Open Access Article
Open Access ArticleSilver derivatives of multi-donor heterocyclic thioamides as antimicrobial/anticancer agents: unusual bio-activity against methicillin resistant S. aureus, S. epidermidis, and E. faecalis and human bone cancer MG63 cell line†
Jaspreet K. Aulakha,
Tarlok S. Lobana *a,
Henna Soodb,
Daljit S. Arorab,
Raminderjit Kaurc,
Jatinder Singhc,
Isabel Garcia-Santos
*a,
Henna Soodb,
Daljit S. Arorab,
Raminderjit Kaurc,
Jatinder Singhc,
Isabel Garcia-Santos d,
Manpreet Kaure and
Jerry P. Jasinskie
d,
Manpreet Kaure and
Jerry P. Jasinskie
aDepartment of Chemistry, Guru Nanak Dev University, Amritsar-143 005, India. E-mail: tarlokslobana@yahoo.co.in; Fax: +91-183-2258820
bDepartment of Microbiology, Guru Nanak Dev University, Amritsar-143 005, India
cDepartment of Molecular Biology and Biochemistry, Guru Nanak Dev University, Amritsar-143 005, India
dDepartamento de Quimica Inorganica, Facultad de Farmacia, Universidad de Santiago, 15782-Santiago, Spain
eDepartment of Chemistry, Keene State College, Keene, NH 03435-2001, USA
First published on 17th May 2019
Abstract
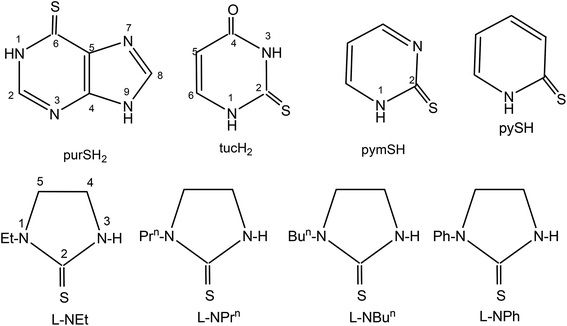
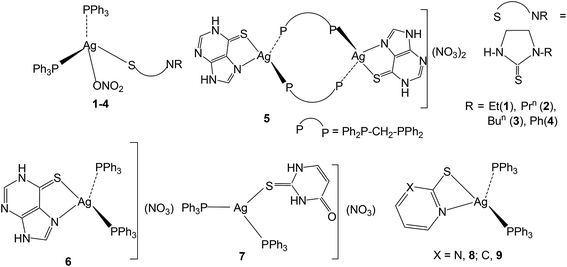
The basic aim of this study pertains to the synthesis of silver nitrate complexes and the study of their antimicrobial and anticancer bio-activity. A series of new silver(I) derivatives of N-substituted-imidazolidine-2-thiones (L-NR, R = Et, Prn, Bun, Ph), purine-6-thione (purSH2), 2-thiouracil (tucH2), pyrimidine-2-thione (pymSH) and pyridine-2-thione (pySH) of composition [Ag(S-L-NR)(PPh3)2(ONO2)] {R = Et (1), Prn (2), Bun (3), Ph (4)}, [Ag2(N,S-purSH2)2(μ-dppm)2](NO3)2·2H2O (5) (dppm = Ph2P–CH2–PPh2), [Ag(L)(PPh3)2](NO3) {L = N,S-purSH2 (6); S-tucH2 (7)}, [Ag(N,S-pymS)(PPh3)2](CH3OH) (8), and [Ag(N,S-pyS)(PPh3)2] (9) have been synthesized and structurally characterized. These new and some previously reported complexes {[Ag2(L-NH)4(PPh3)2](NO3)2 (10), [Ag(L-NMe)2(PPh3)](NO3) (11), and [Ag(S-bzimSH)2(PPh3)2](OAc) (12), L-NH = 1,3-imidazolidine-2-thione; L-NMe = 1-methyl-3-imidazolidine-2-thione and bzimSH2 = benzimidazoline-2-thione)} have shown moderate to high anti-microbial activity against Gram positive bacteria, namely methicillin resistant Staphylococcus aureus (MRSA) and Staphylococcus aureus (MTCC 740), and Gram negative bacteria, namely Staphylococcus epidermidis (MTCC 435), Enterococcus faecalis (MTCC 439), Shigella flexneri (MTCC 1457) and a yeast Candida albicans (MTCC 22). These complexes have also been found to be bio-safe as studied using MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide] assay. The anti-tumor study of silver complexes against human osteosarcoma cell line (MG63) has shown IC50 values in the range of 6–33 μM.
Introduction
Silver is an important metal known since ancient times for its antiseptic and antibacterial properties. Silver compounds are used to control bacterial growth in a variety of medical applications.1,2 For example, silver nitrate was introduced in therapeutic applications in 1884 for the prevention of ophthalmia neonatorum,3 and in eye drops against eye infections.1,2,4 Further, silver(I) sulfadiazine is used to treat severe burns to protect them from bacterial infection and catalyzes skin renewal,5 while a silver sulfadiazine–chlorhexidine mixture is used against catheter infections in vivo.6,7 Silver is even present in the human body (absorbed through the lungs, gastrointestinal tract, mucus membranes and skin) and any excess amount is eliminated from the body through the urine and faeces.2,8 Polymeric silver compounds of ethanediyl bis(isonicotinate) have also been used in coatings of implant materials for killing bacterial infections while remaining biocompatible for cell growth.2,9 In view of the above uses, the investigation of silver(I) chemistry becomes exceptionally appealing, which is bolstered by the ability of this metal to form complexes of variable geometry and nuclearity.10–16 Literature reports reveal that silver(I) complexes of sulfur containing ligands have found application in medicines, analytical chemistry and polymer industry.10–12,17,18In the literature, coordination chemistry of silver(I) with several heterocyclic-2-thiones, namely imidazolidine-2-thiones (L-NH, L-NMe), imidazoline-2-thiones, N-methylbenzothiazole-2-thione, pyrimidine-2-thione, tetrahydropyrimidine-2-thione, pyridine-2-thione, benzoxazoline-2-thione, thiazolidine-2-thione, and benz-imidazoline-2-thione, using different anions (NO3−, ClO4−, F3CSO3−, NCS−, SO42−, BF4−) as well as halides has been reported.13–44 Among them several silver complexes have found use as anti-microbial,23,30,35,37,40–42 anti-tumor,17,18,36,38,39 anti-inflammatory18,23 and anti-malarial agents.39 In regard to the antimicrobial activity, complexes of silver(I) salts with the thio-ligands, namely, imidazolidine-2-thione,30,40 thiazolidine-2-thione,41 benzo-thiazoline-2-thione,41 N-methyl-imidazoline-2-thione,42 pyrimidine-2-thiones,23 4,6-diamino-5-hydroxypyrimidine-2-thione,35 pyridine-2-thione,30 diazinane-2-thione,30 and 2-mercapto-nicotinic acid37 have shown activity against Escherichia coli,23,30,37,40 Pseudomonas aeruginosa,30,35,40 Bacillus subtilis,23,37 Bacillus cereus,23 Staphylococcus aureus,23,35,37 Pseudomonas aeruginosa,37 molds {Aspergillus niger,35,37 Penicillium citrinum,30,35,37, Rhizopus stolonifer,37 Aureobasidium pullulans,37 Fusarium moniliforme,34 Cladosporium sphaerospermum34} and yeasts {Candida albicans,26,32,34 and Saccharomyces cerevisiae30}. It is pointed out here that the investigations merely report antimicrobial activity without any mention of their biosafety.23,30,35,37,39,41,42 The anti-cancer/tumor activity of complexes of silver(I) with pyrimidine-2-thiones,36 tetrahydropyrimidine-2-thione,17 pyridine-2-thiones,36 thiazolidine-2-thione,39 5-chloro-2-mercaptobenzothiazole,18 and 2-mercaptonicotinic acid,18 pertains to in vitro cytotoxic activity against murine leukemia (L1210),17,36 human T-lymphocyte (Molt4/C8 and CEM) cells,17,36 breast cancer cell lines (MDA-MB-231, MCF-7),39 colon cancer cell line (HT-29),39 leiomyosarcoma cancer cells (LMS),18 and breast cancer cell line (MDA-MB-231),38 some of which have shown activity even higher than that of the anti-cancer drug cisplatin.
From our laboratory, coordination chemistry of coinage metals (Cu, Ag) with heterocyclic-2-thiones has been investigated which showed interesting chemical activity such as rupture of C–S bond45–49 and formation of a variety of coordination complexes.50–57 As a part of our interest to pursue bioactivity of metal-heterocyclic-2-thione complexes, we recently reported synthesis and antimicrobial activity of several copper(I)/silver(I) halide complexes of imidazolidine-2-thiones (L-NR, R = H, Me, Et, Prn, Bun, Ph) with or without triphenylphosphine (PPh3) as a co-ligand.44,58,59 These complexes showed moderate to high activity against various bacteria, Staphylococcus aureus, methicillin resistant Staphylococcus aureus (MRSA), Staphylococcus epidermidis, Enterococcus faecalis, Klebsiella pneumoniae, Salmonella typhimurium, Shigella flexneri, Escherichia coli, Candida albicans and Candida tropicalis, but only a few of them were biosafe.44,58,59
Keeping in view the high profile medicinal activity of silver(I) compounds with nitrate as anion as well as in continuation of our interest in this area,44,58,59 in this investigation, a series of silver derivatives of thio-ligands, N-substituted-imidazoldine-2-thiones, (L-NR, R = Et, Prn, Bun, Ph), purine-6-thione (purSH2), 2-thiouracil (tucH2), pyrimidine-2-thione (pymSH), and pyridine-2-thione (pySH) (Chart 1) are synthesized and structurally characterized. These complexes (1–9) as well as a few previously reported ones, namely, [Ag2(L-NH)4(PPh3)2](NO3)2, [Ag(L-NMe)2(PPh3)](NO3), and [Ag(bzimSH)2(PPh3)2](OAc) (10–12) (L-NH = 1,3-imidazolidine-2-thione; L-NMe = 1-methyl-3-imidazolidine-2-thione and bzimSH2 = benz-imidazoline-2-thione)43 have been evaluated for their in vitro antimicrobial activity against various microorganisms {Gram positive bacteria: Staphylococcus aureus (MTCC 740), methicillin resistant Staphylococcus aureus (MRSA) Staphylococcus epidermidis (MTCC 435), Enterococcus faecalis (MTCC 439), Gram negative bacteria Shigella flexneri (MTCC 1457), Escherichia coli (MTCC 119) and a yeast Candida albicans (MTCC 227). Apart from antimicrobial studies, as an extension of bioactivity, some studies are explored in the use of complexes for their anticancer activity against MG63 – a bone cancer cell line.
