Open Access Article
Open Access ArticleCircular RNA SMARCA5 inhibits gastric cancer progression through targeting the miR-346/ FBXL2 axis
Quanying Li†
,
Hongna Tang†,
Fangfang Hu and
Changjiang Qin *
*
Department of General Surgery, Huaihe Hospital of Henan University, No. 8 of Baobei Road, Kaifeng 475000, China. E-mail: hhyychangjiangqin@163.com; Fax: +86-371-23906957; Tel: +86-371-23906957
First published on 10th June 2019
Abstract
Circular RNA (circRNA) SMARCA5 (circSMARCA5) is a cancer-related circRNA that has been observed to be involved in the progression of several types of cancer. However, the role of circSMARCA5 in gastric cancer has not been reported. In the present study, we aimed to explore the function and mechanism of circSMARCA5 in gastric cancer. Our results showed that circSMARCA5 expression was significantly decreased in human gastric cancer tissues and cell lines. Further in vitro investigations demonstrated that overexpression of circSMARCA5 in SGC7901 cells inhibited cell proliferation, migration and invasion. Luciferase reporter assays proved that circSMARCA5 acted as a sponge for miroRNA-346 (miR-346) and regulated the expression of F-Box and leucine rich repeat protein 2 (FBXL2). Furthermore, transfection of miR-346 mimics into cells overexpressing circSMARCA5 blocked the function of circSMARCA5. Finally, we found that knockdown FBXL2 significantly reversed the effects of miR-346 inhibitor on gastric cell proliferation, migration and invasion. Collectively, circSMARCA5 exhibited a tumor suppressor-like activity in gastric cancer via regulating the miR-346/FBXL2 axis.
1. Introduction
Gastric cancer is one of the most common malignancies in the digestive system.1 Gastric cancer remains the second highest in frequency and mortality rate of all cancers throughout the world.1 In recent years, the incidence of gastric cancer is gradually increasing in young people. Because of the lack of specific signs and markers of early gastric cancer, most patients (>70%) develop advanced stages of the disease. Therefore, patients with gastric cancer exhibit high incidence, metastasis rate, and mortality rate, as well as low early diagnosis rate, radical resection rate, and 5 year survival rate.2 The main treatments for gastric cancer are surgical resection, and the combination of neoadjuvant chemoradiotherapy, molecular-targeted therapy and immunotherapy.2 In recent years, increasing research has been performed to improve the prognosis of gastric cancer and develop new therapeutic approaches.Circular RNAs (circRNAs) is a type of RNA which contains a covalently closed continuous loop linking the 3′ and 5′ end.3 CircRNAs have been categorized as noncoding RNA; however, a growing body of evidence indicates that circRNAs possess numerous properties via acting as cytoplasmic microRNA sponges, protein-like modulators, or transcriptional regulators.4 The accumulating researches prove that dysregulation of circRNAs is involved in various human diseases, including cancer.5–7
It has been documented that a large number of circRNAs are differentially expressed in gastric cancer.8,9 Further investigations demonstrate that several circRNAs are implicated in the pathogenesis of gastric cancer. For instance, circRNA PVT1 (circPVT1) expression is upregulated in gastric cancer tissues.10 CircPVT1 is a novel proliferative factor and prognostic marker in gastric cancer. Circ_0000096 has been found to be significantly downregulated in gastric cancer tissues and cell lines.11 Knockdown of circ_0000096 inhibits cell proliferation and migration in vitro and in vivo, indicating that circ_0000096 might be used as a potential novel biomarker for gastric cancer. CircSMARCA5 is a cancer-related circRNA that has been observed to be involved in several types of cancers, such as prostate cancer,12 cervical cancer,13 hepatocellular carcinoma (HCC),14 and glioblastoma multiforme (GBM).15 However, the role of circSMARCA5 in gastric cancer and the mechanism has not been fully investigated. In the current study, we explored the functional mechanism of circSMARCA5 in gastric cancer.
