Open Access Article
Open Access ArticleEffects of dietary tuna dark muscle enzymatic hydrolysis and cooking drip supplementations on growth performance, antioxidant activity and gut microbiota modulation of Bama mini-piglets†
Jun Zhou‡
ab,
Mingyuan Yang‡ab,
Jiaojiao Hanab,
Chenyang Lu*ab,
Ye Liab and
Xiurong Su *ab
*ab
aState Key Laboratory for Quality and Safety of Argo-products, Ningbo University, 818 Fenghua Road, Ningbo, China. E-mail: luchenyang@nbu.edu.cn; suxiurong_public@163.com; Fax: +86-0574-87608368; Tel: +86-0574-87608368
bSchool of Marine Science, Ningbo University, Ningbo, China
First published on 12th August 2019
Abstract
Experiments were conducted to evaluate the enzymatic hydrolysis of tuna dark muscle (EH-TDM) and cooking drip (EH-TCD) as nutrition supplements in Bama mini-piglets. Our results showed that EH-TDM treatment produce considerable benefits for the Bama mini-piglets in terms of the feed efficiency, the prevention of oxidative damage and the control of blood lipid levels. The EH-TCD treatment also improved the growth performance, whereas all other aspects deteriorated. The analyses of the gut microbiota revealed an increased proportion of bacteria involved in energy metabolism and protein utilization in these two groups. Furthermore, short chain fatty acid producing bacteria were increased, as well as the genera Lactobacillus and Bifidobacterium, which are involved in intestinal dysfunction regulation. This study will provide valuable information for the development of marine protein hydrolysates as feed ingredients for the swine industry.
Introduction
Feed ingredients support a healthy intestinal development to prevent bacterial infections and enhance the growth of livestock animals. However, the sustained price increases for protein meal feed commodities and the competition between humans and animals have forced the swine industry to consider the dietary inclusion of cost-effective alternative feedstuffs.1 The positive effect of animal protein sources compared with traditional plant proteins on growth performance for piglets is well known, many fish products and by-products rich in protein have been evaluated in diets for swine.2 In addition, the use of highly digestible and palatable feedstuffs is important for the growth of piglets, especially the challenge of handling diarrhoea problems after weaning.3,4 As undigested protein serves as a nutrient supplement for potential pathogens, the use of highly digestible feed ingredients should improve growth performance and disease resistance.5,6 Otherwise, the marine-derived bioactive peptides provide extra beneficial functions for growing performance and intestinal health in the piglets.7,8 Therefore, marine protein hydrolysates may be an effective alternative compared to traditional protein meal feed.Tuna dark muscle (TDM) is a dark band of tissue under the entire body and near the backbone. It is considered to be a by-product within the tuna industry because it has a dark color, easily oxidized and produces an unpleasant smell.9 Another by-product, tuna cooking drip (TCD), is a mass of condensed liquid that is produced during the steaming and drying processes in the manufacture of canned tuna, which is usually disposed or only used as bacteria medium.10 However, both the enzymatic hydrolysis of tuna dark muscle (EH-TDM) and tuna cooking drip (EH-TCD) are rich in essential fatty acids, proteins (peptides) and minerals. However, there is a lack of evidence to support whether EH-TDM or EH-TCD can be used as nutritional supplements to improve swine growth performance and pork quality.
The gut microbiome is composed of the various microbial species and their secretions and is established shortly after birth and eventually achieves a dynamic balance.11 The gut microbiota can be considered as an invisible ‘‘organ’’ that profoundly affects a number of physiological, nutritional, developmental and immune processes in the host, which in turn affect growth performance and health.12 Additionally, there is growing evidence that the gut microbiota is altered by diet treatment and contributes to energy metabolism and immune functioning in the host.13,14 For example, functional diets interact with the gut microbiota to change the microbial metabolites, including short-chain fatty acids (SCFAs), lipopolysaccharide and bile acids, and these metabolites in turn provide substrates for the metabolic processes within the intestine, mitigate inflammation, and regulate the nutrients that influence satiety.15,16 In particular, a previous finding suggests that the gut microbiota acts by integrating the signaling pathways of the host to regulate energy storage and promote leanness.17 Furthermore, an appropriate reduction in the concentration of dietary protein can improve the structure of the gut microbiota in swine, especially in regards to the proportion of bacteria from the family Peptostreptococcaceae.18
In this study, the efficacy and safety of EH-TDM and EH-TCD were investigated in Bama mini-piglets as the nutritional supplements. The main nutritional components of EH-TDM and EH-TCD were firstly identified, and the growth performance, serum biochemical indicators, pork quality and gut microbiota modulation resulting from the use of EH-TDM and EH-TCD were examined. The results obtained in this study may serve as a valuable reference for screening safe and effective feed ingredients.
Materials and methods
Chemicals
Two-month-old male Bama mini-piglets (Sus scrofa domestica) and standard chow diet were purchased from Zhejiang Kaihua Hongxing Co., Ltd (Hangzhou, China). The certificate number for the pigs is SYXK (ZHE 2008-0110). TDM and TCD were provided by Zhejiang Ningbo Jinri Co., Ltd (Ningbo, China). Proteases (alcalase and flavor proteinase) were purchased from Novozymes Co., Ltd (Bagsvaerd, Denmark). The reagent that was used for the analysis of the nutritional components was purchased from Sigma Co., Ltd (St Louis, MO, USA). All kits that were used for measuring serum indices were purchased from Nanjing Jiancheng Co., Ltd (Nanjing, China).The preparation of the supplements
The optimal conditions for the hydrolysis of TDM and TCD using a mixture of enzymes (alcalase and flavor proteinase at a ratio of 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) were determined. The enzyme mixture and the substrate were then combined at a ratio of 1
1) were determined. The enzyme mixture and the substrate were then combined at a ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 120 and agitated for 8 h at 120 rpm, after which the mixture was incubated at the optimal temperature of 55 °C in order to facilitate enzymatic hydrolysis; the enzyme was subsequently deactivated at a temperature of 85 °C. The percentage of hydrolyzed product for the TDM and TCD reactions reached 57.21% and 53.17%, respectively. The enzyme hydrolysates of EH-TDM and EH-TCD were freeze-dried in the presence of a vacuum for 35 h (−20 °C, 0.013 mBar).