Results and discussion
Synthesis and IR NMR spectroscopy
In this section, comments regarding synthesis of complexes (Chart 2) and IR spectral studies are described. In a typical reaction, to the solution of silver(I) nitrate in methanol was added two m mole of solid L-NEt ligand followed by stirring for 1 h, and to the clear solution was added two m mole of solid PPh3 and the solution was further stirred for 15 minutes. The colorless contents were transferred into a culture tube for crystallization and after about one week crystalline product was obtained whose analytical data supported the composition as [Ag(S-L-NEt)(PPh3)2(ONO2)] 1. Similarly, other complexes, namely, [Ag(S-L-NR)(PPh3)2(ONO2)] (R = Prn, 2; Bun, 3; Ph, 4) were synthesized and bonding established using X-ray crystallography (vide infra). The crystalline pale yellow complex, [Ag2(N,S-purSH2)2(dppm)2](NO3)2 5, was obtained using same procedure from a reaction of silver nitrate with purine-6-thione (purSH2) and bis(diphenylphosphino)methane (dppm). Complexes [Ag(N,S-purSH2)(PPh3)2](NO3) 6, [Ag(S-tucH2)(PPh3)2](NO3)·H2O 7 and. [Ag(L)(PPh3)2] (L = N,S-pymS, 8; N,S-pyS 9) were prepared from the reactions of silver(I) nitrate with two m mole of solid PPh3 in methanol and the respective thio-ligands, purSH2, tucH2, pymSH and pySH with the only difference in case of the latter two complexes, where in Et3N Lewis base was added.The significant IR bands of complexes 1–9 are given in Table 1. The free thio-ligand, L-NEt shows ν(N–H) band of thioamide group at 3240 m, 3191 s cm−1, which shifts to 3238 m, 3175 m cm−1 in complex 1, and this shift supports that the thio-ligand L-NEt is coordinating to silver(I) ion as a neutral ligand. Similarly, low/high energy shift of ν(N–H) band of other thio-ligands, L-NPrn, L-NBun, L-Ph, purSH2, tucH2, is noted in complexes 2–7 (see Table 1). However, the absence of ν(N–H) band in complexes 8 and 9 shows that the thio-ligands (pymSH, pySH) in these two complexes are coordinating as anionic ligands (pymS−, pyS−). The presence of nitrate is recognized from the splitting of ν3 mode in 1–4 complexes which reveal coordination by nitrate and its lack of splitting in 5–7 complexes reveal the presence of ionic nitrate, which are supported by X-ray crystal structure determination. In literature,60 four fundamental IR spectral bands due to the thioamide functional group, –N(H)–C(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) S)–, classified as thioamide band I, near 1500 cm−1 {δ(N–H) + δ(C–H) + ν(C
S)–, classified as thioamide band I, near 1500 cm−1 {δ(N–H) + δ(C–H) + ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N)}, thioamide band II in the region, 1200–1300 cm−1 {ν(C–N) + δ(N–H) + δ(C–H) + νC
N)}, thioamide band II in the region, 1200–1300 cm−1 {ν(C–N) + δ(N–H) + δ(C–H) + νC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) S)}, thioamide band III, near 1000 cm−1 {ν(C–N)+ ν(C–S)}, and finally, thioamide band IV in the region, 700–850 cm−1 {ν(C–S)} were observed.60 In complex 1, thioamide bands III and IV (major contribution from νC–S + νC–N) underwent red shifts of 10 and 45 cm−1 respectively, whereas other two bands (I and II) remained less influenced (major contribution from δN–H + δC–H + νC
S)}, thioamide band III, near 1000 cm−1 {ν(C–N)+ ν(C–S)}, and finally, thioamide band IV in the region, 700–850 cm−1 {ν(C–S)} were observed.60 In complex 1, thioamide bands III and IV (major contribution from νC–S + νC–N) underwent red shifts of 10 and 45 cm−1 respectively, whereas other two bands (I and II) remained less influenced (major contribution from δN–H + δC–H + νC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N + νC
N + νC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) S). It showed the thio-ligand, L-NEt, is binding to metal through sulfur only. Similar trends of these bands were seen in complexes 2–4 and 7 (see Table 1). Complexes 5, 6, 8 and 9 containing both Ag–S and Ag–N bonds have undergone a red shift of band I (16–50 cm−1), slight blue shifts of bands II and III of 5–13 and 4–13 cm−1 respectively, while band IV undergoes red shift of 20–39 cm−1 with lowering of intensity. The appearance of ν(P–CPh) bands in the region 1092–1095 cm−1 (weak to medium in intensity) confirm the presence of coordinated PPh3 in the complexes 1–9.
S). It showed the thio-ligand, L-NEt, is binding to metal through sulfur only. Similar trends of these bands were seen in complexes 2–4 and 7 (see Table 1). Complexes 5, 6, 8 and 9 containing both Ag–S and Ag–N bonds have undergone a red shift of band I (16–50 cm−1), slight blue shifts of bands II and III of 5–13 and 4–13 cm−1 respectively, while band IV undergoes red shift of 20–39 cm−1 with lowering of intensity. The appearance of ν(P–CPh) bands in the region 1092–1095 cm−1 (weak to medium in intensity) confirm the presence of coordinated PPh3 in the complexes 1–9.
| Complex/free thio-ligand | ν(N–H) | ν(P–CPh) | Band I | Band II | Band III | Band IV | NO3− bandsc |
|---|---|---|---|---|---|---|---|
a See ESI for detailed spectral bands. s = strong, m = medium, w = weak.b Band I has contributions from δN–H + δC–H + νC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N; Band II has contributions from νC–N + δN–H + δC–H + νC N; Band II has contributions from νC–N + δN–H + δC–H + νC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) S; band III has contributions from νC–N + νC–S and band IV has contributions from νC–S.c ν1 mode of nitrate could not be assigned owing to the bands of thio-ligand and PPh3 ligand in this region. S; band III has contributions from νC–N + νC–S and band IV has contributions from νC–S.c ν1 mode of nitrate could not be assigned owing to the bands of thio-ligand and PPh3 ligand in this region. |
|||||||
| [Ag(S-L-NEt)(PPh3)2](NO3) 1 | 3238 m | 1093 s | 1507 s | 1277 s | 1027 m | 747 w | ν2, 834 w |
| 3175 m | ν3,1323 s, 1259 w | ||||||
| ν4, 722 s | |||||||
| Ag(S-L-NPrn)(PPh3)2(κ1:O–NO2)] 2 | 3051 m | 1094 s | 1509 m | 1281 w | 1027 m | 745 s | ν2, 851 w |
| ν3, 1384 s, 1349 m | |||||||
| ν4, (obscured by band IV) | |||||||
| Ag(S-L-NBun)(PPh3)2(κ1:O–NO2)] 3 | 3183 m | 1095 m | 1518 s | 1285 s | 1029 w | 747 s | ν2, 858 w; |
| ν3, 1384 s, 1310 m; | |||||||
| ν4, (obscured by band IV) | |||||||
| [Ag(S-L-NPh)(PPh3)2](NO3) 4 | 3128 m | 1094 s | 1519 s | 1244 s | 1027 m | 743 s | ν2, 849 w |
| ν3, 1335 m, 1292 s | |||||||
| ν4, (obscured by band IV) | |||||||
| [Ag2(N,S-purSH2)2(μ-dppm)2]-(NO3)2 5 | 3259 m | 1095 m | 1507 m | 1282 m | 1027 w | 748 m | ν2, 858 w |
| ν3, 1382 m | |||||||
| ν4, 722 s | |||||||
| [Ag(N,S-purSH2)(PPh3)2](NO3) 6 | 3133 w | 1094 m | 1479 m | 1288 s | 1026 w | 743 m | ν2, 852 m |
| ν3, 1384 s | |||||||
| ν4, 722 m | |||||||
| [Ag(S-tucH2)(PPh3)2](NO3) 7 | 3139 m | 1092 s | 1565 s | 1239 m | 1001 w | 723 w | ν2, obscured |
| ν3, 1434 s | |||||||
| ν4, 723 w | |||||||
| [Ag(N,S-pymS)(PPh3)2] 8 | — | 1092 m | 1435 s | 1270 w | 1027 w | 744 s | — |
| [Ag(N,S-pyS)(PPh3)2] 9 | — | 1094 m | 1480 m | 1256 w | 1027 w | 723 w | — |
| L-NEt | 3240 m, | ||||||
| 3191 s | — | 1513 s | 1279 m | 1037 m | 792 m | — | |
| L-NPrn | 3207 s | — | 1514 s | 1280 s | 1037 w | 806 w | — |
| L-NBun | 3196 s | — | 1514 s | 1284 s | 1041 w | 803 w | — |
| L-NPh | 3203 s | — | 1518 s | 1288 m | 1040 m | 757 s | — |
| purSH2 | 3096 m | — | 1529 m | 1275 m | 1014 m | 782 w | — |
| tucH2 | 3135 m | — | 1567 s | 1240 m | 1004 w | 761 w | — |
| pymSH | 3186 w | — | 1472 w | 1275 s | 1014 s | 783 m | — |
| pySH | 3162 s | — | 1494 m | 1255 m | 1023 w | 743 s | — |
| PPh3 | — | 1088 s | — | — | — | — | — |
| dppm | — | 1096 m | — | — | — | — | — |
NMR spectroscopy
Proton NMR signals of coordination compounds 1–9, free thio-ligands and co-ligands (PPh3, dppm) are given in Table 2. The thio-ligand, L-NEt, shows NH proton signal at 6.39 ppm which significantly shifted to low field at 9.25 ppm in its complex 1 and this shift suggests that the thio-ligand is coordinating to the silver(I) ion through its S-donor atom, as a neutral ligand. The ring and side chain protons, C4H2, C5H2 and N–CH2 of thio-ligand L-NEt showed an unresolved multiplet at 3.59(m; 6H), while –CH3 proton signal occurred at 1.14 ppm as a triplet. In complex 1, the C4H2, C5H2 and N–CH2 proton signals were partially resolved into two multiplets, one at 3.72 ppm due to ring protons (4H, C4H2/C5H2) in the low field region and another at 3.59 (m; 2H, N–CH2), while –CH3 signal at 1.21 ppm (t; 3H, CH3) shifted to a little high field region. The proton NMR signal of the thio-ligand and PPh3 suggest coordination to silver through S and P donor atoms of the respective ligands. In other similar complexes 2–4, the –NH protons signals at 5.91, 6.02 and 6.38 ppm shifted significantly to low fields at 7.87, 8.40 and 9.40 ppm respectively. Other ring, N-R side chain and PPh3 phenyl ring protons all showed variable trends in complexes 2–4 relative to the un-coordinated ligands (Table 2).| Complex/free thio-ligand | –NH protons | Ring protons of thio-ligand and co-ligands (PPh3/dppm) |
|---|---|---|
| a See ESI for spectra.b (N–CH2) and (ring proton C4H2 or C5H2) got merged.c CDCl3.d DMSO.e N-Ph protons obscured by phenyl ring of PPh3. | ||