2. Materials and methods
2.1 Tissue collection and cell culture
Clinical tissues were obtained from 23 patients who were pathologically diagnosed with primary gastric cancer. The gastric cancer tissues and paired surrounding normal tissues were collected during surgery therapy from September 2015 to January 2017 in Huaihe Hospital of Henan University (Kaifeng, China). The sample were frozen in liquid nitrogen and then stored at −80 °C. All experiments were performed in compliance with the Ethics Committee of Huaihe Hospital of Henan University and approved by the Ethics Committee of Huaihe Hospital of Henan University. Informed consent was obtained for any experimentation with human subjects.2.2 Cell culture and transfection
Human normal gastric mucosa epithelial cell GES-1 and four human gastric cancer cell lines including MGC803, SGC7901, N87 and AGS cells (Institute of Biochemistry and Cell Biology, SIBS, CAS, Beijing, China) were cultured in RPMI cell culture medium (Hyclone, South Logan, UT, USA) supplemented with 10% FBS (Hyclone) and 1% penicillin/streptomycin. Cells were maintained in a humidified atmosphere containing 5% CO2 at 37 °C.SGC7901 cells were transfected with small interfere RNA (siRNA) targeting F-Box and leucine rich repeat protein 2 (si-FBXL2) or control siRNA (si-NC) using siRNA-Mate (GenePharma, Shanghai, China). The overexpression vector of circSMARCA5 (pLCDH-circSMARCA5) and empty vector (pLCDH) were constructed by Geneseed Co., Ltd. (Guangzhou, China). MiR-346 mimic, control mimic (miR-NC), miR-346 inhibitor, and control inhibitor (NC-inhibitor) were all purchased from RiboBio Co., Ltd. (Guangzhou, China). Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) was used for the transfection with pLCDH-SMARCA5/pLCDH and miR-346/miR-NC. The experiment was performed in triplicate.
2.3 Cell proliferation assay
After 6 h post transient transfection, cells were seeded on the 96-well plates (5 × 104 cells per well) and incubated for 24, 48, and 72 h. Then, CCK-8 solution (Dojindo, Kumamoto, Japan) was added into each well and incubated for 1 h. Optical density (OD) at wavelength of 450 nm was detected using a microplate reader (Bio-Tek, Winooski, VT, USA). The experiment was performed in triplicate.2.4 Cell migration and invasion assays
Cell migration and invasion were measured using transwell assay with Transwell chambers (8 μm-pore; BD Biosciences, San Jose, CA, USA). After 48 h post transfection, cells were collected and suspended in serum free medium at a density of 1 × 106 cells per ml. After that, 100 μl cell suspension was inoculated in the upper chamber. For the invasion assay, the membranes were coated with Matrigel (BD Biosciences). The lower chamber was added with 600 μl RPMI containing 10% FBS. Then the cells were incubated for 24 h. The non-invading cells were removed by scraping, while the invaded cells were mixed with paraformaldehyde for 15 min, and then stained with crystal violet for 10 min. The stained cells were detected under an inverted microscope. Five random fields were selected and the numbers of cells were counted. The experiment was performed in triplicate.2.5 Quantitative real-time PCR (qRT-PCR)
