Open Access Article
Open Access ArticleFormation of multicomponent 2D assemblies of C2v-symmetric terphenyl tetracarboxylic acid at the solid/liquid interface: recognition, selection, and transformation†
Jie Wangab,
Li-Mei Wangb,
Cheng Lub,
Hui-Juan Yan *b,
Shao-Xu Wang*a and
Dong Wang
*b,
Shao-Xu Wang*a and
Dong Wang b
b
aCollege of Environmental and Chemical Engineering, Dalian Jiaotong University, Dalian 116028, P. R. China. E-mail: kdwangsx@126.com
bCAS Key Laboratory of Molecular Nanostructure and Nanotechnology, CAS Research and Education Center for Excellence in Molecular Sciences, Institute of Chemistry, Chinese Academy of Sciences (CAS), Beijing 100190, People's Republic of China. E-mail: yanhj@iccas.ac.cn; Fax: +86 10 82616935
First published on 15th April 2019
Abstract
We report on the two-dimensional self-assembly of C2v-symmetric [1,1′:3′,1′′-terphenyl]-3,3′′,5,5′′-tetracarboxylic acid (TPTA) at the solid/liquid interface by using scanning tunneling microscopy (STM). Two kinds of different self-assembly structure, i.e. a close-packed and porous rosette structure, are formed by TPTA molecules through intermolecular hydrogen bonds. When adding coronene (COR) as a guest into the TPTA assembly, structural transformation from a densely packed row structure to a rosette network structure is observed. It was found that two kinds of cavities with different sizes in the rosette network structure can be used to realize the selective co-adsorption of guest molecules with appropriate shape and size. Three-component 2D host–guest structures were successfully constructed by using 1,2,3,4,5,6-hexakis(4-bromophenyl)benzene (HBPBE) and copper phthalocyanine (CuPc) as guest molecules.
1. Introduction
Two-dimensional supramolecular self-assembly has received a great deal of attention owing to its potential applications in nano-patterning and host–guest structures on surfaces at the molecular level over recent years.1–6 The hydrogen bonding between carboxylic acid groups is the most extensively studied assembly motif for construction of supramolecular assemblies.7–9 Compared with other supramolecular interaction, hydrogen bonding exhibits both high selectivity and directionality, and is suitable to create novel nano-patterning structures with high predictivity.10–12 So far, a variety of molecules with a rigid skeleton and carrying symmetrically distributed carboxyl groups have been designed as model systems to study their assembly architectures.7,13,14 For example, trimesic acid (TMA) can form a series of different molecular networks based on a dimeric motif and trimeric motif between carboxylic acid groups.15–18 Similarly, the hexagonal network connected through hydrogen bonding can be formed by using other C3-symmetric monomers, such as 1,3,5-tris(4-carboxyphenyl)benzene (BTB)19–22 and D3h-symmetric hexa-peri-hexabenzocoronene substituted by three carboxylic acid group (HBC–COOH) molecules.23 Rosei groups reported that the 3,4′,5-biphenyltricarboxylic acid (H3BHTC), which is a symmetry-reduced tricarboxylic acid, can form an offset zigzag chain structure.24 Wuest groups illustrated that a D2h-symmetric tetracarboxylic acid can self-assemble into a Kagome network.25 The change of the symmetry of carboxyl groups in molecules can form a rich library of nanopatterned molecular nanostructures.The nanoporous networks based on hydrogen bonding between carboxyl groups provide an important platform to study surface host–guest chemistry.5,15,26 The incorporation of guest molecules into the nanoporous network on the basis of shape matching has been widely exploited to obtain multi-component pattern at the solid/liquid interface.27–31 On the other hand, the structure transformation between different assemblies, or so-call adaptive assembly, has also been observed. For example, the isophthalic acid can form assembled structure stabilized by dimer hydrogen bonds.32–34 Furthermore, the isophthalic acid can form cyclic trimer motif when accommodating coronene as guest molecule.33 The co-assembly of guest molecules inducing the reorganization of host assemblies has been reported in few systems.24,27,35,36
Herein, we explore the assembly of [1,1′:3′,1′′-terphenyl]-3,3′′,5,5′′-tetracarboxylic acid (TPTA) which consists of a central benzene ring connected to two isophthalic acid units with an angle of 120°, on graphite surface. So far, there is only few reports about the assembly of C2v-symmetric molecule.37 Scheme 1 shows the chemical structure of TPTA. To the best of our knowledge, that is the first study on the self-assembly structure of V-shaped skeleton with benzoic carboxylic end groups. The assembly of TPTA on graphite surface results in two kinds of different self-assembly structures at the solid/liquid interface. As shown in Scheme 1a and b, TPTA form dimeric building blocks via intermolecular hydrogen bonding, which then form a pseudo close-packed structure (pattern A) and a porous rosette structure (pattern B), which features a rare double pore structure. We demonstrate that the assembly of TPTA exhibit both adaptive assembly and host–guest assembly in response to different guest molecules. The introduction of guest molecule coronene (COR) favours the cyclic building motif, as a result, the adaptive structural transformation of pattern A into pattern B is observed. Furthermore, the larger size guest molecules such as copper phthlocyanine (CuPc) and 1,2,3,4,5,6-hexakis(4-bromophenyl)benzene (HBPBE) can be trapped in to the larger size pores to form three-component supramolecular structures, as shown in Scheme 1d and e.