| [Ag(S-L-NEt)(PPh3)2(O–NO2)]c1 | 9.25 (s, 1H, NH) | 3.72(m, 4H, C4H2/C5H2); 3.59 (q, 2H, N–CH2); 1.21 (t, 3H, CH3); 7.51 (m, 12H, o-H), 7.40 (m, 18H, m-H, p-H, PPh3) |
| [Ag(S-L-NPrn)(PPh3)2(O–NO2)]c2 | 7.87 (s, 1H, NH) | 3.48 (t, 2H, C4H2); 3.39 (t, 2H, N–CH2,); 3.29 (t; 2H, C5H2); 1.51 (m; 2H, CH2); 0.85 (t; 3H, CH3); 7.32 (m, 12H, o-H), 7.23 (m, 18 H, m-H, p-H, PPh3) |
| [Ag(S-L-NBun)(PPh3)2(O–NO2)]c3 | 8.40 (s; 1H, N3H) | 3.62(dt; 2H, C4H2)b; 3.62 (dt; 2H, N–CH2)b; 3.50 (t; 2H, C5H2);1.54 (quint; 2H, CH2);1.35 (m; 2H, CH2); 0.95 (t; 3H, CH3); 7.39 (m, 12H, o-H), 7.33 (m, 18H, m-H, p-H, PPh3) |
| [Ag(S-L-NPh)(PPh3)2](NO3)c4 | 9.40 (s, 1H, NH) | 4.18 (t, 2H, C4H2);3.86 (t, 2H, C5H2), 7.36 (m, 21H, o-H, p-H, PPh3+ L-NPh)e, 7.26 (m, 14H, m-H, PPh3+ L-NPh)d |
| [Ag2(N,S-purSH2)2(μ-dppm)2] (NO3)c5 | 13.78 (s, 2H, N1,9H) | 8.29 (s, 1H, C8H); 7.73 (s, 1H, C2H); 3.79 (s, 2H, P–CH2–P, dppm), 7.36 (d, 8 H, o-H), 7.20 (t, 4H, p-H), 7.06 (t, 8H, m-H, dppm) |
| [Ag(N,S-purSH2)(PPh3)2](NO3)c6 | 13.78 (s, 2H, N1,9H) | 8.40 (s, 1H, C8H); 8.33 (s, 1H, C2H); 7.41(m, 12H, o-H), 7.27 (m, 18 H, m-H, p-H, PPh3) |
| [Ag(S-tucH2)(PPh3)2](NO3) d7 | 8.28 (s; 2H, N1,3H) | 7.13 (d; 1H, C6H); 5.55 (d; 1H, C5H); 7.37 (m, 12H, o-H), 7.25 (m, 18H, m-H, p-H, PPh3) |
| [Ag(N,S-pymS)(PPh3)2]d8 | — | 7.85 (d; 2H, C4,6H); 6.50 (t; 1H, C5H); 7.34 (m, 12H, o-H), 7.24 (m, 18 H, m-H, p-H, PPh3) |
| [Ag(N,S-pyS)(PPh3)2]d9 | — | 7.39 (t, 1H, C3H); 7.24 (m, 1H, C4H); 6.81 (t, 1H, C5H); 7.57 (m, 13H, C6H +o-H, PPh3); 7.51 (m, 18H, m-H, p-H, PPh3) |
| L-NEtc | 6.39 (s, 1H, NH) | 3.59(m, 6H, C4H2/C5H2/N–CH2)b;1.14 (t, 3H, CH3) |
| L-NPrn,c | 5.91 (s, 1H, NH) | 3.71 (t, 2H, C4H2); 3.58 (m, 2H, N–CH2,); 3.46 (t, 2H, C5H2); 1.65 (m, 2H, CH2); 0.97 (t, 3H, CH3) |
| L-NBun,c | 6.02 (s, 1H, NH) | 3.64(m, 2H, C4H2)b; 3.53 (m, 4H, C5H2/N–CH2)b; 1.53 (m, 2H, CH2); 1.33 (m, 2H, CH2); 0.91 (t, 3H, CH3) |
| L-NPhc | 6.38 (s, 1H, NH) | 4.20 (t, 2H, C4H2); 3.76 (t, 2H, C5H2); 7.60 (m, 2H, o-H),7.42 (m, 2H, m-H), 7.27 (m, H, p-H) |
| purSH2d | 13.70 (s, 2H, N1,9H) | 8.33 (s, 1H, C8H); 8.14 (s, 1H, C2H) |
| tucH2d | 12.37 (s; 2H, N1,3H) | 7.36 (d; 1H, C6H); 5.78 (d; 1H, C5H) |
| pymSHd | 8.23 (s; 1H, N1H) | 8.65 (d; 1H, C4H); 7.32 (m; 1H, C6H); 6.79 (t; 1H, C5H) |
| pySHd | 13.46 (s; 1H, N1H) | 7.6 (sb, 1H, C6H); 7.35 (t; 1H, C3H); 7.25 (t; 1H, C4H); |
| 6.69 (t; 1H, C5H) | ||
| PPh3c | — | 7.32 (m; o-H, m-H,p-H; 15H) |
| dppmc | — | 2.18 (s; 2H, P–CH2–P); 7.44 (m, 8H, o-H), 7.30 (m, 12H, m-H, p-H) |
Purine-6-thione has shown NMR signals at 13.70 ppm due to –N1H– and –N9H– protons, which marginally shifted to low field at 13.78 ppm in complex 6, and likewise other two signals at 8.33 and 8.14 ppm due to –C8H and –C2H groups shifted to low field at 8.40 and 8.30 ppm respectively, however, the magnitude of shift was very small. The purine-6-thione signals in complex 5 showed similar trend with respect to –N1H/N9H protons, but there was slightly up-field move with respect to the –C8H and –C2H protons. The 2-thiouracil showed proton NMR signals at 12.37, 7.36 and 5.78 ppm due to N1,3H, C6H and C5H moieties and in complex 7 these shifted to 8.28, 7.13 and 5.55 ppm respectively. The pyrimidine-2-thione showed NMR signals due to –N1H proton at 8.23 ppm which disappeared in its complex 8, and signals due to C4H, C6H and C5H protons showed mixed trend –low field/upfield in complex relative to the free thio-ligand. Similarly, in complex 9, there was no signal due to –NH proton which was at 13.46 ppm in uncoordinated pySH (Table 2) and other signals due to C–H protons of thio-ligand showed minor variations in the same way as in case of complex 8. Thus proton NMR clearly supported that thio-ligands pymSH and pySH were coordinating as anions in complexes 8 and 9, while in other complexes, 1–7, the thio-ligands were coordinating as neutral ligands.
The uncoordinated PPh3 showed a multiplet at 7.32 ppm due to o-, m-, and p-H hydrogens of phenyl rings and in complexes 1–4, 6–9, this multiplet separated into ortho-hydrogen and meta-/para-hydrogen signals. For example, in complex 1 new multiplets appeared at low field, 7.52 ppm (m, o-H) and 7.40 ppm (m, m-and p-H). Other complexes 2–4, 6–9 showed similar trends. In dppm complex 5, the –CH2– moiety shows a signal at 3.79 ppm which is low field relative to the –CH2– moiety of free dppm, at 2.18 ppm. The phenyl ring protons of dppm are resolved into ortho, meta and para hydrogen signals in its complex 5. Thus proton NMR shows coordination by PPh3 and dppm in their complexes.
The 13C NMR signals of silver(I) coordination compounds 1, 3, 5–8, free thio-ligands and PPh3/dppm co-ligands are given in Table 3. In complex 1, the C2 signal of thio-ligand L-NEt (179.9 ppm) has undergone a significant upfield shift relative to the free ligand (182.9 ppm). This shift indicates that the thio-ligand (L-NEt) is coordinating to silver(I) ion through S-donor atom and this leads to drift of electron density of nitrogen atoms with lone pairs towards C2 carbon, leading to its enhanced shielding. Similarly, in complexes 3 and 6, the C2 carbon signal has undergone upfield shift to 180.4 and 163.6 ppm, relative to the free ligands (L-Bun, purSH2 = 183.4, 171.4 respectively). Complexes of purine-6-thione (5), 2-thiouracil (7) pyrimidine-2-thione (8) showed small lowfield shifts of C2 carbon (171.6 6, 163.6 7, 183.2 8 ppm), relative to the free ligands (purSH2, tucH2, pymSH = 171.4, 161.54, 181.9 ppm respectively). Further, the ring C4, C5 and side chain N-R carbons in complexes 1 and 3 as well as the C4–6 carbons of complex 8 showed only minor variations as compared to the free thio-ligands. In complex 5, the ring C2 and C4/C8 carbon signals (145.9, 133.3 ppm) of purSH2 have undergone up-field shift in complex 5 relative to the free ligand (152, 145 ppm), due to the deshielding of these carbons. Similar trends are seen in complex 7 in comparison to the free ligand tucH2. The ipso, ortho, meta and para carbon signals of coordinated PPh3 in complexes 1, 3 and 6–8, undergo variations as follows: (i) ipso-carbons have shown upfield shifts in all the complexes, ortho-carbons have shown upfield shifts in complexes 1, 3, 6, 7 and lowfield shift in complex 8, meta- and para-carbons showed only minor variations in either direction in all the complexes. Whereas, complex 5, having dppm as a co-ligand, ipso-, and ortho- carbons showed upfield shift and meta-, para-carbons of P-Ph moiety along with carbon atoms of P–CH2–P group have shown low field shift relative to the free co-ligand. All these variations showed that it is the ipso carbon that is most affected in all the coordination compounds and hence the coordination of the PPh3 to the silver(I) is through P donor atom. This upfield shift is attributed to electromeric effect involving movement of p-C and m-C phenyl ring electrons towards i-C–P bond axis leading to shielding of this carbon. The 1J (13C–31P) coupling constant did not show regular trend.
| Complex/free ligand | Carbons of thio-ligands | Carbons of co-ligands (PPh3/dppm) | |
|---|---|---|---|
| C2/C6 | Other ring/chain carbons | i-C, o-C, m-C, p-C | |
| a See ESI for spectra.b CDCl3.c DMSO. | |||
| [Ag(S-L-NEt)(PPh3)2(O–NO2)]1b | 179.9 | 48.26 (C5), 42.06 (N–CH2), 41.06(C4), 12.23 (CH3) | 133.91 (i-C, JC–P = 60 Hz), 132.47 (o-C, JC–P = 135 Hz), 129.98 (p-C), 128.70 (m-C, JC–P = 40 Hz) |
| [Ag(S-L-NBun)(PPh3)2(O–NO2)]3b | 180.4 | 48.80 (C5), 46.72 (N–CH2), 41.89 (C4), 29.24 (CH2), 19.88 (CH2), 13.78 (CH3) | 133.87 (i-C, JC–P = 70 Hz), 132.78 (o-C, JC–P = 90 Hz), 130.01 (p-C), 128.80 (m-C, JC–P = 35 Hz) |
| [Ag2(N,S-purSH2)2(μ-dppm)2](NO3)2 5c | 171.6 | 145.91(C2), 133.34 (C4/C8), 128.75(C5) | 131.89 (i-C), 131.31 (o-C, J13C–P = 174 Hz), 128.93 (p-C), 128.74 (m-C, JC–P = 52 Hz), 49.13 (CH2) |
| [Ag(N,S-purSH2)(PPh3)2](NO3) 6c | 163.6 | 152.75(C2), 133.63 (C4/C8), 129.25(C5) | 132.60 (i-C, JC–P = 8 Hz), 132.51 (o-C, JC–P = 40 Hz), 129.37 (p-C), 129.25 (m-C) |
| [Ag(S-tucH2)(PPh3)2](NO3) 7c | 163.6 | 164.92(C4), 133.64 (C6), 100.76(C5) | 132.60 (i-C, JC–P = 12 Hz), 132.01 (o-C, JC–P = 40 Hz), 129.36 (p-C), 129.24 (m-C) |
| [Ag(N,S-pymS)(PPh3)2] 8c | 183.2 | 156.16(C4H/C6H), 114.16 (C5H) | 134.29 (i-C, JC–P = 52 Hz), 133.88 (o-C, JC–P = 68 Hz), 130.31 (p-C), 129.27 (m-C, JC–P = 32 Hz) |
| L-NEtb | 182.9 | 47.97 (C5), 41.67 (N–CH2), 41.39 (C4), 12.21 (CH3) | PPh3a: 137.14 (i-C, JC–P = 40 Hz), 133.83 (o-C, JC–P = 76 Hz), 128.85 (p-C), 128.60 (m-C, JC–P = 28 Hz) |
| L-NBun,b | 183.4 | 48.61 (C5), 46.77 (N–CH2), 41.44 (C4), 29.26 (CH2), 19.99 | |
| (CH2), 13.89 (CH3) | dppma: 138.88 (i-C, JC–P = 40 Hz), 132.92 (o-C, JC–P = 84 Hz), 128.80 (p-C), 128.48 (m-C, JC–P = 28 Hz), | ||
| 28.12 (CH2) | |||
| purSH2c | 171.4 | 152.08 (C2), 145.04 (C4/C8), 128.76 (C5) | — |
| tucH2 c | 161.5 | 176.17 (C4), 142.63 (C6), 106.80 (C5) | — |
| pymSH c | 181.9 | 154.69 (C6), 159.08 (C4), 110.01 (C5H) | — |
Molecular structures
The crystal data of complexes 2, 3, 5–8 are given Table 4 which is placed in the experimental section, and their important bond parameters are given in Table 5. Complexes 3, 5 and 6 crystallized in monoclinic crystal system in space groups P21/c (3), C2/c (5) and P21/n (6), while each of complexes 2, 7 and 8 crystallized in triclinic crystal system in space group P![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) .