Total RNA was extracted from the clinical samples and cell lines with TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the instructions. The cDNA was synthesized by One Step PrimeScript cDNA Synthesis Kit (Takara, Shiga, Japan). QRT-PCR was performed by Premix Ex Taq II kit (Takara), on an ABI 7600 fast Real-time PCR System (Applied Biosystems). The All-in-One™ miRNA qRT-PCR Detection Kit (GeneCopoeia, Rockville, MA, USA) was used for the determination of miR-346. The relative expression fold changes were calculated using the 2−ΔΔCt method. GAPDH or U6 were tested as the internal controls. CircSMARCA5 5′-ACA ATG GAT ACA GAG TCA AGT GTT-3′, 5′-CCA CAA GCC TCC CTT TTG TTT T-3′; GAPDH 5′-TGT TCG TCA TGG GTG TGA AC-3′, 5′-ATG GCA TGG ACT GTG GTC AT-3′; miR-346 5′-TGT CTG CCC GCA TGC CTG CCT CT-3′; U6 5′-CTC GCT TCG GCA GCA CA-3′, 5′-AACGCTTCACGAATTTGCGT-3′. The experiment was performed in triplicate.2.6 Western blot analysis
SGC7901 cells were collected and lysed in radioimmunoprecipitation assay (RIPA) buffer (Beyotime Biotechnology, Shanghai, China). The protein concentration in cell lysates was measured using bicinchoninic acid (BCA) kit (KeyGen Biotechnology, Nanjing, Jiangsu, China). Then equal amounts of protein samples were mixed with loading buffer and boiled for 10 min for denaturation. The samples were subjected to 10% SDS-PAGE and transferred onto a polyvinylidene fluoride (PVDF) membrane (Millipore, Bedford, MA, USA). After blocking in 5% skim milk for 1 h, the 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000 diluted specific primary antibodies against FBXL2 and GAPDH (Abcam, Cambridge, MA, USA) were added onto membranes, followed by incubation at 4 °C overnight. On the next day, the 1
1000 diluted specific primary antibodies against FBXL2 and GAPDH (Abcam, Cambridge, MA, USA) were added onto membranes, followed by incubation at 4 °C overnight. On the next day, the 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5000 diluted horseradish peroxidase (HRP)-labeled goat anti-rabbit secondary antibody (Abcam) was added onto the membranes and incubated for 1 h at 37 °C. The enhanced chemiluminescence (ECL) reagent was used to develop the bands. The optical density of targeted bands was determined using Image J software (NIH, Bethesda, MD, USA). The experiment was performed in triplicate.
5000 diluted horseradish peroxidase (HRP)-labeled goat anti-rabbit secondary antibody (Abcam) was added onto the membranes and incubated for 1 h at 37 °C. The enhanced chemiluminescence (ECL) reagent was used to develop the bands. The optical density of targeted bands was determined using Image J software (NIH, Bethesda, MD, USA). The experiment was performed in triplicate.
2.7 Luciferase reporter assay