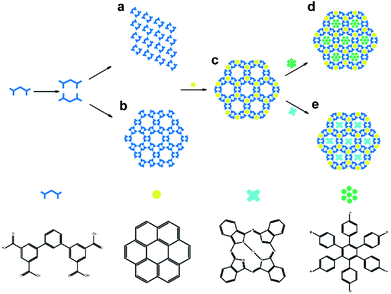 | ||
| Scheme 1 Schematic illustration of the formation of two different self-assembly structures of TPTA molecule and the co-adsorption of guest molecules. | ||
2. Experimental section
2.1. Materials
[1,1′:3′,1′′-Terphenyl]-3,3′′,5,5′′-tetracarboxylic acid (TPTA) and 1,2,3,4,5,6-hexakis(4-bromophenyl)benzene (HBPBE) were purchased from Extension Scientific. Coronene (COR), copper phthalocyanine (CuPc), and n-octanoic acid were purchased from J&K Scientific. All these materials were used without further purification. The STM measurements were performed on a Nanoscope IIIA (Bruker, USA). All the images were recorded using the constant current mode and are shown without further processing. As STM probes, mechanically cut Pt/Ir tips (90/10) were used. The assemblies were prepared by subsequent deposition of the components onto a freshly cleaned HOPG (quality ZYB, Digital Instruments, Santa Barbara, CA) surface.2.2. Method
The saturated solution of chemicals in n-octanoic acid was prepared, and the concentrations of all the solutions were less than 1 × 10−5 mol L−1. Typically, a droplet (2 μL) of TPTA molecules saturated solution was deposited on the freshly cleaved surface of highly oriented pyrolytic graphite (HOPG) to prepare the assembled monolayer structure. The host–guest co-adsorption assemblies were prepared by using the mixture solution of TPTA and appropriate guest molecules (containing compound: COR, HBPBE, CuPc) by volume ratio 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. All STM observations of the monolayers were performed at the interface between n-octanoic and graphite at room temperature.
1. All STM observations of the monolayers were performed at the interface between n-octanoic and graphite at room temperature.