120 and agitated for 8 h at 120 rpm, after which the mixture was incubated at the optimal temperature of 55 °C in order to facilitate enzymatic hydrolysis; the enzyme was subsequently deactivated at a temperature of 85 °C. The percentage of hydrolyzed product for the TDM and TCD reactions reached 57.21% and 53.17%, respectively. The enzyme hydrolysates of EH-TDM and EH-TCD were freeze-dried in the presence of a vacuum for 35 h (−20 °C, 0.013 mBar).
Design of the animal experiments
After allowing for six days of adaptation to a standard commercial diet, 9 Bama mini-piglets with an average body weight of 2.44 ± 0.14 kg were allocated into 3 treatment groups. During the 58-d experiment, the Bama mini-piglets were fed one of three diets: a chow diet (control group); a diet supplemented with 6% EH-TDM (EH-TDM group); and a diet supplemented with 6% EH-TCD (EH-TCD group). They were all reared in temperature-controlled (24–28 °C) and humidity-controlled (60–70%) ambient conditions. We allowed the pigs to freely acquire feed and water. Their body weight was measured every three days, and their feed consumption was recorded in order to determine the average daily feed intake (ADFI), average daily gain (ADG) and feed efficiency (gain![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) feed ratio, G
feed ratio, G![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) F) during the entire course of growth.
F) during the entire course of growth.
Sample collection
After 58 d of feeding, all of the Bama mini-piglets were anesthetized via the injection of sodium barbital and blood samples were obtained using venipuncture. The blood samples were clotted at 37 °C for 15 min and centrifuged at 3000 × g at 4 °C for 10 min. The serum was then separated and preserved at −20 °C for later analysis. Subsequently, the Bama mini-piglets were euthanized by exsanguination. Their organs were removed and immediately placed in liquid nitrogen and stored at −80 °C for later experiments. The visceral index was calculated using the following formula: organ weight/body weight (g kg−1). Fecal samples were collected for the analysis of gut microbiota.Nutritional components analysis
The nutritional composition of the feeds and the Longissimus dorsi muscles obtained from the Bama mini-piglets were analyzed according to the following methods.The protein, fat, ash and moisture content were determined using standard methods obtained from the Association of Official Analytical Chemists (AOAC) as previously described (methods 990.03, 920.39, 942.05 and 930.15 for protein, fat, ash and moisture, respectively).19
The amino acid analysis samples were digested with 0.02 N HCl (dilution 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10) using microwave digestion. The supernatants were processed to obtain the phenylthiocarbamyl derivatives and analyzed by a high-performance liquid chromatography (HPLC) instrument with a variable UV detector at 338 nm using a Zorbax Eclipse AAA column (4.6 mm × 150 mm, 5 μm) (Agilent 1200 Series, NYSE: A, Palo Alto, USA). The mobile phase consisted of a mixture of solvent A (40 mmol L−1 NaH2PO4, pH = 7.8) and solvent B (C2H3N
10) using microwave digestion. The supernatants were processed to obtain the phenylthiocarbamyl derivatives and analyzed by a high-performance liquid chromatography (HPLC) instrument with a variable UV detector at 338 nm using a Zorbax Eclipse AAA column (4.6 mm × 150 mm, 5 μm) (Agilent 1200 Series, NYSE: A, Palo Alto, USA). The mobile phase consisted of a mixture of solvent A (40 mmol L−1 NaH2PO4, pH = 7.8) and solvent B (C2H3N![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CH3OH
CH3OH![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O = 45
H2O = 45![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 45
45![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10). Elution was initiated using constant 0% solvent B for 2 min and then was increased using a linear gradient from 0 to 57% of solvent B over the course of 16 min, then a linear gradient from 57 to 100% of solvent B over the course of 4 min, followed by a linear gradient from 100 to 0% of solvent B for 3 min, and finally re-equilibration to the initial conditions for 1 min, at a flow rate of 2 mL min−1. The absolute content was calculated according to the external standard method.
10). Elution was initiated using constant 0% solvent B for 2 min and then was increased using a linear gradient from 0 to 57% of solvent B over the course of 16 min, then a linear gradient from 57 to 100% of solvent B over the course of 4 min, followed by a linear gradient from 100 to 0% of solvent B for 3 min, and finally re-equilibration to the initial conditions for 1 min, at a flow rate of 2 mL min−1. The absolute content was calculated according to the external standard method.
After the extraction and dissolution of the crude fat, a previously described method was used for the esterification of fatty acids.20 Briefly, the fatty acids were separated using saponification with methanolic sodium hydroxide and then methylated with boron trifluoride-methanol reagent in a nitrogen atmosphere. The obtained esterified fatty acids were extracted with hexane and the recovery was completed via the addition of a salt solution. The degree of esterification of the fatty acids was determined using an ADB-WAX column (60 m × 250 μm × 0.25 μm) with Agilent GC (7890A, NYSE: A, Palo Alto, USA) and PERSEE MS (M7-80EI, Purkinje, Beijing, China) systems using a previously described method.21 The relative contents in percentage form were calculated using an area normalization method.