Wide type circSMARCA5 (circSMARCA5-WT), circSMARCA5 mutant (circSMARCA5-MUT), wide type FBXL2 (FBXL2-WT) and FBXL2 mutant (FBXL2-MUT) were constructed into pGL3-Basic luciferase vector (Promega, Madison, WI, USA). The generated reporter plasmids, along with miR-346 mimic/miR-NC were co-transfected into SGC7901 cells using Lipofectamine 2000 (Invitrogen). Then luciferase activity was detected using dual-luciferase Reporter assay system (Promega) at 48 h post-transfection. The experiment was performed in triplicate.2.8 Statistical analysis
Statistical analysis was performed using SPSS 19.0 software (SPSS, Chicago, IL, USA). Data were expressed as the mean ± standard deviation (SD). Student's t-test or one-way analysis of variance (ANOVA) was used to analyze differences between two experimental groups or among more than two groups, respectively. P values less than 0.05 were considered statistically different.3. Results
3.1 CircSMARCA5 was lowly expressed in human gastric cancer tissues and cell lines
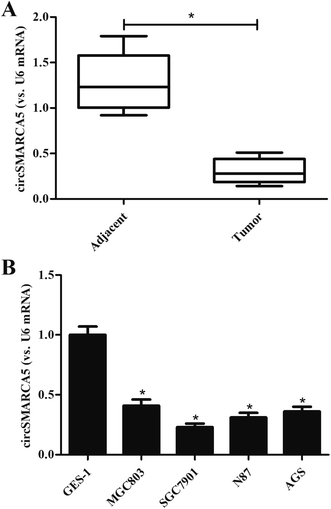
To investigate the role of circSMARCA5 in gastric cancer, we detected the expression of circSMARCA5 in gastric cancer tissues and adjacent normal tissues. As shown in Fig. 1A, the circSMARCA5 expression was markedly downregulated in gastric cancer tissues as compared to adjacent normal tissues. Then we evaluated the circSMARCA5 expression in normal gastric mucosa epithelial cell GES-1 and gastric cancer cell lines. As indicated in Fig. 1B, the expression level of circSMARCA5 in GES-1 cells was significant higher than that in human gastric cancer cell lines including MGC803, SGC7901, N87 and AGS cells. SGC7901 cell line exhibited the lowest circSMARCA5 expression, thus, this cell line was selected in the following experiments.3.2 Overexpression of circSMARCA5 inhibited the proliferation, migration and invasion of gastric cancer cells
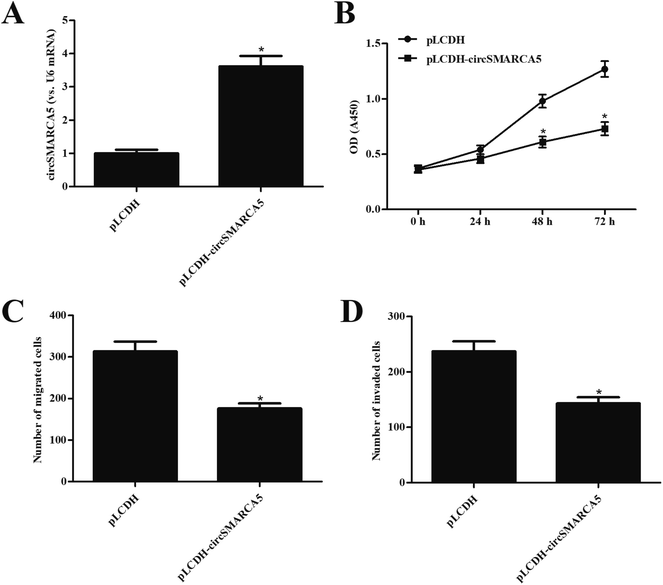
In order to investigate the function of circSMARCA5 in gastric cancer, the circSMARCA5 overexpressing vector, pLCDH-circSMARCA5, was transfected into SGC7901 cells. Transfection efficiency assay showed that circSMARCA5 expression was markedly increased after transfection with pLCDH-circSMARCA5 when compared with pLCDH-transfected SGC7901 cells (Fig. 2A). Next, we detected the effect of circSMARCA5 overexpression on proliferation, migration and invasion. CCK-8 assay demonstrated that circSMARCA5 overexpression dramatically suppressed the cell proliferation of SGC7901 cells (Fig. 2B). In addition, cell migration and invasion were also reduced by circSMARCA5 overexpression (Fig. 2C and D).3.3 MiR-346 mimic reversed the inhibitory effects of circSMARCA5 on gastric cancer cells
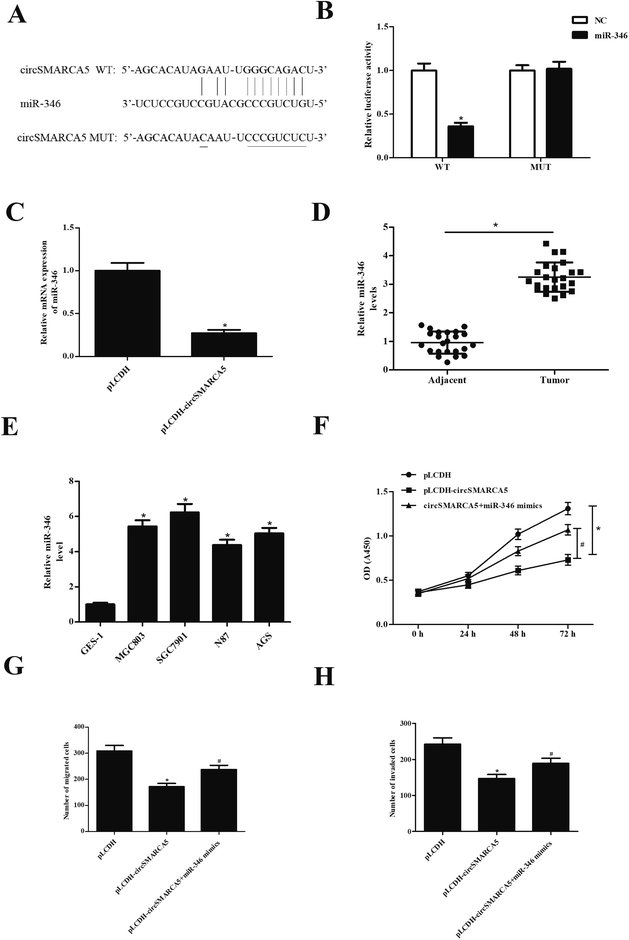