3. Results and discussion
3.1. The assembly of TPTA on HOPG
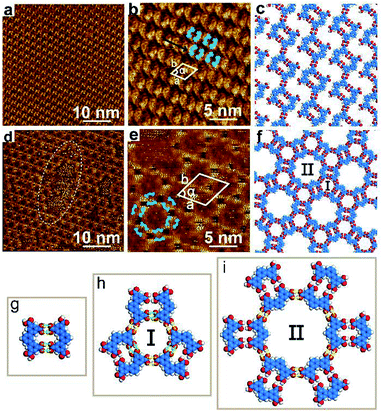
After depositing a droplet of TPTA solution on HOPG, we observed two different structure domains at the interface between n-octanoic acid and graphite at room temperature, as illustrated in Fig. 1a and d. It can be seen from the large scale STM image in Fig. 1a that TPTA molecules self-assemble into densely packed row structure. The high-resolution STM image in Fig. 1b demonstrated that along the densely-packed molecular rows as indicated by black arrow, TPTA molecules show bend contours, which are very consistent with the V-shape chemical structures of TPTA molecule as marked by blue line on STM image.A close observation (Fig. 1b) revealed that the neighboured TPTA molecules are arranged with face-to-face or back-to-back orientation along the densely-packed molecular rows. Considering the typical hydrogen bonding interactions between carboxylic acid groups, TPTA molecules of V-shaped skeleton with carboxylic end groups easily form dimers which are stabilized by four hydrogen bonds between two neighbouring molecules with face-to-face orientation (marked as the green bright line in Fig. 1g). It can be seen from Fig. 1g that the TPTA dimers are composed of two molecules face-to-face rotated by 180° with respect to each other. The V-shaped skeleton of TPTA makes two carboxylic groups in perfect parallel configuration and favours the formation of face-to-face dimers. Therefore, the molecular V-shape skeleton is a prerequisite for the formation of face-to-face dimer. Variable-temperature 1H NMR spectroscopic can demonstrate the existence of intermolecular hydrogen bonding (Fig. S5†). The high-resolution STM image in Fig. 1b can be deduced that the face-to-face TPTA dimers, as building block, are aligned along the densely packed molecular rows and form a well-ordered densely-packed structure on HOPG surface. The close-packed structures correspond to pattern A mentioned in Scheme 1a. The unit cell parameters of pattern A are measured to be a = b = 2.2 ± 0.1 nm, α = 60° as shown by white parallelogram in Fig. 1b. A molecular model on the basis of STM observations for the pattern A is proposed in Fig. 1c. It can be seen from Fig. 1c that each dimer forms a rectangular cavity. The neighbouring molecular rows are connected through intermolecular hydrogen bonding between the neighboured face-to-face dimers building blocks. We note that not all carboxylic group are saturated via hydrogen bonding in this structure.
The large scale STM image of Fig. 1d shows another periodically arranged rosette structure formed by TPTA molecules, which correspond to the pattern B structure mentioned above in Scheme 1b. Compared with pattern A, the coverage of pattern B is less than 10%. Defects are locally observed in the rosette structure (highlighted by the white dotted oval curve in Fig. 1d). It is inferred that the compact structures of pattern A are more stable than pattern B at the n-octanoic acid and graphite interface.
Fig. 1e shows a high-resolution STM image of pattern B. Pattern B show a six-fold symmetry and features a triangle shaped cavity and a hexagon cavity. The small cavities, named as I-type, are measured with a diameter of 1.2 nm. The comparatively large cavities, named as II-type. The inner diameter of the II-type cavity is measured to be about 2.2 nm. The unit cell parameters of pattern B are measured to be a = b = 4.6 ± 0.1 nm, α = 60°, as shown in Fig. 1e. The Fig. 1f is the corresponding molecular model on the basis of STM observations. It can be seen from the model in Fig. 1f that both of the two kinds of cavities are composed of hexamers. Comparatively small I-type cavity is a cyclic hexamer which is composed of three-pair building blocks of face-to-face dimers connected by side-to-side. Three-pair hydrogen bonds are provided by the interactions between carboxylic acid groups within the face-to-face dimers (marked as the green bright line in Fig. 1h), and the other three-pair hydrogen bonds are provided by adjacent dimers (marked as the yellow bright line in Fig. 1h). All interactions between hydrogen bonds are equal. Similarly, large II-type cavity is also a cyclic hexamer which is composed of six-pair building blocks of the face-to-face dimers connected through six-pair hydrogen bonds. All the six-pair hydrogen bonds are provided by adjacent face-to-face dimers (marked as the yellow bright line in Fig. 1i). There are six I-type cavities at the edges of the II-type cavity. Every I-type cavity and the neighboring II-type cavity share a pair of hydrogen bonds.