Serum analysis
The indices of superoxide dismutase (SOD), glutathione peroxidase (GSH-Px) and malondialdehyde (MDA) were determined in order to demonstrate the antioxidant capacity of the blood serum. The indices of total cholesterol (TC), total triglycerides (TG), high-density lipoprotein cholesterol (HDL-C) and low-density lipoprotein cholesterol (LDL-C) are the hallmark determinants of lipid metabolism. The indices of alanine aminotransferase (ALT), aspartate transaminase (AST), alkaline phosphatase (AKP) and total bilirubin (TBILI) are indicative of liver functioning. In addition, the indices of urea nitrogen (BUN), creatinine (CRE), glucose (GLU) and albumin (ALB) are key indicators of metabolism. All measurements of the indices were conducted according to the manufacturer's instructions.The analysis of fecal microbiota
The total bacterial genomic DNA was extracted from the fecal samples using a DNA kit. The concentration and purity of the extracted DNA was determined using a Thermo NanoDrop 2000C (Thermo Fisher Scientific, USA). The amplification of the microbial 16S rDNA within the V3–V4 region was conducted using universal primers with indexes and adaptors (341F: ACTCCTACGGGAGGCAGCAG, 806R: GGACTACHVGGGTWTCTAAT). The KAPA HiFi Hotstart high fidelity enzyme was used for PCR to ensure the accuracy and validity of the products derived from the genomic DNA templates. The quality of the PCR products was determined via sequencing of 250 bp paired-end reads using a Sangon Biotech Illumina HiSeq platform (Shanghai, China).A barcode sequence was used to determine the original datum for each sample and was then removed. The longer tags consisted of the paired-end reads (https://sourceforge.net/project/flashpage/); quality filtering was used to remove tags with a length < 50 bp, an average quality score < 20, or tags with containing simple repeated sequences (https://prinseq.soursefoge.net/). After removing the sequences that did not meet the quality requirements, the UPARSE was used to cluster the high-quality tags into operational taxonomic units (OTUs) with a similarity threshold of 0.97.
Only filtered, high-quality sequences were utilized for the data analysis. The QIIME was used to calculate the richness and diversity indices. Multivariate one-way analysis of variance was used to evaluate the statistical significance of the inter-group separation in order to obtain the graph of the weighted UniFrac principal coordinate analysis (PCoA) scores. The longest sequence for each cluster with a similarity threshold of 0.97 was chosen as the representative sequence, and the Ribosomal Database Project (RDP) classifier was then used to determine the phylogenic taxonomic classification. The redundancy analysis (RDA) model was used to analyze the log-converted relative abundance of each OTU and find the core OTUs that were different between two animal groups. Heat maps were generated using R for the cluster analysis.
Statistical analysis
All data were analyzed using one-way analysis of variance (ANOVA) (SPSS, version 19.0, Chicago, IL, USA) and represented as mean values ± standard errors of the mean (SEM). The differences between the various groups were assessed post-hoc using the Tukey test and were regarded to be statistically significant when P < 0.05.Results
Nutritional composition of the diets
The amount of crude protein, fat, and ash in the diet of EH-TDM and EH-TCD groups were significantly increased compared with control group (P < 0.05), whereas no differences were found between EH-TDM and EH-TCD groups (Table 1). All essential amino acids were present in greater amounts in the EH-TDM and EH-TCD groups than in the control group. Among them, the levels of arginine, histidine, leucine and lysine were found to be significantly increased (P < 0.05) in the EH-TDM group compared with the EH-TCD group. However, the EH-TCD group contained higher contents of phenylalanine and threonine than the EH-TDM group (P < 0.05) (Table 2). The concentration of polyunsaturated fatty acids (PUFAs) in both the EH-TDM and EH-TCD groups were increased compared with that of the control group, whereas the concentration of saturated fatty acids (SFAs) was reduced. There were no differences between the EH-TDM and EH-TCD groups in the levels of PUFAs or SFAs (Table S1†).| Chemical composition (%) | Diets | ||
|---|---|---|---|
| Control | EH-TDM | EH-TCD | |
| Crude protein | 20.53a ± 0.55 | 23.58b ± 0.58 | 24.77b ± 0.20 |
| Crude fat | 7.03a ± 0.22 | 7.57b ± 0.16 | 7.59b ± 0.24 |
| Ash | 4.96a ± 0.11 | 5.50b ± 0.10 | 5.42b ± 0.27 |
| Moisture | 10.76a ± 0.34 | 9.08b ± 0.38 | 9.33b ± 0.24 |
| Essential amino acids (mg g−1) | Diets | ||
|---|---|---|---|
| Control | EH-TDM | EH-TCD | |
| Arginine | 0.75a ± 0.01 | 1.09b ± 0.01 | 0.95c ± 0.01 |
| Histidine | 0.10a ± 0.00 | 0.17b ± 0.00 | 0.11a ± 0.00 |
| Isoleucine | 0.81a ± 0.01 | 1.20b ± 0.01 | 1.16b ± 0.01 |
| Leucine | 1.18a ± 0.01 | 1.90b ± 0.01 | 1.51c ± 0.01 |
| Lysine | 0.12a ± 0.00 | 0.30b ± 0.00 | 0.17a ± 0.00 |
| Methionine | — | — | — |
| Phenylalanine | 0.18a ± 0.00 | 0.48b ± 0.00 | 0.67c ± 0.00 |
| Threonine | 0.63a ± 0.01 | 0.83b ± 0.00 | 1.09c ± 0.01 |
| Tryptophan | 0.11a ± 0.00 | 0.21b ± 0.00 | 0.18b ± 0.00 |
| Valine | 1.02a ± 0.01 | 1.34b ± 0.01 | 1.33b ± 0.01 |
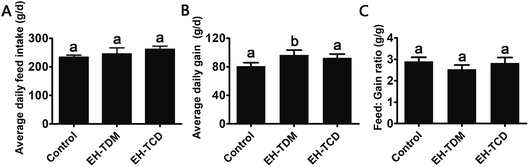
Effects of the EH-TDM and EH-TCD supplements on growth performance
Compared with the control group, the EH-TDH treatment group had significantly increased average daily gain (ADG) and reduced feed efficiency (feed![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) gain ratio, F
gain ratio, F![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) G ratio) (P < 0.05), whereas no differences in the ADG and F
G ratio) (P < 0.05), whereas no differences in the ADG and F![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
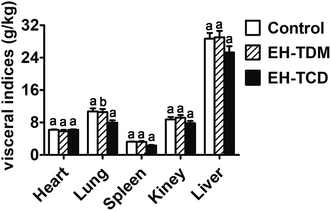
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) G ratio were found with the EH-TCD group (Fig. 1). In addition, compared with the control group, the visceral indices for the EH-TDM treatment group showed no differences, whereas EH-TCD supplementation significantly reduced the lung index (P < 0.05) (Fig. 2).