CircRNAs have been found to function as miRNA sponges to regulate gene expression. Therefore, we predicted the downstream target miRNAs of circSMARCA5 using the Circular RNA Interactome database and Targetscan. The results showed that miR-346 might be a target gene of circSMARCA5 (Fig. 3A). To confirm the prediction, circSMARCA5-WT/circSMARCA5-MUT and miR-346 mimic/miR-NC were co-transfected into SGC7901 cells. The results showed that luciferase activity was obviously decreased after cotransfection with circSMARCA5-WT and miR-346 mimic (Fig. 3B). Additionally, miR-346 expression was significantly decreased after transfection with pLCDH-circSMARCA (Fig. 3C). Furthermore, the levels of miR-346 were significantly up-regulated in gastric cancer tissues and cell lines (Fig. 4D and E). Transfection with miR-346 mimic prevented circSMARCA5-caused inhibitory effects on proliferation, migration and invasion in SGC7901 cells (Fig. 3F–H).3.4 Knockdown of FBXL2 reversed the tumor-suppression effects of miR-346 inhibitor in gastric cancer cells
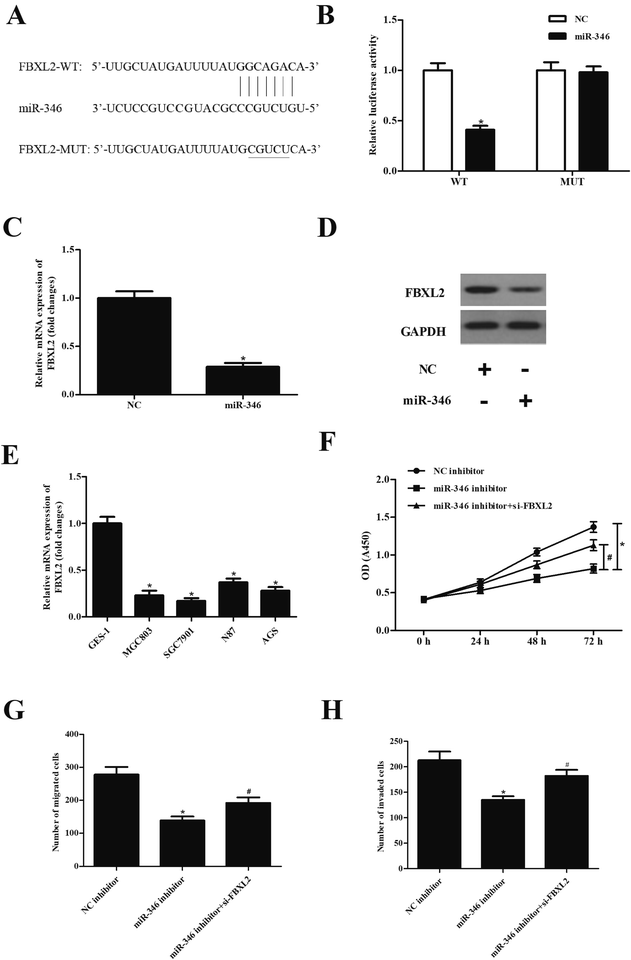
To explore the mechanism of miR-346, the target genes of miR-346 were predicted using Targetscan. As shown in Fig. 4A, FBXL2 was found to be a target gene of miR-346. In order to further confirm the predicted results, FBXL2-WT/FBXL2-MUT and miR-346 mimic/miR-NC were co-transfected into SGC7901 cells. The results in Fig. 4B showed that luciferase activity in SGC7901 cells co-transfected with FBXL2-WT and miR-346 mimic was dramatically decreased as compared to other cells (Fig. 4B). Transfection with miR-346 mimic significantly suppressed the expression of FBXL2 (Fig. 4C and D). We also examined the expression of FBXL2 in gastric cancer cell lines, and the results showed that FBXL2 expression levels were greatly down-regulated in gastric cancer cell lines, as compared with the GES-1 cell line. Additionally, knockdown of FBXL2 mitigated the decreased SGC7901 cells proliferation, migration and invasion caused by miR-346 inhibitor (Fig. 4F–H).4. Discussion
Emerging evidence has shown circRNAs are involved in the progression of many different kinds of cancers, either acting as oncogene or tumor suppressor. Previous study reported that circSMARCA5 exerts a promising druggable tumor suppressor role in GBM.16 Besides, GBM patients with low circSMARCA5 expression have lower overall and progression free survival rates than those patients with higher circSMARCA5 expression. CircSMARCA5 regulates angiogenesis in GBM via regulating vascular endothelial growth factor A (VEGFA) mRNA splicing.15 CircSMARCA5 is up-regulated in prostate cancer and acts as an oncogene by promoting cell cycle and inhibiting cell apoptosis of prostate cancer cell lines.12 Another study showed that circSMARCA5 was downregulated in cervical cancer, and overexpression of circSMARCA5 inhibited cervical cancer cell proliferation, migration and invasion, and induced cell cycle arrest.13 Li et al.14 reported that circSMARCA5 is significantly downregulated in HCC tissues. The expression level of circSMARCA5 is significantly associated with tumor differentiation, tumor-node-metastasis (TNM) stage, cancer invasion, and cancer diameter. Overexpression of circSMARCA5 results in decreased proliferation, increased apoptosis and reduced