, Corros. Sci., 2012, 56, 5–16 CrossRef CAS.
- M. Wasim, S. Shoaib, N. M. Mubarak, Inamuddin and A. M. Asiri, Environ. Chem. Lett., 2018, 16, 861–879 CrossRef CAS.
- K. M. Usher, A. H. Kaksonen, I. Cole and D. Marney, Int. Biodeterior. Biodegrad., 2014, 93, 84–106 CrossRef CAS.
- H. C. Flemming, Economical and Technical Overview, Microbial Influenced Corrosion of Materials, Springer, Berlin, 1996 Search PubMed.
- M. Kiani Khouzani, A. Bahrami, A. Hosseini-Abari, M. Khandouzi and P. Taheri, Metals, 2019, 9, 459 CrossRef.
- D. Enning, H. Venzlaff, J. Garrelfs, H. T. Dinh, V. Meyer, K. Mayrhofer, A. W. Hassel, M. Stratmann and F. Widdel, Environ. Microbiol., 2012, 14, 1772–1787 CrossRef CAS PubMed.
- D. Xu, Y. Li, F. Song and T. Gu, Corros. Sci., 2013, 77, 385–390 CrossRef CAS.
- M. Magot, G. Ravot, X. Campaignolle, B. Oillivier, B. K. C. Patel, M.-L. Fardeau, P. Thomas, J.-L. Crolet and J.-L. Garcia, Int. J. Syst. Bacteriol., 1997, 47, 818–824 CrossRef CAS PubMed.
- S. Kato, I. Yumoto and Y. Kamagata, Appl. Environ. Microbiol., 2015, 81, 67–73 CrossRef PubMed.
- T. Zhang, H. H. Fang and B. C. Ko, Appl. Microbiol. Biotechnol., 2003, 63, 101–106 CrossRef CAS PubMed.
- T. Uchiyama, K. Ito, K. Mori, H. Tsurumaru and S. Harayama, Appl. Environ. Microbiol., 2010, 76, 1783–1788 CrossRef CAS PubMed.
- W. De Windt, N. Boon, S. D. Siciliano and W. Verstraete, Environ. Microbiol., 2003, 5, 1192–1202 CrossRef CAS PubMed.
- J. M. McBeth, B. J. Little, R. I. Ray, K. M. Farrar and D. Emerson, Appl. Environ. Microbiol., 2011, 77, 1405–1412 CrossRef CAS PubMed.
- N. Muthukumar, Electrochem. Commun., 2003, 5, 421–425 CrossRef CAS.
- N. Kip and J. A. van Veen, ISME J., 2015, 9, 542–551 CrossRef CAS PubMed.
- B. J. Little and J. S. Lee, Int. Mater. Rev., 2014, 59, 384–393 CrossRef CAS.
- I. B. Beech and J. Sunner, Curr. Opin. Biotechnol., 2004, 15, 181–186 CrossRef CAS PubMed.
- J. Xia, D. Xu, L. Nan, H. Liu, Q. Li and K. Yang, Chin. J. Mater. Res., 2016, 30, 161–170 CAS.
- H. T. Dinh, J. Kuever, M. Mussmann, A. W. Hassel, M. Stratmann and F. Widdel, Nature, 2004, 427, 829–832 CrossRef CAS PubMed.
- R. Jia, D. Yang, J. Xu, D. Xu and T. Gu, Corros. Sci., 2017, 127, 1–9 CrossRef CAS.
- S. Bao, Soil agro-chemistrical analysis, China Agriculture Press, Beijing, 3rd edn, 2000 Search PubMed.
- J. Zhou, Z. Song, D. Yan, Y. Liu, M. Yang, H. Cao and J. Xing, Bioresour. Technol., 2014, 153, 216–222 CrossRef CAS PubMed.
- H. Tamaki, C. L. Wright, X. Li, Q. Lin, C. Hwang, S. Wang, J. Thimmapuram, Y. Kamagata and W. T. Liu, PLoS One, 2011, 6, e25263 CrossRef CAS PubMed.
- H. Li, J. Qu, T. Li, J. Li, Q. Lin and X. Li, Front Microbiol., 2016, 7, 758 Search PubMed.
- J. G. Caporaso, J. Kuczynski, J. Stombaugh, K. Bittinger, F. D. Bushman, E. K. Costello, N. Fierer, A. G. Peña, J. K. Goodrich, J. I. Gordon, G. A. Huttley, S. T. Kelley, D. Knights, J. E. Koenig, R. E. Ley, C. A. Lozupone, D. McDonald, B. D. Muegge, M. Pirrung, J. Reeder, J. R. Sevinsky, P. J. Turnbaugh, W. A. Walters, J. Widmann, T. Yatsunenko, J. Zaneveld and R. Knight, Nat. Methods, 2010, 7, 335–336 CrossRef CAS PubMed.
