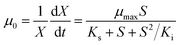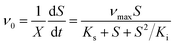Open Access Article
Open Access ArticleSuperior performance and mechanism of chlorobenzene degradation by a novel bacterium
Shihan Zhangab,
Zanyun Yinga,
Juping Youa,
Jiexu Ye *ab,
Zhuowei Chengab,
Dongzhi Chenab and
Jianmeng Chenab
*ab,
Zhuowei Chengab,
Dongzhi Chenab and
Jianmeng Chenab
aCollege of Environment, Zhejiang University of Technology, Hangzhou, 310014, China. E-mail: yejiexu@zjut.edu.cn; Fax: +86-571-88320882; Tel: +86-571-88320853
bKey Laboratory of Microbial Technology for Industrial Pollution Control of Zhejiang Province, Zhejiang University of Technology, Hangzhou, 310014, China
First published on 14th May 2019
Abstract
A newly isolated strain was identified as Ochrobactrum sp. by 16S rRNA sequence analysis and named as ZJUTCB-1. The strain was able to degrade mono-chlorobenzene (CB) as the sole carbon and energy source under aerobic conditions. This study is the first to report the degradation of CB by the genus Ochrobactrum. The degradation rate of CB reached 170.9 μmol L−1 h−1, which is at least 6 times higher than the previously reported data. The strain can efficiently degrade CB under a rang of temperatures (30–40 °C) and pH (6.0–7.0) with optimum at 40 °C and pH 7.0. Salt concentration higher than 0.05 mol L−1 remarkably reduced the biodegradation capability. Moreover, true oxic condition was not an essential element for biodegradation given that the CB degradation rate of 210.4 μmol L−1 h−1 was obtained under microaerobic condition. Based on the Haldane kinetic model, the maximum specific growth rate was 0.895 h−1, which is the highest in ever described CB-degrading strains. According to GC-MS analysis and enzymatic assay, CB was degraded via the meta-cleavage pathway by using 2,3-dioxygenase and 2-chlorophenol as the main intermediates, producing CO2 and Cl− as the final products. The great performance of CB degradation by Ochrobactrum sp. ZJUTCB-1 provided an alternative for development of more effective and reliable biotreatment process.
Introduction
Mono-chlorobenzene (CB) is not only a common industrial solvent but also a widely used intermediate for the manufacture of pesticides and dyes.1 The United States Environmental Protection Agency (US EPA) listed CB as a priority pollutant with a maximum contaminant level of 0.1 ppm due to its persistence and potential toxicity. CB has high volatility and is released into the air, water, and soil, leading to serious environmental issues and health risks.2In recent years, various physical–chemical technologies have been proposed to eliminate CB. These technologies include combustion, adsorption, absorption, photocatalysis, catalytic oxidation, electrochemical reduction, membrane separation, and plasma.3–10 However, these methods require large amounts of energy and are expensive. Compared with physical–chemical technologies, biological technology can be operated under milder conditions and thus result in lower cost.11–14 It has been established that the aerobic biodegradability of CB is thermodynamically feasible.15 However, CB is a recalcitrant substrate, and its performance in biological degradation relies on effective microbial strains. As a result, isolation of a bacterium with excellent CB-degrading capability is critical and has attracted considerable interest.
Scholars have isolated several CB-degrading bacteria, such as Rhodococcus phenolicus sp. G2PT, Pseudomonas aeruginosa, Ralstonia pickettii L2, Acidovorax avenae, Escherichia hermanii, Planococcus sp. strain ZD22, Bacillus sp. TAS6CB, Brevibacillus agri DH-1, and Pandoraea pnomenusa sp. strain MCB032.16–24 These strains obtained aerobic degradation rate of 0.318–3.66 mg L−1 h−1.18,21,25–27 However, their microbial activities remain insufficient to meet the demand for effective large-scale applications. Therefore, microorganisms with better CB-degrading ability are still urgently required. In addition, since microorganisms might expose to low dissolved oxygen due to the insufficient aeration, it is necessary to pay attention to the degradation rate of bacteria under microaerobic conditions.