G ratio were found with the EH-TCD group (Fig. 1). In addition, compared with the control group, the visceral indices for the EH-TDM treatment group showed no differences, whereas EH-TCD supplementation significantly reduced the lung index (P < 0.05) (Fig. 2).
Effects of EH-TDM and EH-TCD supplements on the quality of pork
Small dietary alterations will lead to changes in the nutritional composition of the longissimus dorsi muscle. The crude protein and muscle moisture levels in the EH-TDM and EH-TCD groups were significantly increased compared to those of the control (P < 0.05), whereas the crude fat content was significantly decreased (P < 0.05). The EH-TCD treatment significantly enhanced the crude protein level compared to the EH-TDM treatment (P < 0.05) (Table S2†). Compared with the control, the EH-TDM group had significantly increased levels of threonine and histidine (P < 0.05), and the EH-TCD group had significantly increased leucine, methionine, arginine and isoleucine levels (P < 0.05) (Table S3†). In addition, compared with the control, the EH-TDM group had enhanced levels of total PUFAs and SFAs, whereas the EH-TCD group had reduced levels of total PUFAs and increased levels of total SFAs (Table S4†).Biochemical indices in the serum
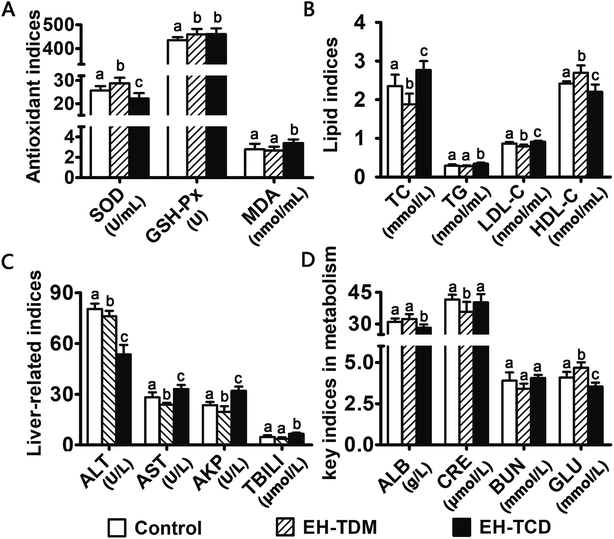
The activity levels of the ROS-scavenging enzymes SOD and GSH-Px in the EH-TDM group was significantly increased (P < 0.05), while the MDA levels were reduced compared with those of the control group (P > 0.05). However, the EH-TCD group showed significantly reduced activity for SOD (P < 0.05) and enhanced activity for GSH-Px (P < 0.05) and increased levels of MDA when compared to the control group (Fig. 3A). The EH-TDM group showed a reduction in the TC (P < 0.05), TG (P > 0.05) and LDL-C (P < 0.05) levels and enhancement of the levels of HDL-C (P < 0.05); whereas, the EH-TCD group showed opposite effects on blood lipid levels compared to those of the EH-TDM group (Fig. 3B). Compared to the control group, EH-TDM decreased all liver-related indices, whereas the activity levels of AST, AKP, and TBILI in the EH-TCD group were significantly increased (P < 0.05), and ALT was significantly decreased (P < 0.05) (Fig. 3C). No significant differences in the CRE and BUN levels were found in the EH-TDM and EH-TCD groups, which indicated that the kidneys were healthy. In addition, EH-TDM increased the ALB (P > 0.05) and GLU (P < 0.05) levels, whereas the EH-TCD group had decreased levels (P < 0.05) (Fig. 3D).Overall structural modulation of gut microbiota
There were 21![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 846, 41
846, 41![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 171 and 42
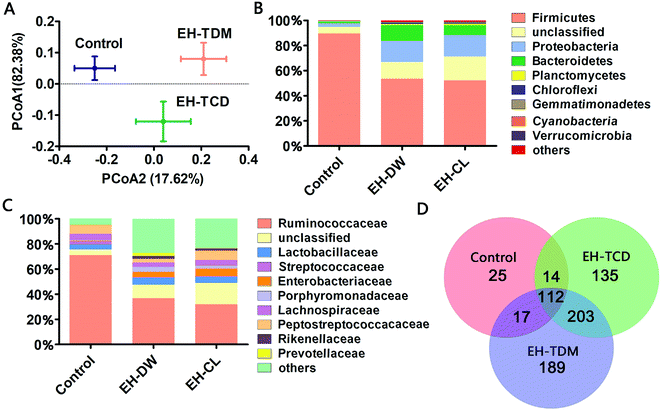
171 and 42![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 129 valid reads obtained from the control, EH-TDM and EH-TCD groups, respectively (Table S5†). Using a 97% sequence similarity cutoff, 5599 operational taxonomic units (OTUs) were identified from the total valid reads. The coverage ratio of each group revealed that this sequencing depth was deep enough to cover the microbial diversity present within the samples (Table S5†). The alpha diversity of the sequencing analysis indicated an increased richness (indicated by ACE and Chao1 indices) and increased diversity (indicated by Shannon and Simpson indices) within EH-TDW and EH-TCD groups when compared with the control group; in particular, EH-TDM treatment resulted in higher richness and diversity indices than the EH-TCD treatment (Table S5†). In addition, the Weight UniFrac Principal Coordinate Analysis (PCoA) showed a clear separation of the EH-TDW and EH-TCD groups from the control group (Fig. 4A).
129 valid reads obtained from the control, EH-TDM and EH-TCD groups, respectively (Table S5†). Using a 97% sequence similarity cutoff, 5599 operational taxonomic units (OTUs) were identified from the total valid reads. The coverage ratio of each group revealed that this sequencing depth was deep enough to cover the microbial diversity present within the samples (Table S5†). The alpha diversity of the sequencing analysis indicated an increased richness (indicated by ACE and Chao1 indices) and increased diversity (indicated by Shannon and Simpson indices) within EH-TDW and EH-TCD groups when compared with the control group; in particular, EH-TDM treatment resulted in higher richness and diversity indices than the EH-TCD treatment (Table S5†). In addition, the Weight UniFrac Principal Coordinate Analysis (PCoA) showed a clear separation of the EH-TDW and EH-TCD groups from the control group (Fig. 4A).