invasion, indicating circSMARCA5 may serve as a monitor biomarker and therapeutic target for HCC. In the present study, we found that circSMARCA5 expression was decreased in human gastric cancer tissues and cell lines. In vitro investigations demonstrated that overexpression of circSMARCA5 inhibited the proliferation, migration and invasion of gastric cancer cells.Increasing evidence indicates that a large number of miRNAs have been found to take vital parts in the pathogenesis and progression of cancers. MiR-346 promotes the biological functions of breast cancer cells, including cell proliferation, colony formation, migration and invasion.17 Guo et al.18 proved that miR-346 contributes to cervical cancer cell malignancy via up-regulating argonaute 2 (AGO2) expression. Additionally, miR-346 acts as an oncogenic miRNA in non-small cell lung cancer (NSCLC) since it can facilitate cell growth and metastasis, and suppress cell apoptosis in NSCLC cell lines.19 It has been shown that circRNAs usually acts as miRNA sponges to regulate gene expression. Ning and his colleagues reported that circFBLIM1 acts as a competing endogenous RNA (ceRNA) to promote the progression of HCC by sponging miR-346.20 Herein, we found that circSMARCA5 directly binds to miR-346. Overexpression of circSMARCA5 caused significant decrease in miR-346 expression. Transfection with miR-346 mimic prevented the inhibitory effects of circSMARCA5 on proliferation, migration and invasion in gastric cancer cells. These results suggested that circSMARCA5 executed its effects via sponging miR-346 in gastric cancer cells.
FBXL2 is a highly conserved member of F-box protein family. FBXL2 is a component of Skp-Cullin-F box (SCF) ubiquitin E3 ligase, playing critical role in ubiquitination of proteins, thereby regulating diverse processes. FBXL2 has been found to inhibit tumorigenesis by targeting and ubiquitinating several oncoproteins. FBXL2 exerts human lung tumor suppressor because it can cause cell cycle arrest through ubiquitin-mediated degradation of cyclin D3, which is a cell cycle-related protein.21 Besides, FBXL2 inhibits gastric cancer proliferation and invasion by ubiquitin-mediated degradation of forkhead box M1 (FoxM1) transcription factor in gastric cancer cells.22 Chen et al. reported that FBXL2 is ectopically expressed in a transformed murine lung epithelial (MLE) cell line. Ectopic expression of FBXL2 induces cyclin D2 degradation, whereas knockdown of FBXL2 results in significant increase in cyclin D2 expression. Moreover, FBXL2 inhibits cell proliferation and induces apoptosis in leukemic cells.23 Our results showed that FBXL2 was a target gene of miR-346. The expression of FBXL2 was markedly suppressed after transfection with miR-346 mimic. Additionally, knockdown of FBXL2 blocked the inhibitory effects of miR-346 inhibitor on SGC7901 cells proliferation, migration and invasion. These findings implied that miR-346 served as an oncogenic miRNA in gastric cancer via targeting FBXL2.