3.2. Bicomponent supramolecular assembly: TPTA–COR
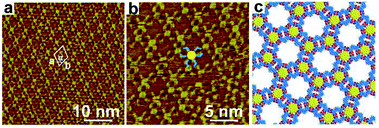
When a droplet of n-octanoic acid solution containing TPTA and COR were added to the HOPG, we observed that the assembled structure of TPTA–COR covers the entire graphite surface, as shown in the large-scale STM image of Fig. 2a. The detailed information of the TPTA–COR host–guest structure can be seen from the high-resolution STM image Fig. 2b. The big hollow hexagonal structures and the shape of TPTA molecules are well-resolved in the STM images (marked as a blue line with 120° in Fig. 2b). The COR molecules (marked as a yellow bright point in Fig. 2b) are immobilized in I-type cavities of the pattern B, because the size and shape of the COR molecule fit the I-type cavities formed by the small cyclic hexamer which are composed of three-pair building block of face-to-face dimer. Interestingly, there is no COR molecules trapped by II-type cavities of the rosette structure. Furthermore, even when an excess of COR was added into the assembly structure of TPTA, we can only observe the COR molecules immobilized at I-type cavity. Based on previous studies, the highly symmetric structure of COR and strict size match render it a suitable size and shape to fit the hexagon pores formed by isophthalic acid.33 However, the size and shape of COR molecule is not matching with the hexagon II-type cavity. Hence, it was thought that the TPTA networks structure could be considered as a site-selective template of COR.In addition, the domain of the pattern A disappeared with the addition of the guest COR molecules into this system. The introducing of COR induces the transformation of host structure from densely packed row structure to rosette network structure. In another word, the introduction of guest molecule enhances the stability of pattern B. The unit cell parameters are measured a = b = 4.6 ± 0.1 nm, α = 60° as shown in Fig. 2a.
3.3. Three component supramolecular assembly
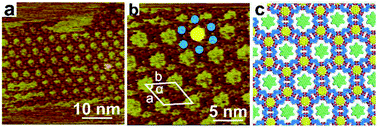
Bicomponent supramolecular assembly structure of TPTA–COR system could serve as a two-dimensional template and offers open cavity II for the different guest molecules to form three-component self-assemblies structures. The rosette network structure conducts size selectivity, that is, guest molecules smaller than the cavity can be accommodated, whereas the bigger ones are excluded. Hence, we chose HBPBE and CuPc molecules with appropriate size as guest to fit the cavities of the bicomponent assembly structure of TPTA–COR system.When depositing a droplet of n-octanoic acid solution containing TPTA, COR and HBPBE to the freshly cleaved surface of HOPG, the assembled structure of TPTA–COR–HBPBE at the interface was formed, and the obtained large-scale STM image is shown in Fig. 3a. The large scale STM image reveal that a highly ordered three component network was formed by two kinds of guest molecules in the periodically porous network. The high-resolution STM image Fig. 3b presents that the two kinds of guest molecules are selectively co-adsorbed to form the highly ordered three component network. The HBPBE molecules can be immobilized in II-type cavities but not in I-type cavities. Big circular bright spot corresponded to single HBPBE molecule (marked as a yellow circular in Fig. 3b), and small circular dim spot corresponded to single COR molecule (marked as a blue circular in Fig. 3b). Hexagonal cavity was constructed by six small circular dim spots and the center is occupied by a big circular bright spot. TPTA molecule is not clearly resolve in the high-resolution STM, presumably due to the high electronic density in COR and HBPBE molecules. Fig. 3c suggests molecular model for the TPTA–COR–HBPBE host–guest structure. The unit cell parameters are measured a = b = 4.7 ± 0.1 nm, α = 60° as shown in Fig. 3b.
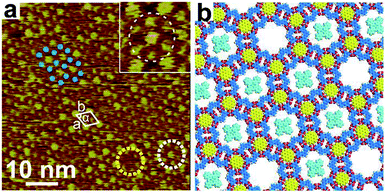
When depositing a droplet of n-octanoic acid solution containing TPTA, COR and CuPc to HOPG, the assembled structure of TPTA–COR–CuPc was formed at the interface, as shown in the STM image of Fig. 4a. It was found that CuPc molecules can be immobilized in II-type cavities by randomly, as illustrated by the small yellow and white dotted line. Small circular bright spots are ascribed to COR molecule, as marked by a blue circular in Fig. 4a. And big circular bright spots are ascribed to CuPc molecule. The CuPc molecules are surrounded by six small circular bright spots, as illustrated by the small yellow and white dotted line. An inset image clearly shows the detailed co-assembled structure in Fig. 4a. Fig. 4b suggests molecular model for the TPTA–COR–CuPc host–guest. The unit cell parameters are measured a = b = 4.7 ± 0.1 nm, α = 60° as shown in Fig. 4a.