The major pathway for CB degradation is initiated with a dioxygenases, TecA.28 The resulting dihydrodiols are rearomatized by dihydrodiol dehydrogenases, encoded by tecB genes,29 to chlorocatechols, which serve as the ring-fission substrate. The ring of chlorocatechols are cleaved in the ortho position by 1,2-dioxygenase (products of clc gene cluster) or in the meta position by 2,3-dioxygenase (products of cbzE genes).30,31 On the other hand, Zhang et al.18 found that R. pickettii L2 degraded CB via initial mono-oxygenation rather than dioxygenation, generating 2-chlorophenol as the first intermediate. Hence, the metabolic pathways are diversity, depending on the type of degrading bacteria. Therefore, the degradation pathway should be determined for a novel strain that can utilize CB as the sole carbon and energy source.
This work focuses on: (1) the isolation and characterization of a new bacterium with superior CB degradation ability, (2) investigating the effect of initial CB concentration, pH, temperature, salt concentration, initial cell concentration, and dissolved oxygen concentration on CB biodegradation, (3) determination of the CB degradation kinetics, and (4) proposing the CB degradation pathway.
Experimental section
Chemicals and medium
CB (≥99%) and ethyl acetate (≥99.5%) were acquired from Shanghai Lingfeng Chemical Reagent Co., Ltd., China. Catechol (≥99.5%) and Brilliant Blue G250 were acquired from Shanghai Aladdin Bio-Chem Technology Co., Ltd., China. Na2HPO4·12H2O and NaH2PO4·2H2O with purity over 99% were provided by Guangdong Guanghua Chemical Factory Co., Ltd., China. (NH4)2SO4, MgSO4, K2SO4, and NaCl with purity over 99% were obtained from Shanghai Shisihewei Chemical Co., Ltd., China. Yeast extract and tryptone were obtained from Nuoyang Biotechnology Company, China. Agar and 2-chlorophenol were obtained from Sinopharm Chemical Reagent, China. All other chemical agents (analytical grade) were obtained from Huipu Chemical Co., Ltd., China.The employed minimal salt medium (MSM) contains (1 L): 0.66 g (NH4)2SO4, 0.11 g MgSO4, 0.12 g K2SO4, 22.20 g Na2HPO4·12H2O, 5.93 g NaH2PO4·2H2O, 0.62 mL vitamin solution, and 1.25 mL trace element solution. The composition of the vitamin solution was based on our previous work.14 The trace element solution includes (1 L): 0.01 g H3BO3, 0.1 g CaCl2·2H2O, 0.01 g CuSO4·5H2O, 0.024 g NiCl2·6H2O, 0.025 g Na2WO4·2H2O, 0.025 g Na2MoO4, 0.1 g CoCl2·6H2O, 0.1 g FeSO4·7H2O, 0.01 g AlK (SO4)2·12H2O, 0.13 g ZnCl2, 0.5 g MnSO4·H2O, 1 g NaCl, 1.5 g NTA and 3 g MgSO4. The pH value of the MSM (without the addition of the trace element and vitamin solution) was set to 7.0 by adding 1 mol L−1 NaOH solution. The MSM was sterilized at 110 °C for 40 min. The trace mineral and vitamin solutions were filtered after being disposed and transferred to MSM. The Luria-Bertani (LB) medium contains (1 L): 5.0 g yeast extract, 10.0 g NaCl, and 10.0 g tryptone. Before use, the LB medium was sterilized for 20 min at 121 °C. All the solid media contain 2 wt% agar.
Isolation of the strain ZJUTCB-1
An activated sludge obtained from a Hangzhou wastewater treatment plant was used to isolate the strain. The activated sludge was inoculated in MSM containing 100 mg L−1 CB at 30 °C and 160 rpm to enrich the bacterium. When the CB concentration was decreased by 50%, the solution was displaced using fresh medium containing 100 mg L−1 CB. After repeating the operation for five times, the diluted enrichment liquid was coated on solid MSM with CB for 3–5 days at 30 °C. Hereafter, a single colony selected from solid MSM was inoculated to the fresh LB liquid medium. The obtained pure culture was added to the fresh MSM with CB to test its capability for CB degradation. The bacterium was designated as strain ZJUTCB-1.The strain ZJUTCB-1 was identified by 16S rRNA analysis. A kit of DNA extraction (Sangon, Shanghai, China, SK8255) was used to extract genomic DNA of the strain according to the manufacturer's instruction. The 16S rRNA sequence was amplified via polymerase chain reaction (PCR) for 30 cycles by using universal forward and reverse primers such as CAGAGTTTGATCCTGGCT and AGGAGGTGATCCAGCCGCA. The PCR program was carried out as follows: denaturing for 4 min at 94 °C and then running 30 cycles of 45 s at 94 °C (denaturation), 45 s at 55 °C (annealing), and 1 min at 72 °C (extension). The resulting product was sequenced by the Shanghai Sangon Biotech Co., Ltd., China. The obtained sequence was in comparison with that reported in the GenBank database via the BLAST program. A phylogenic tree was established using MegAlign software (MEGA 5.1).