.
| 2 | 3 | 5 | |
|---|---|---|---|
| Empirical formula | C42H42AgN3O3P2S | C43H44AgN3O3P2S | C60H56Ag2N10O8P4S2 |
| CCDC | 1878467 | 1878471 | 1878472 |
| M | 838.65 | 852.68 | 1448.88 |
| T/K | 293(2) | 173(2) | 293(2) |
| λ Å | Cu-Kα, 1.54178 | Mo-Kα, 0.71073 | Cu-Kα, 1.54178 |
| Crystal system | Triclinic | Monoclinic | Monoclinic |
| Space group | P![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) |
P21/c | C2/c |
| a (Å) | 10.5641(6) | 13.6628(4) | 25.2895(7) |
| b (Å) | 12.9718(7) | 16.2436(5) | 13.2208(2) |
| c (Å) | 15.3593(8) | 18.8410(6) | 23.5888(7) |
| α (°) | 77.698(4) | 90 | 90 |
| β (°) | 82.402(4) | 105.902(3) | 125.671(4) |
| γ (°) | 79.796(4) | 90 | 90 |
| V (Å3) | 2014.12(19) | 4021.4(2) | 6407.1(4) |
| Z | 2 | 4 | 4 |
| Dcalcd (g cm−3) | 1.383 | 1.408 | 1.502 |
| μ (mm−1) | 5.577 | 0.675 | 6.958 |
| F(000) | 864 | 1760 | 2944 |
| Reflns collected | 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 231 231 |
36![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 999 999 |
12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 856 856 |
| Unique reflns | 7658 (Rint = 0.0443) | 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 487 (Rint = 0.0418) 487 (Rint = 0.0418) |
6067 (Rint = 0.0264) |
| Data/restraints/parameters | 7658/0/480 | 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 487/0/479 487/0/479 |
6067/223/447 |
| Reflecs with [I > 2σ(I)] | 6347 | 9990 | 5309 |
| Final R indices [I > 2σ(I)] | R1 = 0.0548 | R1 = 0.0449 | R1 = 0.0383 |
| wR2 = 0.1389 | wR2 = 0.0962 | wR2 = 0.1040 | |
| Final R indices (all data) | R1 = 0.0668 | R1 = 0.0702 | R1 = 0.0467 |
| wR2 = 0.1469 | wR2 = 0.1121 | wR2 = 0.1101 | |
| Largest diff. peak/hole e Å−3 | 1.720 and −0.671 | 1.193 and −0.482 | 1.160 and −0.441 |
| 6 | 7 | 8 | |
|---|---|---|---|
| Empirical formula | C41H34AgN5O3P2S | C40H36AgN3O5P2S | C41H37AgN2OP2S |
| CCDC | 1878468 | 1878469 | 1878470 |
| M | 846.60 | 840.59 | 775.59 |
| T/K | 293(2) | 293(2) | 173(2) |
| λ, Å | Cu-Kα, 1.54178 | Cu-Kα, 1.54178 | Mo-Kα, 0.71073 |
| Crystal system | Monoclinic | Triclinic | Triclinic |
| Space group | P21/n | P![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) |
P![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) |
| a (Å) | 9.2210(3) | 12.5062(7) | 10.1240(4) |
| b (Å) | 25.4962(8) | 13.1744(7) | 13.4635(6) |
| c (Å) | 17.0609(5) | 15.1280(7) | 14.1829(6) |
| α (°) | 90 | 99.534(4) | 77.466(4) |
| β (°) | 100.830(3) | 103.826(5 | 78.784(4) |
| γ (°) | 90 | 114.897(5) | 78.993(4) |
| V (Å3) | 3939.6(2) | 2092.7(2) | 1828.62(14) |
| Z | 4 | 2 | 2 |
| Dcalcd (g cm−3) | 1.427 | 1.334 | 1.409 |
| μ (mm−1) | 5.728 | 5.408 | 0.730 |
| F(000) | 1728 | 860 | 796 |
| Reflns collected | 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 236 236 |
15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 670 670 |
21![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 420 420 |
| Unique reflns | 7485 (Rint, 0.0407) | 7954(Rint, 0.0586) | 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 042 (Rint, 0.0323) 042 (Rint, 0.0323) |
| Data/restraints/parameters | 7485/0/482 | 7954/0/472 | 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 042/0/436 042/0/436 |
| Reflecs with [I > 2σ(I)] | 6025 | 6920 | 9569 |
| Final R indices [I > 2σ(I)] | R1 = 0.0464 | R1 = 0.0449 | R1 = 0.0393 |
| wR2 = 0.1153 | wR2 = 0.1161 | wR2 = 0.1125 | |
| Final R indices (all data) | R1 = 0.0608 | R1 = 0.0519 | R1 = 0.0575 |
| wR2 = 0.1253 | wR2 = 0.1226 | wR2 = 0.1339 | |
| Largest diff. peak/hole e Å−3 | 1.114 and −0.612 | 0.745 and −0.972 | 0.808 and −0.488 |
| Mononuclear complexes | |||||||
|---|---|---|---|---|---|---|---|
| 2 | 3 | 6 | 8 | 7 | |||
| Ag–S | 2.6161(13) | 2.5340(7) | Ag–S | 2.6478(10) | 2.5502(6) | Ag–S | 2.5464(9) |
| Ag–O | 2.496(4) | 2.4999(19) | Ag–N | 2.468(4) | 2.579(2) | Ag–P | 2.4531(7) |
| Ag–P | 2.4552(11) | 2.4524(6) | Ag–P | 2.4511(8) | 2.4477(6) | Ag–P | 2.4787(8) |
| Ag–P | 2.4450(11) | 2.4554(6) | Ag–P | 2.4301(9) | 2.4528(6) | S–C | 1.688(3) |
| S–C | 1.695(6) | 1.708(3) | S–C | 1.693(4) | 1.722(3) | O1–N | 1.264(5) |
| O1–N | 1.175(7) | 1.232(3) | O1–N | 1.200(6) | — | O2–N | 1.227(5) |
| O2–N | 1.284(7) | 1.249(3) | O2–N | 1.218(6) | — | O3–N | 1.235(6) |
| O3–N | 1.227(7) | 1.233(3) | O3–N | 1.240(5) | — | — | |
| O–N–O | 117.9(5), 121.8(5), 120.1(5) | ||||||
| P–Ag–S | 109.64(4) | 114.81(2) | P–Ag–S | 101.23(3) | 115.58(2) | P–Ag–S | 119.40(3) |
| P–Ag–S | 108.08(4) | 108.88(2) | P–Ag–S | 120.37(3) | 118.61(2) | P–Ag–S | 110.51(3) |
| P–Ag–O1 | 98.00(12) | 101.72(5) | P–Ag–N | 108.14(10) | 105.16(5) | P–Ag–P | 124.53(2 |
| P–Ag–O1 | 109.00(12) | 102.46(2) | P–Ag–N | 105.28(9) | 106.64(5) | O–N–O | 119.5(4) |
| O1–Ag–S | 97.59(13) | 103.09(6) | N–Ag–S | 79.87(9) | 61.89(5) | O–N–O | 121.7(5) |
| P–Ag–P | 129.48(4) | 122.73(2) | P–Ag–P | 130.17(3) | 125.17(2) | O–N–O | 118.8(4) |
| Dinuclear complex 5 | |||||||
|---|---|---|---|---|---|---|---|
| Ag–S | 2.7239(9) | S–C; Ag⋯Ag | 1.685(3) 3.466 (4) | P–Ag–S | 124.79(3) | P–Ag–N | 94.06(6) |
| Ag–P | 2.4273(6) | O1–N | 1.242(6) | P– Ag–S1 | 91.39(2) | S– Ag–N | 77.27(7) |
| Ag–P | 2.4677(7) | O2–N | 1.224(7) | P– Ag–N | 97.98(6) | P– Ag–P | 143.54(2) |
| Ag–N | 2.553(3) | O3–N | 1.264(6) | O–N–O | 122.1(6) | 120.7(5) | 117.1(6) |
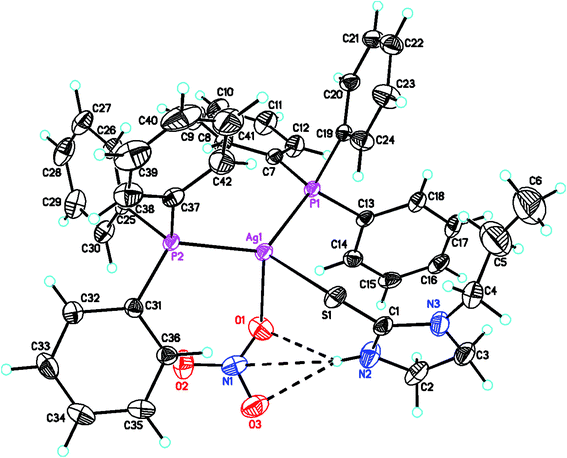
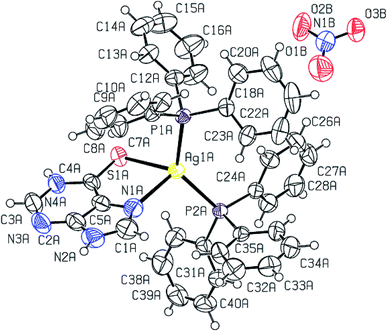
The molecular structures of complexes, [Ag(S-L-NPrn)(PPh3)2(O–NO2)] 2, and [Ag(S-L-NBun)(PPh3)2(O–NO2)] (3) are given in Fig. 1 and 2 respectively. In complex 2, silver(I) is bonded to one S donor atom of the L-NPrn thio-ligand, two P donor atoms of PPh3 ligands and one O donor atom of a nitrate ion at bond distances of 2.6161(13) (S), 2.4552(11), 2.4450(11) (P) and 2.496(4) (O) Å respectively (Table 5). The corresponding bond distances of complex 3 are 2.5340(7)(S), 2.4524(6), 2.4554(6) (P) and 2.4999(19)(O), respectively. These bond distances are less than the sum of the covalent radii of the silver atom and the donor group (Ag–S, 3.50, Ag–P, 3.55, Ag–O, 3.20 Å)61 and thus bonds in these complexes are similar to those found in literature.43 While Ag–P and Ag–O distances in two complexes are similar, the corresponding Ag–S distances are different. The n-butyl group at N atom of thio-ligand L-NBun with positive inductive effect appears to increase lewis basicity of this ligand, which makes relatively short Ag–S bond in complex 3 versus that in 2 where it is longer. The short Ag–S distance in 3 is followed by relatively longer C–S bond distance (1.695(6) Å, 2; 1.708(3) Å, 3). The differences in Ag–S bond distances in these two complexes are reflected in Ag–O distances which is marginally short in 2. The O–N bond distances of ONO2 group differ more and lie in the range, 1.175 to 1.284 Å, while those in complex 3, these distances are 1.232(3) to 1.249(3) Å – short in complex 2 {1.175(7) Å}versus that in 3 {1.232(3) Å}. The bond angles around the Ag metal center are in the ranges 97–129° (2) and 101–123° (3) respectively, suggesting distorted tetrahedral geometry of these complexes. It is found that O1–Ag–S angle is smallest, 97.59(13), while P–Ag–P angle is largest, 129.48(4). It is opposite in complex 3 and these differences are attributed to the steric effect of R groups at N atoms of L-NR moiety.