In conclusion, the present study evaluated the role of circSMARCA5 in gastric cancer and the underlying mechanisms. The results showed that circSMARCA5 inhibited cell proliferation, migration and invasion of gastric cancer cell lines. Furthermore, we demonstrated that circSMARCA5 served as a tumor suppressor in gastric cancer via regulating miR-346/FBXL2 axis.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This study was supported by the Science and Technology Development Project of Henan Province (No. 192102310099 and 182300410359), Key Research Program of Higher Education of China (No. 19A320020), Research Project of Henan Provincial Department of Education (No. 172102310152), Henan Province Medical Science and Technology Breakthrough Project (No. 201601029) and Science and Technology Development Project of Kaifeng City in China (No. 1906010).References
- P. Karimi, F. Islami, S. Anandasabapathy, N. D. Freedman and F. Kamangar, Gastric cancer: descriptive epidemiology, risk factors, screening, and prevention, Cancer Epidemiol., Biomarkers Prev., 2014, 23, 700–713 CrossRef PubMed.
- Z. Song, Y. Wu, J. Yang, D. Yang and X. Fang, Progress in the treatment of advanced gastric cancer, Tumor Biol., 2017, 39, 1010428317714626 Search PubMed.
- K. K. Ebbesen, J. Kjems and T. B. Hansen, Circular RNAs: identification, biogenesis and function, Biochim. Biophys. Acta, 2016, 1859, 163–168 CrossRef CAS PubMed.
- K. Y. Hsiao, H. S. Sun and S. J. Tsai, Circular RNA - new member of noncoding RNA with novel functions, Exp. Biol. Med., 2017, 242, 1136–1141 CrossRef CAS PubMed.
- J. Greene, A. M. Baird, L. Brady, M. Lim, S. G. Gray, R. Mcdermott and S. P. Finn, Circular RNAs: Biogenesis, Function and Role in Human Diseases, Front. Mol. Biosci., 2017, 4, 38–95 CrossRef PubMed.
- P. Shi, J. Wan, H. Song and X. Ding, The emerging role of circular RNAs in gastric cancer, Am. J. Cancer Res., 2018, 8, 1919–1932 Search PubMed.
- S. Lux and L. Bullinger, Circular RNAs in cancer, Adv. Exp. Med. Biol., 2018, 1087, 215–230 CrossRef PubMed.
- P. Li, S. Chen, H. Chen, X. Mo, T. Li, Y. Shao, B. Xiao and J. Guo, Using circular RNA as a novel type of biomarker in the screening of gastric cancer, Clin. Chim. Acta, 2015, 444, 132–136 CrossRef CAS PubMed.
- W. Sui, Z. Shi, W. Xue, M. Ou, Y. Zhu, J. Chen, H. Lin, F. Liu and Y. Dai, Circular RNA and gene expression profiles in gastric cancer based on microarray chip technology, Oncol. Rep., 2017, 37, 1804–1814 CrossRef CAS PubMed.
- J. Chen, 250P Circular RNA profile identifies circPVT1 as a proliferative factor and prognostic marker in gastric cancer, Cancer Lett., 2017, 388, 208–219 CrossRef CAS PubMed.
- P. Li, H. Chen, S. Chen, X. Mo, T. Li, B. Xiao, R. Yu and J. Guo, Circular RNA 0000096 affects cell growth and migration in gastric cancer, Br. J. Cancer, 2017, 116, 626–633 CrossRef CAS PubMed.
- Z. Kong, X. Wan, Y. Zhang, P. Zhang, Y. Zhang, X. Zhang, X. Qi, H. Wu, J. Huang and Y. Li, Androgen-responsive circular RNA circSMARCA5 is up-regulated and promotes cell proliferation in prostate cancer, Biochem. Biophys. Res. Commun., 2017, 493, 1217–1223 CrossRef CAS PubMed.