CB degradation experiments and kinetics experiments
The strain ZJUTCB-1 was pre-grown in MSM with CB at 30 °C to late exponential growth phase and then collected via centrifugation (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm, 3 min, 4 °C). The harvested cells were re-suspended in MSM for further use after washing with MSM thrice.
000 rpm, 3 min, 4 °C). The harvested cells were re-suspended in MSM for further use after washing with MSM thrice.
In a typical test, the degradation experiment was conducted in 340 mL serum vials with 50 mL MSM containing 520–2600 μmol L−1 CB and 24–62 mg DCW L−1 ZJUTCB-1 (160 rpm) under aerobic condition (∼7 mg L−1 DO). N2 with high purity was passed into the serum vials for 30 min to achieve a microaerobic condition (∼0.2 mg L−1 DO). For pH test, the initial pH of MSM was adjusted to 5.0, 6.0, 7.0, 8.0, and 9.0. To evaluate the effect of temperature, we varied the incubation temperature from 25–45 °C. The effect of salt concentration on CB degradation was investigated using MSM containing 0–0.4 mol L−1 NaCl. Each group of experiments set up a blank control group (without bacterium). The samples in the liquid and gas phase were obtained at the certain interval. The degradation kinetics was also investigated under the cell concentration of 39 mg DCW L−1 with different initial CB concentrations (520–2600 μmol L−1) at 160 rpm and 30 °C. The classical Haldane model was used to estimate the kinetics as follows:32
where μ0 and ν0 are the specific cell growth and CB degradation rate, respectively, h−1; μmax and νmax are the maximum specific cell growth and degradation rate, h−1, respectively; S is the concentration of CB, μmol L−1; X is the concentration of strain ZJUTCB-1, mg DCW L−1; Ki is the inhibition constant, μmol L−1, and Ks is the half-saturation constant, μmol L−1.
Enzyme assays
The bacteria were harvested by centrifugation (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm, 10 min, 4 °C), washed twice with 50 mmol L−1 phosphate buffer (pH = 7.2), and suspended in phosphate buffer. The cells were broken by sonication and centrifuged at 12
000 rpm, 10 min, 4 °C), washed twice with 50 mmol L−1 phosphate buffer (pH = 7.2), and suspended in phosphate buffer. The cells were broken by sonication and centrifuged at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm, 4 °C for 30 min. The precipitate was discarded, and the supernatant was kept on ice until use. The reaction system contained 0.3 μmol catechol, 1.0 mL of 50 mmol L−1 phosphate buffer, and cell extract (0.3–0.9 mg of protein) in a final volume of 3 mL. The mixtures were reacted in water bath (30 °C) for 10 min. When cis,cis-muconic acid and 2-hydroxymuconic semialdehyde were formed, the absorbance at 260 and 375 nm increased, respectively.
000 rpm, 4 °C for 30 min. The precipitate was discarded, and the supernatant was kept on ice until use. The reaction system contained 0.3 μmol catechol, 1.0 mL of 50 mmol L−1 phosphate buffer, and cell extract (0.3–0.9 mg of protein) in a final volume of 3 mL. The mixtures were reacted in water bath (30 °C) for 10 min. When cis,cis-muconic acid and 2-hydroxymuconic semialdehyde were formed, the absorbance at 260 and 375 nm increased, respectively.