Microbial shifts in response to supplementation with EH-TDM and EH-TCD based on taxonomic analysis
A taxonomic analysis was performed based on the use of Ribosomal Database Project (RDP) classifiers to express the variation of the gut microbial composition among the Bama mini-piglets receiving the control, EH-TDM or EH-TCD diets. The microbial distributions at the phylum and class levels had changed after 58-d of EH-TDM and EH-TCD feeding (Fig. 4B and C). Firmicutes, Proteobacteria and Bacteroidetes were the most abundant phyla present in the fecal samples, representing 77.02–93.89% of the total sequences (Table S6†). Compared with the control group (89.65%, 1.24% and 3% in Firmicutes, Bacteroidetes and Proteobacteria, respectively), the percentage of the bacteria from the phylum Firmicutes was decreased in the EH-TDM (53.54%) and EH-TCD groups (52.11%), whereas the proportion of the bacteria from the phyla Bacteroidetes (12.32% and 7.85% in EH-TDM and EH-TCD groups, respectively) and Proteobacteria (16.98% and 17.06%) was increased. When comparing the relative abundance of bacteria between the EH-TDW and EH-TCD groups at the phylum level, there were no significant differences except for Bacteroidetes. In addition, the ratio of Bacteroidetes to Firmicutes was significantly reduced in the EH-TDM (4.34) and EH-TCD groups (6.64) compared to the control group (72.30).Bacteria from the families Ruminococcaceae, Lactobacillaceae, Enterobacteriaceae, Lachnospiraceae and Peptostreptococcaceae were the most dominant, making up 53.10–87.43% of the total (Table S7†). Four families, Ruminococcaceae, Peptostreptococcaceae, Lactobacillaceae and Lachnospiraceae, were the most highly represented families within the phylum Firmicutes. When compared with the control (71.06%, 4.94%, 3.9% and 6.78% in Ruminococcaceae, Lachnospiraceae, Lactobacillaceae and Peptostreptococcaceae, respectively), the percentage of Ruminococcaceae and Lachnospiraceae was decreased after the EH-TDM (36.87% and 3.5%) and EH-TCD treatments (31.93% and 4.44%), whereas the proportion of Lactobacillaceae (5.57% and 4.65% in EH-TDM and EH-TCD groups) was increased. The proportion of Peptostreptococcaceae was decreased in the EH-TDM group (2.81%) but increased in the EH-TCD group (7.42%). Porphyromonadaceae, Rikenellaceae and Prevotellaceae were the most highly represented classes from the phylum Bacteroidetes, and their abundances were higher in the EH-TDM group.
In this study, the presence of bacteria from 168, 521 and 464 genera were identified in the control, EH-TDM and EH-TCD groups, respectively. Among all genera, there were 112 genera were detected within three groups. Unique genera accounted for an average of 3.6%, 27.2% and 19.4% of the total genera in the control, EH-TDM and EH-TCD groups, respectively. The genera within the phyla Elusimicrobia, Armatimonadetes, Thermotogae and Crenarchaeota were only found in the EH-TDM group, and the genera within the phyla Lentisphaerae, Thermodesulfobacteria and Deinococcus-thermus were only found in the EH-TCD group. In particular, the unique short-chain fatty acid (SCFA)-producing genera Sutterella was only found in the EH-TDM group, and the other two SCFA-producing genera, Faecalibacterium and Roseburia, were found in both the EH-TDM and EH-TCD groups (Fig. 4D).
According to the criterion of being represented by more than 1% of the total DNA sequences in at least one group, seventeen genera were screened out in our study (Table S8†). These 17 genera plus the unclassified genera made up 94.58%, 70.66% and 75.89% of the total sequences from the control, EH-TDM and EH-TCD groups, respectively. Among them, 9, 5 and 3 genera belong to the phylum Firmicutes, Proteobacteria and Bacteroidetes, respectively. Compared with the control group, the abundance of genera Subdoligranulum (5.28% and 11.14% in EH-TDM and EH-TCD groups), unclassified Lachnospiraceae (2.09% and 1.88%) and Streptococcus (0.19% and 0.35%) was decreased, whereas the frequency of the detection of these genera, except for unclassified Peptostreptococcaceae (2.81% and 7.4%), was increased in the EH-TDM and EH-TCD groups. Bacteria from Lactobacillus, unclassified Ruminococcaceae, Ruminococcus, unclassified Porphyromonadaceae, Alistipes, Pseudomonas, Serratia, unclassified Christensenellaceae, and Prevotella were more abundant in the EH-TDM group, whereas bacteria from Citrobacter, Blautia, Enterobacter and Rhodobacter were more abundant in the EH-TCD group.