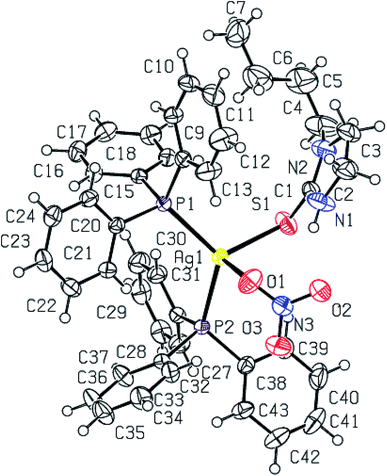
Purine-6-thione chelates to Ag(I) in complex [Ag(N,S-purSH2)(PPh3)2](NO3) 6 (Fig. 3), through its N7, S-donor atoms with Ag–N7 and Ag–S bond distances of 2.468(4) and 2.6478(10) Å respectively. The Ag–P bond distances of 2.4511(8), 2.4301(9) (P) Å are similar to those found in complex 2. The nitrate NO3− ion is not coordinated in complex 6 with O–N bond distances of 1.200(6), 1.218(6) and 1.240(5) Å. It is noted that the average O–N distance of 1.219 (6) Å in complex 6 is less than the similar average O–N distances of 1.229(7) Å in complex 2 and 1.238(3) Å in complex 3. The difference is obviously due to the coordinated nitrate in 2 and 3, and ionic in complex 6 (Fig. 3). The N7–Ag–S bite angle of 79.87(9)° is smallest, while P–Ag–P angle of 130.17(3)° is the largest among angles around the Ag atom. The geometry is severely distorted from the tetrahedron.
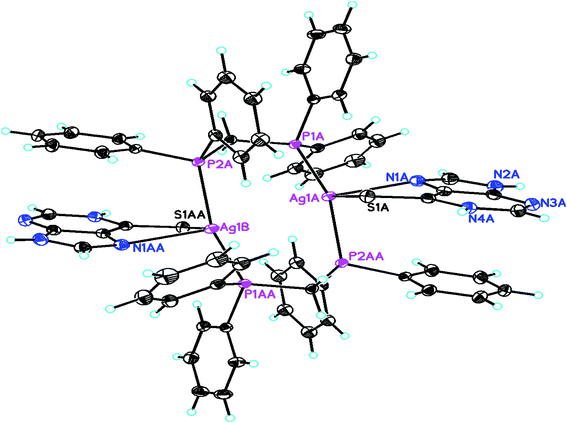
When dppm was used in place of PPh3, purine-6-thione with silver(I) nitrate has yielded a P,P-bridged dinuclear complex, [Ag2(N,S-purSH2)2(μ-P,P-dppm)2](NO3)2·2H2O 5 (Fig. 4). The thio-ligand purSH2 is chelating through its N,7 S-donor atoms with relatively longer, Ag–S and Ag–N bond distances of 2.7239(9) and 2.553(3) Å respectively as compared with that of mononuclear complex 6 (Table 4), the Ag–P distances are however comparable in complexes 5 and 6. The nitrates are lying outside the coordination sphere of silver metal ion. The angles around Ag vary from 77.27(7) to 143.54(2)° and thus geometry is severely distorted from tetrahedral geometry. The central core Ag2P4C2 of complex 5 forms an eight membered ring with Ag···Ag contact distance of 3.466 Å which is close to twice the sum of radius of silver(I) ion, viz., 3.4 Å.61
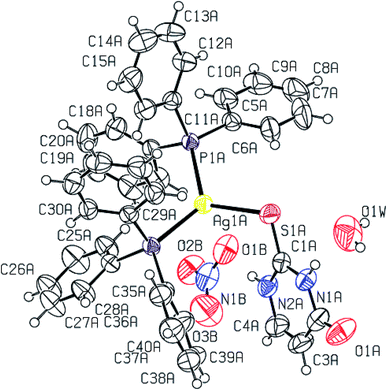
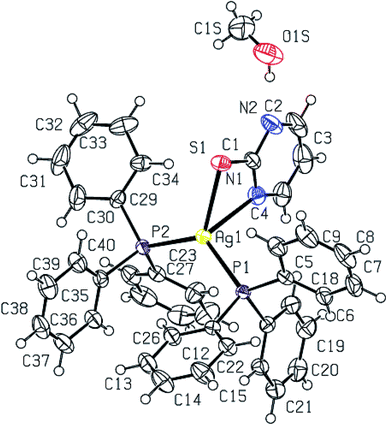
2-Thiouracil has formed a three coordinate complex, [Ag(S-tucH2)(PPh3)2](NO3)·H2O 7. Here, silver(I) is bonded to one S donor atom at Ag–S distance of 2.5464(9) Å and two P-donors at Ag–P bond distances of 2.4531(7) and 2.4787(8) Å (Fig. 5). The angles around the central silver atom in the range of 110–124° suggest distorted trigonal planar arrangement. The nitrate is lying outside the coordination sphere. The pyrimidine-2-thione ligand chelates to the silver atom as an anionic ligand through its N1, S– donor atoms in [Ag(N,S-pymS)(PPh3)2] 8 (Fig. 6). Here, the Ag–S bond distance {2.5502(6) Å} is smaller than that found in the analogous complex [Ag(S-pymSH)(PPh3)2]NO3, owing to pyrimidine-2-thione as anionic ligand in former complex versus neutral ligand in the latter complex.23 Other bond distances, namely, Ag–P and Ag–N bond distances are in same trend as found in complex 6 with Ag–N bonds. The angles around Ag vary in the range, 61–125° with N–Ag–S bite angle of 61.89(5) and P–Ag–P bond angle of 125.17(2)o suggesting distorted tetrahedral geometry of complex 8. The C–S bond lengths in complexes 2, 3, 5–8 fall in the range 1.68–1.72 Å, and are less than the C–S single bond length (1.81 Å) but longer than the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) S double bond length (1.62 Å), suggesting a partial double bond character in the C–S bonds in complexes.61
S double bond length (1.62 Å), suggesting a partial double bond character in the C–S bonds in complexes.61
Antimicrobial studies
| Complex/free thio-ligand/standard drug | MRSAe | S. aureusf | S. epidermidisg | E. faecalish | S. flexnerii | C. albicansj | |
|---|---|---|---|---|---|---|---|
| a All measurements are in mm diameter of the inhibition zone (N.A. indicates no activity).b The standard deviation varied in the range 0–1 based on three readings.c Studies were made in DMSO.d Commercially available antimicrobial agents.e MRSA.f Staphylococcus aureus.g Staphylococcus epidermidis.h Enterococcus faecalis.i Shigella flexneri.j C. albicans.k Gentamicin acts as positive control against bacteria (MRSA, S. aureus, S. epidermidis, E. faecalis, S. flexneri) and amphotericin B acts as positive control against yeast, C. albicans. | |||||||
| Imidazolidine-2-thione (1–4, 10, 11) and benzimidazoline-2-thione (11) complexes | |||||||
| [Ag(S-L-NEt)(PPh3)2(O–NO2)] 1 | 23 | 22 | 30 | 23 | 19 | 22 | |
| [Ag(S-L-NPrn)(PPh3)2(O–NO2)] 2 | 24 | 20 | 24 | 30 | 13 | 21 | |
| [Ag(S-L-NBun)(PPh3)2-(ONO2)]] 3 | 14 | 13 | 15 | 30 | 23 | 14 | |
| [Ag(S-L-NPh)(PPh3)2(O–NO2)] 4 | 22 | 19 | 20 | 19 | 24 | 18 | |
| {[Ag2(L-NH)4(PPh3)2](NO3)2 10 | 30 | 24 | 21 | 22 | 15 | 26 | |
| [Ag(S-L-NMe)2(PPh3)](NO3) 11 | 25 | 23 | 30 | 34 | 20 | 24 | |
| [Ag(S-bzimSH)2(PPh3)2] (OAc) 12 | 17 | NA | 12 | NA | NA | 13 | |
| L-NH | NA | NA | NA | NA | 15 | NA | |
| L-NMe | NA | NA | 13 | 12 | NA | NA | |
| L-NEt | NA | NA | 13 | 12 | NA | NA | |
| L-NPrn | NA | NA | 13 | 12 | NA | NA | |
| L-NBun | 12 | NA | NA | 12 | NA | NA | |
| L-NPh | NA | NA | 13 | 12 | NA | NA | |
| BzimSH | 15 | 15 | NA | NA | 16 | 13 | |
| PPh3 | NA | NA | NA | NA | NA | NA | |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||||||
| Purine-6-thione, 2-thiouracil, pyrimidine-2-thione and pyridinr-2-thione complexes (6–9) | |||||||
| [Ag2(N,S-purSH2)2(μ-dppm)2]-(NO3)2 5 | 13 | 12 | 12 | NA | NA | 17 | |
| [Ag(N,S-purSH2)(PPh3)2]-(NO3) 6 | 15 | 12 | 14 | NA | NA | 18 | |
| [Ag(S-tucH2)(PPh3)2](NO3) 7 | 20 | 14 | 14 | NA | 26 | 20 | |
| [Ag(N,S-pymS)(PPh3)2] 8 | 12 | NA | NA | NA | NA | 13 | |
| [Ag(N,S-pyS)(PPh3)2] 9 | 20 | NA | 14 | 13 | NA | 18 | |
| purSH2 | 18 | 13 | NA | NA | NA | NA | |
| tucH2 | 19 | 16 | NA | NA | NA | 12 | |
| PymSH | 19 | 17 | 17 | NA | 20 | 23 | |
| PySH | 20 | 19 | 18 | NA | 14 | 30 | |
| dppm | NA | NA | NA | NA | NA | NA | |
| Gentamicind,k | 33d | 26d | 25d | 27d | 34.5d | — | |
| Amphotericin Bd,k | — | — | — | — | — | 34d | |
| Complex/standard drug | MRSA | S. Aureus | S. epidermidis | E. faecalis | S. flexneri | C. albicans |
|---|---|---|---|---|---|---|
| a MIC in μg mL−1.b ND-not determined. | ||||||
| Imidazolidine-2-thione (1–4, 10, 11) and benzimidazoline-2-thione (11) complexes | ||||||
| 1 | 10 | 10 | 5 | 10 | 50 | 50 |
| 2 | 50 | 50 | 10 | 7 | ND | 1010 10 |
| 3 | ND | ND | 1000 | 1 | 7 | ND |
| 4 | 10 | 500 | 50 | 500 | 10 | 750 |
| 10 | 5 | 10 | 50 | 50 | 1000 | 5 |
| 11 | 7 | 10 | 7 | 1 | 50 | 10 |
| 12 | 7 | ND | ND | ND | ND | ND |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||||
| Purine-6-thione, 2-thiouracil, pyrimidine-2-thione and pyridinr-2-thione complexes (6–9) | ||||||
| 5 | ND | ND | ND | ND | ND | 50 |
| 6 | 1250 | ND | ND | ND | ND | 50 |
| 7 | 50 | ND | ND | ND | 7 | 10 |
| 8 | ND | ND | ND | ND | ND | ND |
| 9 | 50 | ND | ND | ND | ND | 50 |
| Gentamicin | 10 | 0.5 | 30 | 30 | 5 | — |
| Amphotericin B | — | — | — | — | — | 0.1 |
Complexes 1–3, 10 and 11 have shown activity against the microorganism Gram positive bacteria Enterococcus faecalis (MTCC 439) in the range 22–34 mm. Significantly, three complexes (2, 3 and 11) have more activity (zoi, 30–34 mm) with low MIC values (1–7 μg mL−1) than that of gentamicin (zoi, 27 mm, 30 μg mL−1). The antimicrobial activity against Gram negative bacteria Shigella flexneri (MTCC 1457) in the range 19–24 mm (zoi) has been shown by the complexes 1, 3, 4 and 11 with MIC values in the range 7–50 μg mL−1. This activity is lower than that shown by the standard drug (zoi, 34.5 mm, MIC, 5 μg mL−1). Complexes 1, 2, 10 and 11 are active against yeast Candida albicans (MTCC 227) (21–26 mm) with MIC values in the range 5–50 μg mL−1. This activity is lower than that of the standard drug amphotericin B (zoi, 34 mm; MIC, 0.1 μg mL−1). Only one complex showed activity of 26 mm with MIC value of 5 μg mL−1, which is relatively close to that of the reference compound. Finally, it is added here that these complexes are more active as compared with the uncoordinated thio-ligands.