Specific enzyme activities were calculated using the extinction coefficient of each reaction product: 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 800 L mol−1 cm−1 for cis,cis-muconic acid at 260 nm and 36
800 L mol−1 cm−1 for cis,cis-muconic acid at 260 nm and 36![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 L mol−1 cm−1 for 2-hydroxymuconic semialdehyde at 375 nm.33,34 Specific enzyme activities were expressed in μmol of product formed per min per mg of protein.35 The content of protein in the cell-free extract was determined using the Bradford method.36
000 L mol−1 cm−1 for 2-hydroxymuconic semialdehyde at 375 nm.33,34 Specific enzyme activities were expressed in μmol of product formed per min per mg of protein.35 The content of protein in the cell-free extract was determined using the Bradford method.36
Analytical methods
The concentrations of CO2 and CB in the gas phase were determined using the 6890N gas chromatograph (Agilent Technologies, America). For CB analysis, it was measured by a column of HP-INNOWax and flame ionization detector under air and hydrogen flow rates of 450 and 40 mL min−1, respectively. The inlet was in split mode with N2 as carrier gas (33.4 mL min−1). The temperatures of the injector, detector, and oven were 200, 180, and 100 °C, respectively. CO2 was measured using a thermal conductivity detector and a column of HP-Plot-Q with helium as carrier gas (2 mL min−1). The temperatures of the injector, detector, and oven were 90, 180, and 40 °C.The concentration of Cl− was measured using an ICS-2000 ion chromatograph (DIONEX, America) equipped with the Ionpac AS19-HC column. The eluent was KOH with a concentration of 20 mmol L−1 at a flow rate of 1.00 mL min−1.
The intermediate products were identified by GC-MS (Agilent 7890N/MS 5975, America), using a column of HP-5MS and helium as the carrier gas (1.5 mL min−1). The temperature of the injector was 250 °C. The temperature of the oven was maintained at 35 °C for 10 min, ramped to 190 °C at 10 °C min−1, kept for another 2 min, heated to 225 °C at 6 °C min−1, and kept for another 1 min. The mass spectrometer was operated in the electron impact mode. The electron ionization mass spectra were acquired at 70 eV at ionization source temperature of 230 °C. The intermediate products were determined and compared with the NIST standard library.
The cell concentration was determined by measuring the optical density at 600 nm by using a U-2910 spectrophotometer (HITACHI, Japan). The cell concentration was then calculated according to an established correlation between the absorbance and dry-weight concentration of strain ZJUTCB-1.
Results and discussion
Identification of strain ZJUTCB-1
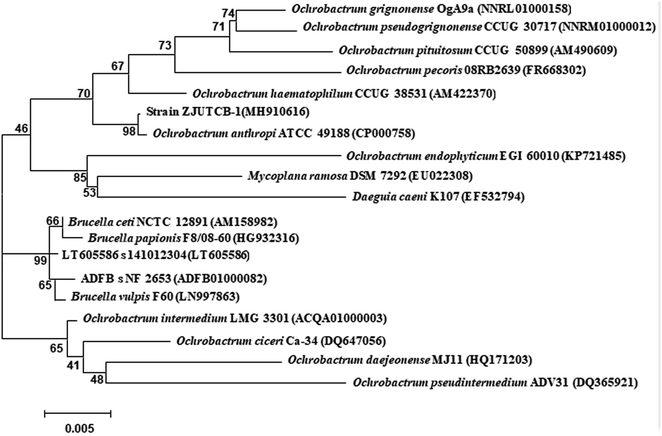
The colonies of ZJUTCB-1 were light yellow with neat edges and smooth surface. The morphology of the bacterium was bacilli with parallel edges and round ends without flagellation. It was a Gram negative bacterium. A sequence of the 16S rRNA gene with 1425-bp fragment was submitted to the GenBank database (accession number: MH910616). As shown in Fig. 1, the strain ZJUTCB-1 was closely similar to the strain of the genus Ochrobactrum. The strain ZJUTCB-1 had the highest similarity (approximately 98%) with Ochrobactrum anthropic ATCC 49188 (GenBank accession No. CP000758). As a result, it was initially assigned as Ochrobactrum sp., and has been deposited in the China Center for Type Culture Collection with accession number of CCTCC M 2018490.Ochrobactrum sp. can decompose various environmental pollutants, including benzo[a]pyrene, di-n-butyl phthalate, phenanthrene, phenol, and tetrabromobisphenol A.37–41 To the best of our knowledge, the present work first described an Ochrobactrum sp. strain with CB degradation capability.