Microbial shifts in response to the supplementation of EH-TDM and EH-TCD based on operational taxonomic unit (OTU) analysis
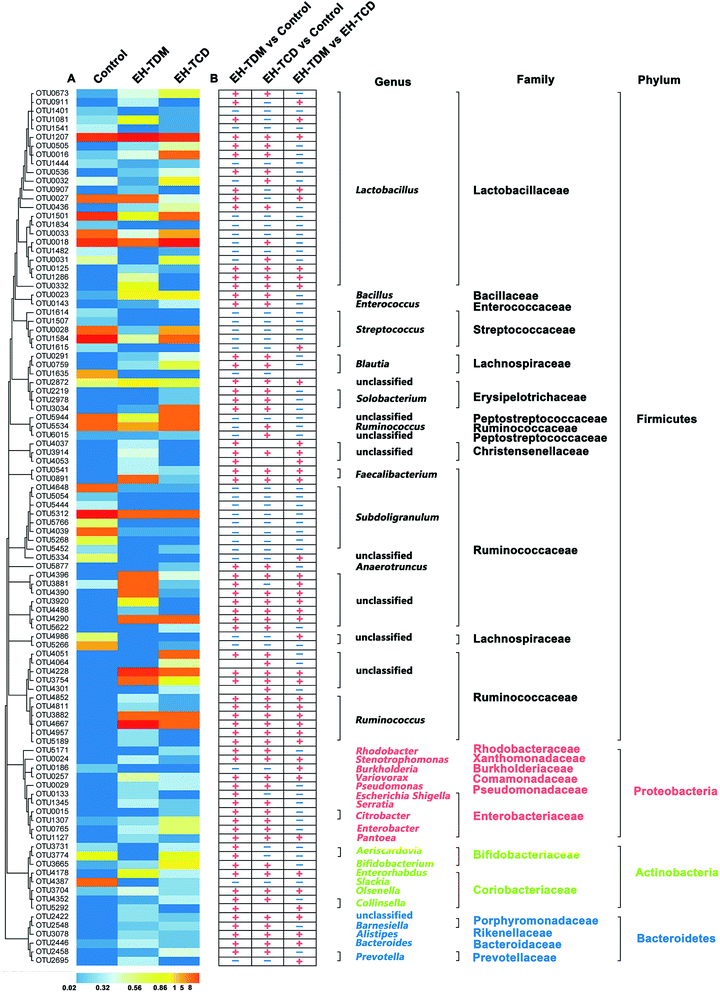
Operational Taxonomic Unit (OTU) analysis is a taxon-independent analysis that was performed for deep ecological analysis of the gut microbial composition. We classified an OTU as having 97% sequence similarity. 1249, 4003 and 3391 OTUs were identified in the control, EH-TDM and EH-TCD groups, respectively. Redundancy analysis (RDA) was used to identify the core OTUs and the changes in their abundance as a result of the EH-TDM and EH-TCD treatments. Among them, 100 core OTUs were detected in this study. The abundances of the 100 core OTUs were visualized hierarchically in a clustered heat map (Fig. 5). Here, we showed that EH-TDM and EH-TCD caused changes in the abundance of 87 OTUs when compared to the control group, 60 of which were increased and 27 decreased. Among them, 34 out of 60 had higher abundance and 22 out of 27 had lower abundance in the EH-TDM group than in the EH-TCD group. Furthermore, compared with the control group, the left 13 OTUs showed opposite change trends between EH-TDM and EH-TCD groups. Among them, 6 and 7 of the OTUs were increased and decreased, respectively, in the EH-TDM group.Discussion
Previous studies have reported that marine protein hydrolysates as nutritional supplements to improve swine growth performance, but the comparison among marine protein hydrolysates and mechanism have not been clearly clarified. In this study, EH-TDM treatment improved the feed efficiency when compared with that of the control, whereas the EH-TCD group showed no increase in the growth performance of piglets (Fig. 1). Thus, EH-TDM treatment was shown to provide a measurement benefit to the Bama mini-piglets. Carcass leanness is associated with dietary fatty acid composition and level, and PUFAs absorbed from the diet specifically inhibit the endogenous synthesis of fatty acids, which improves the lean ratio in pigs. The levels of PUFAs in the EH-TDM and EH-TCD groups were enhanced compared with those of the control group (Table S1†), and dietary PUFA supplements led to a significant increase in moisture and a decrease in the crude fat in the longissimus dorsi muscle in both the EH-TDM and EH-TCD groups (P < 0.05) (Table S4†). The influence of dietary PUFAs on the improvement of the lean ratio has been demonstrated for the use of both EH-TDM and EH-TCD.The health of the Bama mini-piglets was indicated by the biochemical indices that were determined from serum. An imbalance between superoxide radical formation and the antioxidant action of these enzymes leads to oxidative damage, and subsequently lead to neurodegeneration, an increased risk of swine disease.22 The MDA level varies as a consequence of lipid peroxidation,23 and the level of MDA in the EH-TCD group was significantly higher (P < 0.05) than that in the control, whereas the levels of SOD and GSH-Px were significantly increased (P < 0.05) in the EH-TDM group (Fig. 3A). A previous study showed that increased TC, TG, and LDL concentrations and reduced HDL concentrations are common features of hyperlipidemia.24 In addition, dyslipidemia leads to a markedly increased risk of swine disease.25 Thus, these data demonstrated that EH-TDM was effective in preventing oxidative damage and controlling blood lipids, whereas EH-TCD appeared to increase the risk of diseases related to the dysfunction of these processes in pigs. Moreover, hepatocytes are the main facilitators of the distribution of ALT, AST and AKP enzymes, which only enter the serum in large quantities when the hepatocyte membrane is damaged or necrotic.26,27 The extent of liver damage can be sensitively detected by measuring the serum enzyme activity. The concentration of TBILI, a metabolite of bile salt, is considered as an indicator of liver damage when its levels are increased in serum. After EH-TCD treatment, pigs had significantly increased levels of these indicators (Fig. 3C). Thus, EH-TCD supplementation appeared to cause damage to the liver.
As far as we know, EH-TDM and EH-TCD supplements are primarily administered in feed, and the structure of the gut microbiota in pigs was considered as primary object of research during this study. The PCoA results revealed the changes in the overall structure of the gut microbiome that were caused by the EH-TDM and EH-TCD treatments (Fig. 4A). Furthermore, large variations in the abundance of bacteria at several taxonomic levels in the EH-TDM and EH-TCD groups were observed, such as with the two main phylum, Bacteroidetes and Firmicutes. Although Firmicutes was the most abundant in all groups, the EH-TDM group (53.54%) showed a 40.28% reduction and the EH-TCD (52.11%) showed a 41.87% reduction in Firmicutes bacteria compared with the control (89.65%). Conversely, the EH-TDM group (12.32%) had a 11.08% increase in the abundance of Bacteroidetes while the EH-TCD group (7.85%) showed a 6.61% increase compared to the control (1.24%) (Fig. 4B). The large decrease in the ratio of Bacteroidetes/Firmicutes bacteria is a strong indicator of the fact that the microbiome structure modulated the feed efficiency.28,29 Additional evidences indicated that the reduction in Bacteroidetes was associated with the elevation of energy metabolism.30 Consistent with the results showing remarkable increases in body weight gain and feed efficiency, the abundance of Bacteroidetes in the EH-TDM group was reduced by 11.08% compared to the control group and had a lower ratio of Bacteroidetes/Firmicutes bacteria than the EH-TCD group.