4. Conclusions
In summary, we have observed 2D assemblies of a V-shape TPTA molecule at the n-octanoic acid and graphite interface. TPTA form two kinds of different self-assembly structure by hydrogen bonds. One is close-packed (pattern A) and the other one is porous rosette structure (pattern B). We observe both adaptive assembly and host–guest assembly when introducing different guest molecules to co-assemble with TPTA. Be specific, introducing the COR molecule into assembly system induce a structural transformation from densely packed row structure to rosette network structure. By using HBPBE and CuPc molecules with appropriate shape and size, the three-component 2D host–guest structure was constructed with highly selectively molecular recognition on the HOPG surface. The present work demonstrates a novel system showing two-types of nanopores which show different surface supramolecular chemistry in response to different guest molecules.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors gratefully acknowledge the Natural Science Foundation of Liaoning Province under the Grant No. 20180550736, Research Project of Teaching Reform of Graduate Education in Liaoning Province (2017) and the National Natural Science Foundation of China under the Grant No. 21776027, 21473198, 21573252, 21725306 for financial support to this work.Notes and references
- J. V. Barth, G. Costantini and K. Kern, Nature, 2005, 437, 671–679 CrossRef CAS PubMed.
- L. Cheng, X. Peng, S. Zhang, K. Deng, L. Shu, J. Wan and Q. Zeng, Appl. Surf. Sci., 2018, 462, 1036–1043 CrossRef CAS.
- J. A. A. W. Elemans and S. De Feyter, Soft Matter, 2009, 5, 721–735 RSC.
- K. Iritani, K. Tahara, S. De Feyter and Y. Tobe, Langmuir, 2017, 33, 4601–4618 CrossRef CAS PubMed.
- S. Stepanow, M. Lingenfelder, A. Dmitriev, H. Spillmann, E. Delvigne, N. Lin, X. Deng, C. Cai, J. V. Barth and K. Kern, Nat. Mater., 2004, 3, 229 CrossRef CAS PubMed.
- J. A. Theobald, N. S. Oxtoby, M. A. Phillips, N. R. Champness and P. H. Beton, Nature, 2003, 424, 1029–1031 CrossRef CAS PubMed.
- X.-H. Kong, K. Deng, Y.-L. Yang, Q.-D. Zeng and C. Wang, J. Phys. Chem. C, 2007, 111, 17382–17387 CrossRef CAS.
- M. Mura, F. Silly, G. A. D. Briggs, M. R. Castell and L. N. Kantorovich, J. Phys. Chem. C, 2009, 113, 21840–21848 CrossRef CAS.
- G. Velpula, M. Li, Y. Hu, Y. Zagranyarski, W. Pisula, K. Muellen, K. S. Mali and S. De Feyter, Chem.–Eur. J., 2018, 24, 12071–12077 CrossRef CAS PubMed.
- K. S. Mali, J. Adisoejoso, E. Ghijsens, I. De Cat and S. De Feyter, Acc. Chem. Res., 2012, 45, 1309–1320 CrossRef CAS PubMed.
- W. Mamdouh, M. Dong, S. Xu, E. Rauls and F. Besenbacher, J. Am. Chem. Soc., 2006, 128, 13305–13311 CrossRef CAS PubMed.
- T. Rakickas, M. Gavutis, A. Reichel, J. Piehler, B. Liedberg and R. Valiokas, Nano Lett., 2008, 8, 3369–3375 CrossRef CAS PubMed.
- M. Lackinger and W. M. Heckl, Langmuir, 2009, 25, 11307–11321 CrossRef CAS PubMed.
- F. De Marchi, D. L. Cui, J. Lipton-Duffin, C. Santato, J. M. MacLeod and F. Rosei, J. Chem. Phys., 2015, 142, 8 CrossRef PubMed.
- S. Griessl, M. Lackinger, M. Edelwirth, M. Hietschold and W. M. Heckl, Single Mol., 2002, 3, 25–31 CrossRef CAS.
- S. J. H. Griessl, M. Lackinger, F. Jamitzky, T. Markert, M. Hietschold and W. M. Heckl, J. Phys. Chem. B, 2004, 108, 11556–11560 CrossRef CAS.
- Y. Ishikawa, A. Ohira, M. Sakata, C. Hirayama and M. Kunitake, Chem. Commun., 2002, 2652–2653 RSC.