Only complex 9 showed activity (zoi = 13 mm) against Gram positive bacteria Enterococcus faecalis (MTCC 439), while other complexes and free ligands were inactive. Among the free thio-ligands, only pymSH and pySH showed activity of 20 and 14 mm (zoi) against Gram negative bacteria Shigella flexneri (MTCC 1457), but as regards their complexes, only 2-thiouracil complex 7 showed activity of 26 mm (zoi) which was somewhat close to that of standard drug gentamicin (zoi = 34.5 mm) (Table 7). Additionally, their MIC values were comparable (complex 7, MIC = 7 μg mL−1; gentamicin, MIC = 7 μg mL−1). This is an interesting outcome of the study owing to the importance of tucH2 derivatives in the development of metal based drugs.62–64 Finally, as regards activity against the yeast Candida albicans, the thio-ligand purSH2 was inactive, while its complexes 5 and 6 showed moderate activity of 17 mm (5) and 18 mm (6) respectively. Further, while free tucH2 was inactive, its complex 7 was found to be active with (zoi = 20 mm). In contrast, the thio-ligands pymSH (23 mm) and pySH (30 mm) were found to be more active than their corresponding complexes 8 (13 mm) and 9 (18 mm). Further, complex 9 was more active against Candida albicans than the reported analogous complex, [Ag(S-pySH)2(PPh3)]NO3 with pyridine-2-thione (pySH) bonded as neutral ligand.30
Among these complexes, complex 7 showed MIC of 10 μg mL−1, while all other complexes showed high MIC values. Complexes 5–9 and thio-ligands showed lower activity than the standard antibiotics, gentamicin and amphotericin B. This outcome of the study is consequence of systematic scanning of complexes for their antimicrobial activity and cell viability in view of scant reports in literature as highlighted in this paper.
| Complexes | OD values | Cell viability |
|---|---|---|
| a OD value of control = 0.298.b OD value of control = 0.672. | ||
| [Ag(L-NEt)(PPh3)2(O–NO2)] 1 | 0.237a | 79.53% |
| [Ag(L-NBun)(PPh3)2(O–NO2)] 3 | 0.224a | 75.17% |
| [Ag(L-NPh)(PPh3)2(O–NO2)] 4 | 0.288a | 96.31% |
| [Ag(tucH2)(PPh3)2](NO3) 7 | 0.565b | 84% |
| [Ag(pymS)(PPh3)2] 8 | 0.666b | 99.10% |
| [Ag(L-NMe)2(PPh3)](NO3) 11 | 0.271a | 90.94% |
Antitumor activity
A status of anti-cancer/tumor activity of complexes of silver(I) with heterocyclic-2-thiones is given in introduction,17,18,36,38,39 and it was noted that the study pertained to in vitro cytotoxic activity against murine leukemia (L1210), human T-lymphocyte (Molt4/C8 and CEM) cells, breast cancer cell lines (MDA-MB-231, MCF-7), colon cancer cell line (HT-29), and leiomyosarcoma cancer cells (LMS). In this investigation, silver(I) complexes (1–7, 9–10) with the thio-ligands, namely, N-substituted-imidazolidine-2-thiones, benzimidazoline-2-thione, purine-6-thione, pyridine-2-thione and 2-thiouracil have been studied for their in vitro antitumor activity against human bone cancer cell line MG63, using MTT [3-(4,5-dimethylthiazol-2-yl)-2, 5-diphenyl tetrazolium bromide] assay. The antitumor potential of these complexes was evaluated by exposing a fixed number of cancer cells to increasing concentration of complexes. Fig. 7 and 8 depict the effect of complex concentration on the death of the cancerous cells. As an example, complex 2 at concentrations of 6.25, 12.5, 25, 50 and 100 μM, showed the percent killing of cancerous cells up to 40, 65, 75, 85 and 91% respectively. Similar effect was observed when complexes 1, 3–7, 9–10 were studied against MG63 cancerous cells. Further, the cyto-toxic activity of complexes on cancerous cell line is interpreted on the basis of their IC50 (half maximal inhibitory concentration) values (Table 8). The IC50 values of different complexes and the free thio-ligands were calculated from the curves obtained between concentration of complexes and percentage viability of cancerous cells (Fig. 9).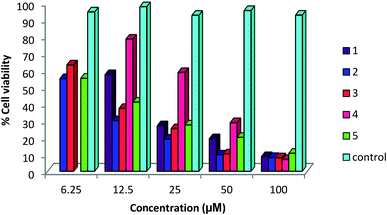 | ||
| Fig. 7 Effect of concentration of complexes 1–5 on viability of MG 63 cells; untreated cells represent control. | ||
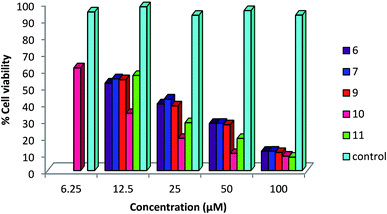 | ||
| Fig. 8 Effect of concentration of complexes 6, 7, 9–11 on viability of MG 63 cancerous cells; where untreated cells represent control. | ||
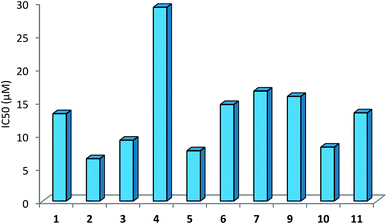
The IC50 values of complexes 1–12 ranged between 6.42-29.22 μM (Table 9). Among the complexes tested, complexes, 2, 3, 5 and 10 have shown IC50 values of 6.42 (2), 9.21 (3), 7.62 (5) and 8.17 (10) μM respectively; other complexes 1, 6, 7, 9 and 11 have IC50 values in close range, 13.22 to 16.61 μM, and finally complex 4 lowest IC50 value of 29.22 μM. The free thio-ligands, L-NR (R = H, Me, Et, Pr, Bu, and Ph), purSH2, tucH, and pymSH are either inactive or have low ant-cancer activity.
| Complexes | IC50 (μM) | IC50 (μM) | |
|---|---|---|---|
| a Ligands: L-NR, R = H, Me, Et, Prn. Bun and Ph; PPh3: IC50 (μM) is >100; purSH2, 24.78, tucH2, 44.26 and pySH, 29.66. | |||
| [Ag(L-NEt)(PPh3)2(O–NO2)] 1 | 13.20 | [Ag(N,S-purSH2)(PPh3)2](NO3) 6 | 14.58 |
| [Ag(L-NPrn)(PPh3)2(O–NO2)] 2 | 6.42 | [Ag(S-tucH2)(PPh3)2](NO3) 7 | 16.61 |
| [Ag(L-NBun)(PPh3)2(O–NO2)] 3 | 9.21 | [Ag(N,S-pyS)(PPh3)2] 9 | 15.80 |
| [Ag(L-NPh)(PPh3)2(O–NO2)] 4 | 29.22 | [Ag2(L-NH)4(PPh3)2]-(NO3)2 10 | 8.17 |
| [Ag2(N,S-purSH2)2(μ-dppm)2]-(NO3)2 5 | 7.62 | [Ag(L-NMe)2(PPh3)](NO3) 11 | 13.33 |
Conclusion
Coordination compounds of N-substituted 1,3-imidazolidine-2-thiones, purine-6-thione, 2-thiouracil, pyrimidine-2-thione and pyridine-2-thione with silver(I) nitrate have shown moderate to high antimicrobial bio-activity against methicillin resistant Staphylococcus aureus (MRSA), Staphylococcus aureus, Staphylococcus epidermidis, Enterococcus faecalis and yeast Candida albicans. Coordination compounds [Ag(S-L-NEt)(PPh3)2(O–NO2)] 1, [Ag(S-L-NPh)(PPh3)2(O–NO2)] 4, [Ag(S-tucH2)(PPh3)2](NO3) 7, [Ag(N,S-pymS)(PPh3)2] 8 and [Ag(S-L-NMe)2(PPh3)](NO3) 11 studied for their in vitro cell viability were found to be non-cytotoxic with respective cell viability of 79.53 (1), 96.31 (4), 84 (7), 99.10 (8) and 90.94% (11) respectively. Specifically complexes of N-substituted imidazoldine-2-thiones have shown unusual bio-activity against methicillin resistant Staphylococcus aureus (MRSA), Staphylococcus epidermidis and Enterococcus faecalis which rivals with gentamicin or even better in some cases. The anti-tumor study of silver complexes against human osteosarcoma cell line (MG63) has shown IC50 values in the range 6–33 μM. The activity of complexes is attributed to the enhanced lipophilic character of complexes, in comparison to silver nitrate which being soluble in water combines with chloride in body system forming insoluble AgCl, thus losing its bioactivity. However, presence of nitrate in ionic/coordinated forms in the present mixed ligand complexes of thio-ligands and co-ligands develop chemical environments which lead to their enhanced lipophilic character responsible for bioactivity. Future investigations might provide more light on mechanistic aspects. And in vivo studies are needed to explore their medicinal potential.Experimental section
Chemicals and techniques
The thio-ligands, namely, purine-6-thione (purSH2), pyrimidine-2-thione (pymSH), pyridine-2-thione (pySH), 2-thiouracil (tucH2) and benz-imidazoline-2-thione (bzimSH2) were procured from Sigma-Aldrich Ltd and used as such, whereas, other thio-ligands such as, imidazolidine-2-thione (L-NH), 1-methyl-imidazolidine-2-thione (L-NMe), 1-ethyl-imidazolidine-2-thione (L-NEt), 1-n-propyl-imidazolidine-2-thione (L-NPrn), 1-n-butyl-imidazolidine-2-thione (L-NBun), and 1-phenyl-imidazolidine-2-thione (L-NPh) were prepared as per the literature methods.65,66 Silver(I) nitrate was purchased from Sigma-Aldrich and used as received. Elemental analysis (C, H, N, S) were carried out using the Thermo Finnigan Flash technique. The melting points were determined with a Gallenkamp electrically heated apparatus. The IR spectra were recorded using KBr pellets on a Varian 660 FT IR Spectrometer in the 4000–400 cm−1 range. The 1H/13C NMR spectra were recorded in CDCl3 and DMSO using Bruker Avance II 400 NMR spectrometer at 400 MHz and Bruker Ascend 500 NMR spectrometer at 500 MHz with TMS as an internal reference. The ESI-mass spectra were recorded in DMSO or CHCl3 solvents using Bruker Daltonik LS-MS high resolution micro TOF-Q II 10356 spectrometer.Synthesis of Silver(I) complexes
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v) yielded colorless crystals of 2 (yield, 0.086 g, 70%, mp 153–156 °C). Elemental analysis for AgC42H42N3SP2O3 (838.65): calcd C 60.09; H 5.00; N 5.00; S 3.81%; found: C 60.35; H 5.25; N 5.37, S 4.12.