- R. C. Edgar, B. J. Haas, J. C. Clemente, C. Quince and R. Knight, Bioinformatics, 2011, 27, 2194–2200 CrossRef CAS PubMed.
- W. Li and A. Godzik, Bioinformatics, 2006, 22, 1658–1659 CrossRef CAS PubMed.
- Q. Wang, G. M. Garrity, J. M. Tiedje and J. R. Cole, Appl. Environ. Microbiol., 2007, 73, 5261–5267 CrossRef CAS PubMed.
- N. Segata, J. Izard, L. Waldron, D. Gevers, L. Miropolsky, W. S. Garrett and C. Huttenhower, Genome Biol., 2011, 12, R60 CrossRef PubMed.
- S. Louca, L. W. Parfrey and M. Doebeli, Science, 2016, 353, 1272–1277 CrossRef CAS PubMed.
- A. S. T. M. D1141-98, 2013.
- P. B. Raja, M. Fadaeinasab, A. K. Qureshi, A. A. Rahim, H. Osman, M. Litaudon and K. Awang, Ind. Eng. Chem. Res., 2013, 52, 10582–10593 CrossRef CAS.
- D. Enning and J. Garrelfs, Appl. Environ. Microbiol., 2014, 80, 1226–1236 CrossRef PubMed.
- R. Liang, D. F. Aktas, E. Aydin, V. Bonifay, J. Sunner and J. M. Suflita, Environ. Sci. Technol., 2016, 50, 4844–4853 CrossRef CAS PubMed.
- N. R. Krieg, J. T. Staley, D. R. Brown, B. P. Hedlund, B. J. Paster, N. L. Ward, W. Ludwig and W. B. Whitman, Bergey's Manual of Systematic Bacteriology, Springer, USA, 2nd edn, 2010 Search PubMed.
- D. R. Lovley, E. J. P. Phillips, D. J. Lonergan and P. K. Widman, Appl. Environ. Microbiol., 1995, 61, 2132–2138 CAS.
- S. A. Haveman, R. J. DiDonato Jr, L. Villanueva, E. S. Shelobolina, B. L. Postier, B. Xu, A. Liu and D. R. Lovley, Appl. Environ. Microbiol., 2008, 74, 4277–4284 CrossRef CAS PubMed.
- A. C. Greene, B. K. Patel and S. Yacob, Int. J. Syst. Evol. Microbiol., 2009, 59, 781–785 CrossRef CAS PubMed.
- A. Vigneron, E. B. Alsop, B. Chambers, B. P. Lomans, I. M. Head, N. Tsesmetzis and H. Nojiri, Appl. Environ. Microbiol., 2016, 82, 2545–2554 CrossRef CAS PubMed.
- K. Dronen, I. Roalkvam, J. Beeder, T. Torsvik, I. H. Steen, A. Skauge and T. Liengen, Environ. Sci. Technol., 2014, 48, 8627–8635 CrossRef PubMed.
- A. Lasa and J. L. Romalde, Genomics Data, 2017, 12, 7–10 CrossRef PubMed.
- H. Al-Awadhi, N. Dashti, M. Kansour, N. Sorkhoh and S. Radwan, Int. Biodeterior. Biodegrad., 2012, 69, 10–16 CrossRef CAS.
- A. Wentzel, T. E. Ellingsen, H. K. Kotlar, S. B. Zotchev and M. Throne-Holst, Appl. Microbiol. Biotechnol., 2007, 76, 1209–1221 CrossRef CAS PubMed.
- C. Holmström and S. Kjelleberg, FEMS Microbiol. Ecol., 1999, 30, 285–293 CrossRef.
- M. K. Schutz, M. L. Schlegel, M. Libert and O. Bildstein, Environ. Sci. Technol., 2015, 49, 7483–7490 CrossRef CAS PubMed.
- M. Moradi, Z. Song, L. Yang, J. Jiang and J. He, Corros. Sci., 2014, 84, 103–112 CrossRef CAS.
- Z. Guo, T. Liu, Y. F. Cheng, N. Guo and Y. Yin, Colloids Surf., B, 2017, 157, 157–165 CrossRef CAS PubMed.
- J. Wu, D. Zhang, P. Wang, Y. Cheng, S. Sun, Y. Sun and S. Chen, Corros. Sci., 2016, 112, 552–562 CrossRef CAS.
- M. A. Malik, M. A. Hashim, F. Nabi, S. A. AL-Thabaiti and Z. Khan, Int. J. Electrochem. Sci., 2011, 6, 1927–1948 CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9ra03386f |
| This journal is © The Royal Society of Chemistry 2019 |