Previous results have showed that long-term supplementation with protein-rich foods leads to an increase in the abundance of Bacteroides.31 OTU2446 (derived from Bacteroides spp.) (Fig. 5) demonstrated a strong response to EH-TDM and EH-TCD supplementation, with significantly enhancement compared with the control group (from 0.09% to 0.35% and 0.22%, respectively). Bacteria from the genus Bacteroides have the ability to remove side chains from bile acids, which allows the bile acids to return to the liver circulation and subsequently convert sugars to fermentation products that are beneficial to humans.32 The presence of bacteria from the genus Escherichia–Shigella, which have crucial effects on protein utilization in animals,18 were more abundant in the TDM groups than in the control group (Table S8†). Of note, most of the undigested protein-derived nutrients can be fermented by gut microbiota, increasing the yield of metabolites such as SCFA. In addition, some amino acids secreted by gut microbiota can be used as precursors for the synthesis of short-chain fatty acids.33 As the main energy source of colorectal cells, SCFAs regulate the absorption of various nutrients and hormone production in the intestine, and participate in energy metabolism extensively.34,35 SCFA-producing bacteria, such as Faecalibacterium, Roseburia, Sutterella, Blauti, Bacteroides, Coprococcus and Ruminococcus, might be associated with swine health.36–38 In this study, these SCFA-producing genera were significantly increased by EH-TDM and EH-TCD treatment. Specially, Sutterella was only detected in the EH-TDM group, and the other two genera, Faecalibacterium and Roseburia, were found in both the EH-TDM and EH-TCD groups (Fig. 5).
Additionally, the genera Lactobacillus and Bifidobacterium exert a range of beneficial health effects. Lactobacillus bacteria secrete bacteriocins to inhibit the growth of certain pathogenic bacteria, as well as lactic acid and hydrogen peroxide.39 Bifidobacterium bacteria regulate intestinal microbial homeostasis, inhibit pathogens and harmful bacteria and convert a number of dietary compounds into bioactive molecules.40 In this study, bacteria from the genus Lactobacillus were significantly enriched in the EH-TDM group (5.56%) compared with the control group (3.9%), whereas their increase in the EH-TCD group (4.64%) was lower than in the EH-TDM group (Table S8†). OTU2446 (Bifidobacterium spp.) showed a higher relative abundance in both supplementation groups (0.35% and 0.22%) than in the control group (0.09%) (Fig. 5).
In conclusion, our findings have indicated that the modulation of the gut microbiota structure induced by EH-TDM and EH-TCD supplementation is linked to improvements in both feed efficiency and animal health. Particularly, EH-TDM treatment significantly improved body weight gain, prevented oxidative damage and controlled blood lipids. However, EH-TCD did cause some damage to the lung and the liver. Simultaneously, EH-TDM and EH-TCD treatments enriched the populations of presumably beneficial bacteria with activities related to energy harvesting, protein utilization and SCFAs production. Our results provide valuable information for the development of safe and effective marine protein hydrolysates as nutritional supplements.
Ethical statement
The experimental and animal care procedures were performed in accordance with the guidelines prepared by the Ningbo University Laboratory Animal Center (affiliated with the Zhejiang Laboratory Animal Common Service Platform, Ningbo, China). All protocols were approved by the Ningbo University Laboratory Animal Center under permit number SYXK (ZHE 2008-0110).Data availability statement
The sequences obtained from the fecal samples from the three groups (control, EH-TDM and EH-TCD groups) have been deposited in the NCBI Sequence Read Archive Database under the accession number SRP151201.Conflicts of interest
The authors declare no competing financial interest.Acknowledgements
This work was sponsored by the National Key Research and Development Program of China (2018YFD0901102), the Natural Science Foundation of Zhejiang Province (LY18C010001 and LY19C010003), Regional Demonstration Project of Marine Economic Innovation and Development in 2014 and 2016, and K. C. Wong Magna Fund of Ningbo University. We thank Nature Research Editing Service for language editing.References
- T. A. Woyengo, E. Beltranena and R. T. Zijlstra, J. Anim. Sci., 2014, 92, 1293 CrossRef CAS PubMed.
- J. L. Tucker, V. D. Naranjo, T. D. Bidner and L. L. Southern, J. Anim. Sci., 2011, 89, 1466–1473 CrossRef CAS PubMed.
- J. C. Kim, C. F. Hansen, B. P. Mullan and J. R. Pluske, Anim. Feed Sci. Technol., 2001, 83, 3–16 Search PubMed.
- J. M. Heo, J. C. Kim, C. F. Hansen, B. P. Mullan, D. J. Hampson and J. R. Pluske, Arch. Anim. Nutr., 2008, 62, 343–358 CrossRef CAS PubMed.
- M. Opheim, M. L. Strube, H. Sterten, M. Overland and N. P. Kjos, Arch. Anim. Nutr., 2016, 70, 44–56 CrossRef CAS PubMed.
- F. O. Opapeju, D. O. Krause, R. L. Payne, M. Rademacher and C. M. Nyachoti, J. Anim. Sci., 2009, 87, 2635–2643 CrossRef CAS PubMed.
- Y. F. Liu, C. Luan, X. Xia, S. An and Y. Z. Wang, Int. J. Pept. Res. Ther., 2011, 17, 175 CrossRef CAS.
- S. K. Kim and I. Wijesekara, J. Funct. Foods, 2010, 2, 1–9 CrossRef CAS.
- E. Sánchezzapata, M. Amensour, R. Oliver, E. Fuenteszaragoza, C. Navarro, J. Fernándezlópez, E. Sendra, E. Sayas and J. A. Pérezalvarez, Food Nutr. Sci., 2011, 2, 22–30 Search PubMed.
- J. I. Choi, H. J. Kim, J. H. Kim, B. S. Song, B. S. Chun, D. H. Ahn, M. W. Byun and J. W. Lee, Radiat. Phys. Chem., 2009, 78, 601–603 CrossRef CAS.