1, v/v) yielded colorless crystals of 2 (yield, 0.086 g, 70%, mp 153–156 °C). Elemental analysis for AgC42H42N3SP2O3 (838.65): calcd C 60.09; H 5.00; N 5.00; S 3.81%; found: C 60.35; H 5.25; N 5.37, S 4.12.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v) yielded colorless crystals of 3 (yield, 0.093 g, 75%, mp 118–120 °C). Elemental analysis for AgC43H44N3SP2O3 (852.68): calcd C 60.56; H 5.16; N 4.92; S 3.75%; found: C 60.77; H 5.56; N 5.06, S 3.67%.
1, v/v) yielded colorless crystals of 3 (yield, 0.093 g, 75%, mp 118–120 °C). Elemental analysis for AgC43H44N3SP2O3 (852.68): calcd C 60.56; H 5.16; N 4.92; S 3.75%; found: C 60.77; H 5.56; N 5.06, S 3.67%.X-ray crystallography
Molecular structures of complexes 2, 3, 5–8 which formed good quality crystals have been solved by X-ray Crystallography; the quality of other complexes was poor. The crystal of a complex was mounted on a polymer loop and data were measured on a Rigaku-Oxford Diffraction (2, 5–8), Agilent, Eos, Gemini diffractometer (3) at 173(2) (3, 8) and 293(2) (2, 5–7) K. The diffractometer was equipped with a graphite monochromator with Mo–Kα radiation (λ = 0.71073 Å; 3, 8), or Cu-Kα radiation (λ = 1.54178 Å; 2, 5–7). The data collection and cell refinement were processed with CrysAlisPro while data reduction was processed with CrysAlisRED (2, 3, 5–8).67 The structures were solved by the direct methods and refined using full-matrix least-squares techniques based on F2 using ShelXL-2008, 2015.68,69 Table 4 contains crystal data.Antimicrobial studies
Antimicrobial screening
A 100 μL of activated test organism (prepared as above) sample was inoculated onto suitable medium plates. Wells A 100 μL of activated test organism (prepared as above) sample was inoculated onto suitable medium plates by the spread plate method. Wells measuring 8 mm in diameter were cut out in the medium using a sterilized stainless steel borer. A stock solution of each test complex of concentration 50 mg mL−1 was prepared in DMSO and each well was filled with 0.1 mL of this test complex and kept for incubation in an upright position for 18–24 h. Sensitivity was measured in terms of a diameter of the zone of inhibition. Any organism with a clear zone of inhibition ≤12 mm was considered to be resistant to the compound.70Minimum inhibitory concentration (MIC)
Minimum inhibitory concentration of the selected coordination compound dissolved in DMSO was worked out by the agar dilution method for their antimicrobial activity against the sensitive microorganisms. From the stock solution, the concentration of a complex in the range (0.001–2 mg mL−1) was mixed with a Muller Hinton agar medium and then poured on plates. These plates were then inoculated with 0.1 mL of the activated bacterial and yeast strains by streaking with a sterile swab. The plates were incubated at 37 °C for bacteria and 25 °C for yeast for 24 h each. The minimum concentration of the extract causing inhibition of the visible growth was taken as MIC. The results were compared with that of DMSO as a control.MTT toxicity assay
In order to check the level of cellular toxicity of the test compounds, MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide] assay was performed71 as follows. Ten milliliter of sheep blood was taken into an injection syringe containing 3 mL of Alsever's solution (anticoagulant) and transferred to sterile centrifuge tube. The blood was centrifuged at 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm at room temperature (25 °C) for 20 min so as to separate the plasma from the cells. The supernatant was discarded and 6 mL phosphate buffer saline (PBS) was added and it was again centrifuged. The blood cells were washed thrice with PBS by centrifugation and the pellet was re-suspended in 6 mL of PBS. Various dilutions of these cells were prepared using PBS and counted with the help of a hemocytometer under a light microscope so as to obtain cells equivalent to 1 × 105 cells per mL. One hundred microliters (100 μL) of this diluted suspension was added in each well and incubated at 37 °C for overnight. The supernatant was removed carefully and 200 μL of the sample solution (contains 10 mg mL−1) was added and incubated further for 24 h. The supernatant was removed again and 20 μL MTT solution (5 mg mL−1) was added to each well and incubated further for 3.5 h at 37 °C on an orbital shaker at 60 rpm. After incubation, the supernatant was removed without disturbing the cells and 50 μL of DMSO was added to each well to dissolve the formazan crystals. The absorbance was measured at 590 nm using an automated microplate reader (Biorad 680-XR, Japan). The wells with untreated cells served as a control. Reduction of MTT can only occur in metabolically active cells, as MTT is converted into formazan crystals and hence the absorbance directly represents the viability of the cells (% viability = (optical density (OD) of treated/OD of control) × 100). If the percent viability of the blood cells is quite high, then the compounds are non-cytotoxic in nature.
000 rpm at room temperature (25 °C) for 20 min so as to separate the plasma from the cells. The supernatant was discarded and 6 mL phosphate buffer saline (PBS) was added and it was again centrifuged. The blood cells were washed thrice with PBS by centrifugation and the pellet was re-suspended in 6 mL of PBS. Various dilutions of these cells were prepared using PBS and counted with the help of a hemocytometer under a light microscope so as to obtain cells equivalent to 1 × 105 cells per mL. One hundred microliters (100 μL) of this diluted suspension was added in each well and incubated at 37 °C for overnight. The supernatant was removed carefully and 200 μL of the sample solution (contains 10 mg mL−1) was added and incubated further for 24 h. The supernatant was removed again and 20 μL MTT solution (5 mg mL−1) was added to each well and incubated further for 3.5 h at 37 °C on an orbital shaker at 60 rpm. After incubation, the supernatant was removed without disturbing the cells and 50 μL of DMSO was added to each well to dissolve the formazan crystals. The absorbance was measured at 590 nm using an automated microplate reader (Biorad 680-XR, Japan). The wells with untreated cells served as a control. Reduction of MTT can only occur in metabolically active cells, as MTT is converted into formazan crystals and hence the absorbance directly represents the viability of the cells (% viability = (optical density (OD) of treated/OD of control) × 100). If the percent viability of the blood cells is quite high, then the compounds are non-cytotoxic in nature.
Antitumor studies
To determine in vitro cytotoxicity of silver(I) complexes of heterocyclic thiones, MTT assay MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide],72 was performed on a human osteosarcoma cell line, MG63. The cell line was procured from National Cell Center, Pune, India and was grown and matured in Dulbecco's Modified Eagle Medium (DMEM; Sigma-Aldrich) supplemented with 10% fetal bovine serum (FBS; Sigma-Aldrich) in tissue culture flask in the incubator supplied with 5% CO2 at 37 °C and in 90% relative humidity provided by the growth medium. These cells were then collected in the confluent phase by treating with Trypsin-Hank's Balanced Salt Solution (HBSS). The cell suspension was centrifuged at 1500 rpm for 15 min. The supernatant was removed and 5 mL of medium (DMEM) was added to the cell pellet. The cell number was counted using hemocytometer under a light microscope and the cell density was made to 40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cell per mL (in DMEM) in the cell suspension.
000 cell per mL (in DMEM) in the cell suspension.
The 100 μL of cell suspension was added to each well of the 96-well plate and the plate was kept in the CO2 incubator with 5% CO2, at 37 °C for 24 h. The culture was further incubated for 24 h in the presence of 100 μL of each of silver(I) complex of varying concentration (100, 50, 25, 12.5 and 6.25 μM) diluted in DMEM. The supernatant was removed from each well and 100 μL of MTT (0.5 mg mL−1 in DMEM) was added to each well, and further incubated at 37 °C for 4 h. After incubation, the supernatant was removed and the dark purple colored MTT formazan crystals were formed at the bottom of each of the well. The MTT formazan crystals thus formed were resuspended in 100 μL of dimethyl sulfoxide and the 96-well plate was shaken in a Labsystem Multiskan EX ELISA reader for 3 minute to dissolve the formazan crystals. The absorbance of the purple colored solution thus formed was then measured at 570 nm. The wells with untreated cells served as control. Reduction of MTT can only occur in metabolically active cells, as MTT is converted into formazan crystals and hence the absorbance directly represents the viability of the cells (% viability = (optical density (OD) of treated/OD of control) × 100). The IC50 values (50% inhibitory concentration), which is defined as the concentration of complex that reduced the number of living cancerous cells by 50%, were calculated from three independent experiments by generating an equation of logarithmic trend line of percentage cell viability against concentration of compounds in Microsoft excel.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
Financial assistance from the Council of Scientific and Industrial Research (CSIR), New Delhi for the Emeritus Scientist Grant [21(0904)/12-EMR-II] to T. S. L. is gratefully acknowledged. J. P. J. acknowledges the NSF-MRI program (Grant No. CHE-1039027) for funds to purchase the X-ray diffractometer.References
- S. Eckhardt, P. S. Brunetto, J. Gagnon, M. Priebe, B. Giese and K. M. Fromm, Chem. Rev., 2013, 113, 4708–4754 CrossRef CAS PubMed.
- K. M. Fromm, Nat. Chem., 2011, 3, 178 CrossRef CAS PubMed.
- P. S. Brunetto and K. M. Fromm, Chimia, 2008, 62, 249–252 CrossRef CAS.
- G. McDonnell and A. D. Russell, Clin. Microbiol. Rev., 1999, 12, 147–179 CrossRef CAS PubMed.
- C. L. Fox and S. M. Modak, Antimicrob. Agents Chemother., 1974, 5(6), 582–588 CrossRef CAS PubMed.
- S. Bassetti, J. Hu, R. B. D'Agostino Jr and R. J. Sherertz, Antimicrob. Agents Chemother., 2001, 45, 1535–1538 CrossRef CAS PubMed.
- S. Shukla and A. P. Mishra, J. Chem., 2013, 1–6 Search PubMed.
- A. T. Wan, R. A. J. Conyers, C. J Coombs and J. P. Masterton, Clin. Chem., 1991, 37, 1683–1687 CAS.
- T. V. Slenters, I. Hauser-Gerspach, A. U. Daniels and K. M. Fromm, J. Mater. Chem., 2008, 18, 5359–5362 RSC.
- P. G. Blower and J. R. Dilworth, Coord. Chem. Rev., 1987, 76, 121–185 CrossRef CAS.
- E. S. Raper, Coord. Chem. Rev., 1994, 129, 91–156 CrossRef CAS.
- E. S. Raper, Coord. Chem. Rev., 1996, 153, 199–255 CrossRef CAS.
- A. Sola, R. Lopez, A. Coxall and W. Clegg, Eur. J. Inorg. Chem., 2004, 4871–4881 CrossRef.
- T. S. Lobana, R. Sharma and R. J. Butcher, Polyhedron, 2008, 27, 1375–1380 CrossRef CAS.