- Y. Ye, W. Sun, Y. Wang, X. Shao, X. Xu, F. Cheng, J. Li and K. Wu, J. Phys. Chem. C, 2007, 111, 10138–10141 CrossRef CAS.
- F. Cometto, K. Frank, B. Stel, N. Arisnabarreta, K. Kern and M. Lingenfelder, Chem. Commun., 2017, 53, 11430–11432 RSC.
- L. Kampschulte, M. Lackinger, A.-K. Maier, R. S. K. Kishore, S. Griessl, M. Schmittel and W. M. Heckl, J. Phys. Chem. B, 2006, 110, 10829–10836 CrossRef CAS PubMed.
- Z. Ma, Y.-Y. Wang, P. Wang, W. Huang, Y.-B. Li, S.-B. Lei, Y.-L. Yang, X.-L. Fan and C. Wang, ACS Nano, 2007, 1, 160–167 CrossRef CAS PubMed.
- J. M. MacLeod, O. Ivasenko, C. Fu, T. Taerum, F. Rosei and D. F. Perepichka, J. Am. Chem. Soc., 2009, 131, 16844–16850 CrossRef CAS PubMed.
- L. Zhang, J. Li, S. Qiu, X. Huang and Z. Zeng, New J. Chem., 2017, 41, 3260–3264 RSC.
- J. M. MacLeod, Z. Ben Chaouch, D. F. Perepichka and F. Rosei, Langmuir, 2013, 29, 7318–7324 CrossRef CAS PubMed.
- H. Zhou, H. Dang, J.-H. Yi, A. Nanci, A. Rochefort and J. D. Wuest, J. Am. Chem. Soc., 2007, 129, 13774–13775 CrossRef CAS PubMed.
- X.-H. Kong, K. Deng, Y.-L. Yang, Q.-D. Zeng and C. Wang, J. Phys. Chem. C, 2007, 111, 9235–9239 CrossRef CAS.
- J. Adisoejoso, K. Tahara, S. Okuhata, S. Lei, Y. Tobe and S. De Feyter, Angew. Chem., Int. Ed., 2009, 48, 7353–7357 CrossRef CAS PubMed.
- T. Balandina, K. Tahara, N. Saendig, M. O. Blunt, J. Adisoejoso, S. Lei, F. Zerbetto, Y. Tobe and S. De Feyter, ACS Nano, 2012, 6, 8381–8389 CrossRef CAS PubMed.
- K. Tahara, K. Kaneko, K. Katayama, S. Itano, N. Chi Huan, D. D. D. Amorim, S. De Feyter and Y. Tobe, Langmuir, 2015, 31, 7032–7040 CrossRef CAS PubMed.
- K. Tahara, K. Nakatani, K. Iritani, S. De Feyter and Y. Tobe, ACS Nano, 2016, 10, 2113–2120 CrossRef CAS PubMed.
- G. Velpula, T. Takeda, J. Adisoejoso, K. Inukai, K. Tahara, K. S. Mali, Y. Tobe and S. De Feyter, Chem. Commun., 2017, 53, 1108–1111 RSC.
- I. Cebula, H. Lu, M. Zharnikov and M. Buck, Chem. Sci., 2013, 4, 4455–4464 RSC.
- S. Lei, M. Surin, K. Tahara, J. Adisoejoso, R. Lazzaroni, Y. Tobe and S. De Feyter, Nano Lett., 2008, 8, 2541–2546 CrossRef CAS PubMed.
- C. Shen, I. Cebula, C. Brown, J. Zhao, M. Zharnikov and M. Buck, Chem. Sci., 2012, 3, 1858–1865 RSC.
- M. Blunt, X. Lin, M. d. C. Gimenez-Lopez, M. Schroder, N. R. Champness and P. H. Beton, Chem. Commun., 2008, 2304–2306 RSC.
- Q.-N. Zheng, L. Wang, Y.-W. Zhong, X.-H. Liu, T. Chen, H.-J. Yan, D. Wang, J.-N. Yao and L.-J. Wan, Langmuir, 2014, 30, 3034–3040 CrossRef CAS PubMed.
- C. Lu, Y. Li, L.-M. Wang, H.-J. Yan, L. Chen and D. Wang, Chem. Commun., 2019, 55, 1326–1329 RSC.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9ra01493d |
| This journal is © The Royal Society of Chemistry 2019 |