- D. Zhang, H. Ji, H. Liu, S. Wang, J. Wang and Y. Wang, Appl. Microbiol. Biotechnol., 2016, 100, 1–13 CrossRef PubMed.
- J. Z. Wang, W. T. Du, Y. L. Xu, S. Z. Cheng and Z. J. Liu, Microb. Pathog., 2017, 105, 122 CrossRef PubMed.
- L. Elmén, Clin. Exp. Immunol., 2015, 179, 363–377 CrossRef PubMed.
- Z. Wang, D. Koonen, M. Hofker and J. Fu, Curr. Opin. Lipidol., 2016, 27, 216–224 CrossRef CAS PubMed.
- A. Koh, F. D. Vadder, P. Kovatcheva-Datchary and F. B. Ckhed, Cell, 2016, 165, 1332–1345 CrossRef CAS PubMed.
- L. Ming, Y. Wang, G. Fan, X. Wang, S. Xu and Y. Zhu, Front. Microbiol., 2017, 8, 2146 CrossRef PubMed.
- F. Bäckhed, H. Ding, T. Wang, L. V. Hooper, Y. K. Gou, A. Nagy, C. F. Semenkovich and J. I. Gordon, Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 15718 CrossRef PubMed.
- P. Fan, P. Liu, P. Song, X. Chen and X. Ma, Sci. Rep., 2017, 7, 43412 CrossRef PubMed.
- J. P. Wang and I. H. Kim, Anim. Sci. J., 2015, 85, 942–950 CrossRef PubMed.
- C. Cui, Y. Li, H. Gao, H. Zhang, J. Han, D. Zhang, Y. Li, J. Zhou, C. Lu and X. Su, PLoS One, 2017, 12, e0186216 CrossRef PubMed.
- H. Zhang, Y. Li, C. Cui, T. Sun, J. Han, D. Zhang, C. Lu, J. Zhou, L. Cheong and Y. Li, Appl. Microbiol. Biotechnol., 2018, 1–11 Search PubMed.
- M. Valko, C. J. Rhodes, J. Moncol, M. Izakovic and M. Mazur, Chem.-Biol. Interact., 2006, 160, 1 CrossRef CAS PubMed.
- R. Turk, D. Juretić, D. Geres, A. Svetina, N. Turk and Z. Flegarmestrić, Anim. Reprod. Sci., 2008, 108, 98–106 CrossRef CAS PubMed.
- G. D. Kolovou, K. K. Anagnostopoulou and D. V. Cokkinos, Postgrad. Med. J., 2005, 81, 358–366 CrossRef CAS PubMed.
- A. Alonso, X. Yin, N. S. Roetker, J. W. Magnani, R. A. Kronmal, P. T. Ellinor, L. Y. Chen, S. A. Lubitz, R. L. Mcclelland and D. D. Mcmanus, J. Am. Heart Assoc., 2014, 3, e001211 Search PubMed.
- J. Yang, L. Yan and W. Fang, J. Agric. Food Chem., 2010, 58, 6525–6531 CrossRef CAS PubMed.
- H. Sadia, Q. S. Akter, R. Afroz and T. Siddika, J. Bangladesh Soc. Physiol., 2016, 11, 23 CrossRef.
- R. E. Ley, F. Bäckhed, P. Turnbaugh, C. A. Lozupone, R. D. Knight and J. I. Gordon, Proc. Natl. Acad. Sci. U. S. A., 2005, 102, 11070–11075 CrossRef CAS PubMed.
- E. Jami, B. A. White and I. Mizrahi, PLoS One, 2014, 9, e85423 CrossRef PubMed.
- V. K. Ridaura, J. J. Faith, F. E. Rey, J. Cheng, A. E. Duncan, A. L. Kau, N. W. Griffin, V. Lombard, B. Henrissat and J. R. Bain, Science, 2013, 341, 1241214 CrossRef PubMed.
- G. D. Wu, J. Chen, C. Hoffmann, K. Bittinger, Y. Y. Chen, S. A. Keilbaugh, M. Bewtra, D. Knights, W. A. Walters and R. Knight, Science, 2011, 334, 105–108 CrossRef CAS PubMed.
- M. Blaut, Aktuel. Ernaehrungsmed., 2015, 40(01), 43–49 CrossRef CAS.
- E. P. Neis, C. H. Dejong and S. S. Rensen, Nutrients, 2015, 7, 2930–2946 CrossRef CAS PubMed.
- J. M. Allaire, S. M. Crowley, H. T. Law, S. Y. Chang, H. J. Ko and B. A. Vallance, Trends Immunol., 2018, 39, 677–696 CrossRef CAS PubMed.
- S. Miquel, R. Martín, O. Rossi, L. G. Bermúdezhumarán, J. M. Chatel, H. Sokol, M. Thomas, J. M. Wells and P. Langella, Curr. Opin. Microbiol., 2013, 16, 255–261 CrossRef CAS PubMed.
- K. Takahashi, A. Nishida, T. Fujimoto, M. Fujii, M. Shioya, H. Imaeda, O. Inatomi, S. Bamba, M. Sugimoto and A. Andoh, Digestion, 2016, 93, 59–65 CAS.
- F. Wang, T. Yu, G. Huang, D. Cai, X. Liang, H. Su, Z. Zhu, D. Li, Y. Yang, P. Shen, R. Mao, L. Yu, M. Zhao and Q. Li, J. Microbiol. Biotechnol., 2015, 25, 1195–1204 CrossRef CAS PubMed.
- H. Xu, W. Huang, Q. Hou, L. Y. Kwok, Z. Sun, H. Ma, F. Zhao, Y. K. Lee and H. Zhang, Sci. Bull., 2017, 62, 31–38 CrossRef.
- Z. K. Wang, Y. S. Yang, A. T. Stefka, G. Sun and L. H. Peng, Aliment. Pharmacol. Ther., 2014, 39, 751–766 CrossRef CAS PubMed.
- B. Mayo and D. V. Sinderen, Bifidobacteria Genomics & Molecular Aspects, 2010 Search PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9ra02594d |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2019 |