- G. A. Bowmaker, N. Chaichit, C. Pakawatchai, B. W. Skelton and A. H. White, Can. J. Chem., 2009, 87, 161–170 CrossRef CAS.
- A. Cingolani, F. Marchetti, C. Pettinari, R. Pettinari, B. W. Skelton and A. H. White, Inorg. Chem., 2002, 41, 1151–1161 CrossRef CAS PubMed.
- P. C. Zachariadis, S. K. Hadjikakou, N. Hadjiliadis, S. Skoulika, A. Michaelides, J. Balzarini and E. D. Clercq, Eur. J. Inorg. Chem., 2004, 1420–1426 CrossRef CAS.
- L. Kyros, N. Kourkoumelis, M. Kubicki, L. Male, M. B. Hursthouse, I. I. Verginadis, E. Gouma, S. Karkabounas, K. Charalabopoulos and S. K. Hadjikakou, Bioinorg. Chem. Appl., 2010, 2010, 1–12 CrossRef PubMed.
- J. S. Casas, E. G. Martinez, A. Sanchez, A. S. Gonzalez, J. Sordo, U. Casellato and R. Graziani, Inorg. Chim. Acta, 1996, 241, 117–123 CrossRef CAS.
- P. Aslanidis, A. G. Hatzidimitriou, E. G. Andreadou, A. A. Pantazaki and N. Voulgarakis, Mater. Sci. Eng., C, 2015, 50, 187–193 CrossRef CAS PubMed.
- P. Aslanidis, P. Karagiannidis, P. D. Akrivos, B. Krebs and M. Lage, Inorg. Chim. Acta, 1997, 254, 277–284 CrossRef CAS.
- W. McFarlane, P. D. Akrivos, P. Aslanidis, P. Karagiannidis, C. Hatzisymeon, M. Numan and S. Kokkou, Inorg. Chim. Acta, 1998, 281, 121–125 CrossRef CAS.
- P. A. Papanikolaou, A. G. Papadopoulos, E. G. Andreadou, A. Hatzidimitriou, P. J. Cox, A. A. Pantazakic and P. Aslanidis, New J. Chem., 2015, 39, 4830–4844 RSC.
- G. A. Bowmaker, C. Pakawatchai, B. W. Skelton, Y. Thanyasirikul and A. H. White, Z. Anorg. Allg. Chem., 2008, 634, 2583–2588 CrossRef CAS.
- G. Giusti, G. Geier, A. Currao and R. Nesper, Acta Crystallogr., Sect. C: Cryst. Struct. Commun., 1996, 52, 1914–1917 CrossRef.
- G. Christofidis, P. J. Cox and P. Aslanidis, Polyhedron, 2012, 31, 502–505 CrossRef CAS.
- F. Jian, P. Sun, H. Xiao and K. Jiao, Chin. J. Inorg. Chem., 2003, 9, 979–982 Search PubMed.
- R. Sultana, T. S. Lobana, R. Sharma, A. castineiras, T. Akitsu, K. Yahagi and Y. Aritake, Inorg. Chim. Acta, 2010, 363, 3432–3441 CrossRef CAS.
- P. Aslanidis, P. J. Cox, S. Divanidis and P. Karagiannidis, Inorg. Chim. Acta, 2004, 357, 2677–2686 CrossRef CAS.
- S. Nawaz, A. A. Isab, K. Merz, V. Vasylyeva, N. Metzler-Nolte, M. Saleem and S. Ahmad, Polyhedron, 2011, 30, 1502–1506 CrossRef CAS.
- P. Aslanidis, S. Divanidis, P. J. Cox and P. Karagiannidis, Polyhedron, 2005, 24, 853–863 CrossRef CAS.
- P. A. Perez-Jourido, J. A. Garcia-Vazquez, J. Romero, M. S. Louro, A. Sousa, Q. Chen, Y. Chang and J. Zubieta, J. Chem. Soc., Dalton Trans., 1996, 2047–2054 RSC.
- W. Su, M. Hong, J. Weng, Y. Liang, Y. Zhao, R. Cao, Z. Zhou and A. S. C. Chan, Inorg. Chim. Acta, 2002, 331, 8–15 CrossRef CAS.
- W. Su, R. Cao, M. Hong, W. T. Wong and J. Lu, Inorg. Chem. Commun., 1999, 2, 241–243 CrossRef CAS.
- S. I. Mostafa and N. Hadjiliadis, Transition Met. Chem., 2008, 33, 529–534 CrossRef CAS.
- S. K. Hadjikakou, I. I. Ozturk, M. N. Xanthopoulou, P. C. Zachariadis, S. Zartilas, S. Karkabounas and N. Hadjiliadis, J. Inorg. Biochem., 2008, 102, 1007–1015 CrossRef CAS PubMed.
- K. Nomiya, S. Takahashi and R. Noguchi, J. Chem. Soc., Dalton Trans., 2000, 2091–2097 RSC.
- S. A. Elsayed, B. J. Jean-Claude, I. S. Butler and S. I. Mostafa, J. Mol. Struct., 2012, 1028, 208–214 CrossRef CAS.
- N. R. F. M. Sofyan, F. J. Nordin, M. R. M. A. Razak, S. N. A. Halim, N. A. F. M. Khir, A. Muhammad, N. F. Rajab and R. Sarip, J. Chem., 2018, 2018, 1–10 CrossRef.
- N. O. Al-Zamil, K. A. Al-Sadhan, A. A. Isab, M. I. M. Wazeer and A. R. A. Al-Arfaj, Spectroscopy, 2007, 21, 61–67 CrossRef CAS.
- L. Kyros, C. N. Banti, N. Kourkoumelis, M. Kubicki, I. Sainis and S. K. Hadjikakou, J. Biol. Inorg. Chem., 2014, 19, 449–464 CrossRef CAS PubMed.
- I. Sainis, C. N. Banti, A. M. Owczarzak, L. Kyros, N. Kourkoumelis, M. Kubicki and S. K. Hadjikakou, J. Inorg. Biochem., 2016, 160, 114–124 CrossRef CAS PubMed.
- T. S. Lobana, R. Sultana, R. J. Butcher, J. P. Jasinski and T. Akitsu, Z. Anorg. Allg. Chem., 2014, 640, 1688–1695 CrossRef CAS.
- J. K. Aulakh, T. S. Lobana, H. Sood, D. S. Arora, V. A. Smolinski, C. E. Duff and J. P. Jasinski, J. Inorg. Biochem., 2018, 178, 18–31 CrossRef CAS PubMed.
- T. S. Lobana, R. Sultana and R. J. Butcher, Dalton Trans., 2011, 40, 11382–11384 RSC.
- T. S. Lobana, R. Sultana, G. Hundal and R. J. Butcher, Dalton Trans., 2010, 39, 7870–7872 RSC.
- T. S. Lobana, R. Sultana, R. J. Butcher, A. Castineiras, T. Akitsu, F. J. Fernandez and M. C. Vega, Eur. J. Inorg. Chem., 2013, 5161–5170 CrossRef CAS.
- T. S Lobana and R. Sultana, J. Chem. Sci., 2012, 124, 1261–1268 CrossRef.
- R. Sultana, T. S. Lobana and A. Castineiras, RSC Adv., 2015, 5, 100579–100588 RSC.
- T. S. Lobana, R. Sultana, A. Castineiras and R. J. Butcher, Inorg. Chim. Acta, 2009, 362, 5265–5270 CrossRef CAS.
- T. S. Lobana, R. Sultana, R. J. Butcher, J. P. Jasinski, J. A. Golen, A. Castineiras, K. Pröpper, F. J. Fernandez and M. C. Vega, J. Organomet. Chem., 2013, 745–746, 460–469 CrossRef CAS.
- T. S. Lobana, R. Sultana and G. Hundal, Polyhedron, 2008, 27, 1008–1016 CrossRef CAS.
- T. S. lobana, R. Sultana, G. Hundal and A. Castineiras, Polyhedron, 2009, 28, 1573–1577 CrossRef CAS.
- T. S. Lobana, R. Sharma, R. Sharma and R. J. Butcher, Z. Anorg. Allg. Chem., 2008, 634, 1785–1790 CrossRef CAS.
- T. S. Lobana, R. Sharma, E. Bermejo and A. Castineiras, Inorg. Chem., 2003, 42, 7728–7730 CrossRef CAS PubMed.
- T. S. Lobana, R. Sharma, G. Hundal and R. J. Butcher, Inorg. Chem., 2006, 45, 9402–9409 CrossRef CAS PubMed.
- T. S. Lobana, R. Sharma, R. Sharma, S. Mehra, A. Castineiras and P. Turner, Inorg. Chem., 2005, 44, 1914–1921 CrossRef CAS PubMed.
- J. K. Aulakh, T. S. Lobana, H. Sood, D. S. Arora, I. Garcia-Santos, G. Hundal, M. Kaur, V. A. Smolenski and J. P. Jasinski, Dalton Trans., 2017, 46, 1324–1339 RSC.
- T. S. Lobana, J. K. Aulakh, H. Sood, D. S. Arora, I. Garcia-Santos, M. Kaur, C. E. Duff and J. P. Jasinski, New J. Chem., 2018, 42, 9886–9900 RSC.
- B. Singh and K. P. Thakur, J. Inorg. Nucl. Chem., 1974, 36, 1735–1737 CrossRef CAS.
- J. E. Huheey, E. A. Keiter and R. L. Keiter, Inorganic Chemistry: Principles of Structure and Reactivity, Harper Collins, New York, 4th edn, 1993 Search PubMed.
- G. W. Anderson, I. F. Halverstadt, W. H. Miller and R. O. Roblin Jr, J. Am. Chem. Soc., 1945, 67, 2197–2200 CrossRef CAS PubMed.
- A. F. Eweas, Q. A. Abdallah and E. I. Hassan, J. Appl. Pharm. Sci., 2014, 4, 102–111 Search PubMed.
- C. S. W. Harker, E. R. T. Tiekink and M. W. Whitehouse, Inorg. Chim. Acta, 1991, 181, 23–30 CrossRef CAS.
- J. A. Garcia-Vazquez, A. Sousa-Pedrares, M. Carabel, J. Romero and A. Sousa, Polyhedron, 2005, 24, 2043–2054 CrossRef CAS.
- W. Walia, S. Kaur, J. Kaur, A. K. Sandhu, T. S. Lobana, G. Hundal and J. P. Jasinski, Z. Anorg. Allg. Chem., 2015, 641, 1728–1736 CrossRef.
- Oxford Diffraction, CrysAlisPro CCD and CrysAlisPro, RED Oxford Diffraction Ltd., Abingdon, UK, 2010 Search PubMed.
- G. M. Sheldrick, Acta Crystallogr., Sect. A: Found. Crystallogr., 2008, 64, 112–122 CrossRef CAS PubMed.
- G. M. Sheldrick, Acta Crystallogr., Sect. C: Cryst. Struct. Commun., 2015, 71, 3–8 CrossRef PubMed.
- D. S. Arora and J. Kaur, Int. J. Antimicrob. Agents, 1999, 12, 257–262 CrossRef CAS PubMed.
- J. G. Onsare and D. S. Arora, J. Appl. Microbiol., 2014, 118, 313–325 CrossRef PubMed.
- T. Mosmann, J. Immunol. Methods, 1983, 65, 55–63 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Detailed IR spectra, 1H/13C-NMR spectra, and ESI-MS isotopic patterns. CCDC 1878467, 1878471, 1878472, 1878468, 1878469 and 1878470. For ESI and crystallographic data in CIF or other electronic format see DOI: 10.1039/c9ra01804b |
| This journal is © The Royal Society of Chemistry 2019 |