- J. D. C. Tian and L. Liang, Involvement of circular RNA SMARCA5/microRNA-620 axis in the regulation of cervical cancer cell proliferation, invasion and migration, Eur. Rev. Med. Pharmacol. Sci., 2018, 22, 8589–8598 Search PubMed.
- Z. Li, Y. Zhou, G. Yang, S. He, X. Qiu, L. Zhang, Q. Deng and F. Zheng, Using circular RNA SMARCA5 as a potential novel biomarker for hepatocellular carcinoma, Clin. Chim. Acta, 2019, 492, 37–44 CrossRef CAS PubMed.
- D. Barbagallo, A. Caponnetto, D. Brex, F. Mirabella, C. Barbagallo, G. Lauretta, A. Morrnoe, F. Certo, G. Barbagallo, R. Caltabiano, G. M. Barbagaool, V. Spina-Purrello, M. Ragusa, C. Di Pietro, T. B. Hansen and M. Purrello, CircSMARCA5 regulates VEGFA mRNA splicing and angiogenesis in glioblastoma multiforme through the binding of SRSF1, Cancers, 2019, 11, E194–E205 CrossRef PubMed.
- D. Barbagallo, A. Caponnetto, M. Cirnigliaro, D. Brex, C. Barbagallo, F. D'Angeli, A. Morrone, R. Caltabiano, G. M. Barbagallo, M. Ragusa, C. Di Pietro, T. B. Hansen and M. Purrello, CircSMARCA5 Inhibits Migration of Glioblastoma Multiforme Cells by Regulating a Molecular Axis Involving Splicing Factors SRSF1/SRSF3/PTB, Int. J. Mol. Sci., 2018, 19, E480–E492 CrossRef PubMed.
- F. Yang, L. J. Luo, L. Zhang, D. D. Wang, S. J. Yang, L. Ding, J. Li, D. Chen, R. Ma, J. Z. Wu and J. H. Tang, MiR-346 promotes the biological function of breast cancer cells by targeting SRCIN1 and reduces chemosensitivity to docetaxel, Gene, 2017, 600, 21–28 CrossRef CAS PubMed.
- J. Guo, J. Lv, M. Liu and H. Tang, miR-346 Up-regulates Argonaute 2 (AGO2) Protein Expression to Augment the Activity of Other MicroRNAs (miRNAs) and Contributes to Cervical Cancer Cell Malignancy, J. Biol. Chem., 2015, 290, 30342–30350 CrossRef CAS PubMed.
- C. C. Sun, S. J. Li, Z. P. Yuan and D. J. Li, MicroRNA-346 facilitates cell growth and metastasis, and suppresses cell apoptosis in human non-small cell lung cancer by regulation of XPC/ERK/Snail/E-cadherin pathway, Aging, 2016, 8, 2509–2524 CAS.
- N. Bai, E. Peng, X. Qiu, N. Lyu, Z. Zhang, Y. Tao, X. Li and Z. Wang, circFBLIM1 act as a ceRNA to promote hepatocellular cancer progression by sponging miR-346, J. Exp. Clin. Cancer Res., 2018, 37, 172–205 CrossRef PubMed.
- B. B. Chen, J. R. Glasser, T. A. Coon and R. K. Mallampalli, F-box protein FBXL2 exerts human lung tumor suppressor-like activity by ubiquitin-mediated degradation of cyclin D3 resulting in cell cycle arrest, Oncogene, 2012, 31, 2566–2579 CrossRef CAS PubMed.
- L. Q. Li, D. Pan, H. Chen, L. Zhang and W. J. Xie, F-box protein FBXL2 inhibits gastric cancer proliferation by ubiquitin-mediated degradation of forkhead box M1, FEBS Lett., 2016, 590, 445–452 CrossRef CAS PubMed.
- B. B. Chen, J. R. Glasser, T. A. Coon, Z. Chunbin, H. L. Miller, F. Moon, J. F. Mcdyer, B. Michael and R. K. Mallampalli, F-box protein FBXL2 targets cyclin D2 for ubiquitination and degradation to inhibit leukemic cell proliferation, Blood, 2012, 119, 3132–3141 CrossRef CAS PubMed.
Footnote |
| † These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2019 |