CB degradation by Ochrobactrum sp. ZJUTCB-1
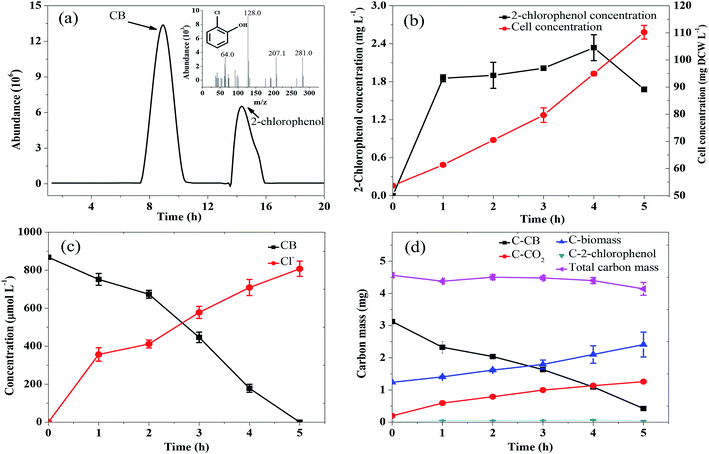
Fig. 2 shows the CB degradation performance with different initial substrate concentrations under aerobic condition. As shown in Fig. 2a, the degradation of CB encountered a lag phase and then gradually increased within the timeframe of the operation, indicating that the enzyme pertinent to the CB degradation was an induced enzyme rather than a constitutive enzyme. Correspondingly, the cell concentration growth also encountered a lag phase with the feeding of CB (Fig. 2b). Moreover, it could be found that CB at all tested concentrations was completely degraded within 26 h. The highest average degradation rate of 170.9 μmol L−1 h−1 was obtained at 1216 μmol L−1 CB degradation, which was at least 6 times of the reported data shown in Table 1. Ochrobactrum sp. ZJUTCB-1 exhibited the best CB degradation activity in ever described microorganisms.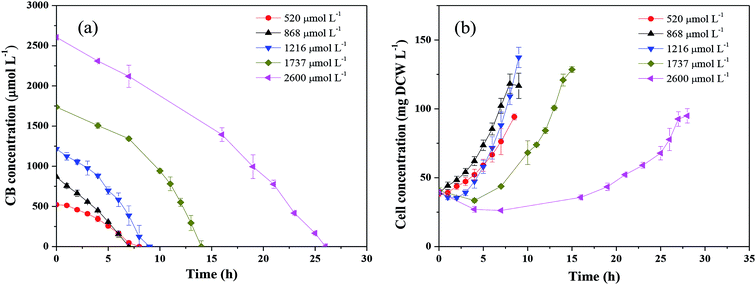 | ||
| Fig. 2 Degradation of CB by Ochrobactrum sp. ZJUTCB-1 with different initial CB concentrations under aerobic condition (pH = 7.0, T = 30 °C, and [cell]0 = 39 mg DCW L−1). | ||
| Culture | Condition | CB concentration (mg L−1) | Reaction time | Removal efficiency (%) | Average degradation rate (mg L−1 h−1) | Temperature (°C) | Ref. |
|---|---|---|---|---|---|---|---|
| Ralstonia pickettii L2 | Aerobic | 220 | 75 h | 100 | 2.933 | 30 | 18 |
| Acidovorax avenae | Aerobic | 7.65 | 24 h | 100 | 0.318 | 25 | 19 |
| Escherichia hermanii | Aerobic | 394 | 300 h | 100 | 1.313 | 30 | 20 |
| Planococcus sp. ZD22 | Aerobic | 112.5 | 120 h | 67.3 | 0.63 | 20 | 21 |
| Mixed fungal–bacterial consortium | Aerobic | 220 | 60 h | 100 | 3.666 | 30 | 25 |
| Ralstonia sp. DSM 8910 | Aerobic | 50 | 21.6 h | 93 | 2.155 | 20 | 26 |
| Labrys portucalensis F11 | Aerobic | 56 | 10 d | 0.5 ± 0.03 | 25 | 27 | |
| Enrichment culture | Anaerobic | 225 | 50 d | 100 | 0.1875 | 30 | 42 |
| Sediment culture | Anaerobic | 78.75 | 140 h | 100 | 0.5625 | 43 | |
| Ochrobactrum sp. ZJUTCB-1 | Aerobic | 136.75 | 9 h | 100 | 19.23 (170.9 μmol L−1 h−1) | 30 | This work |
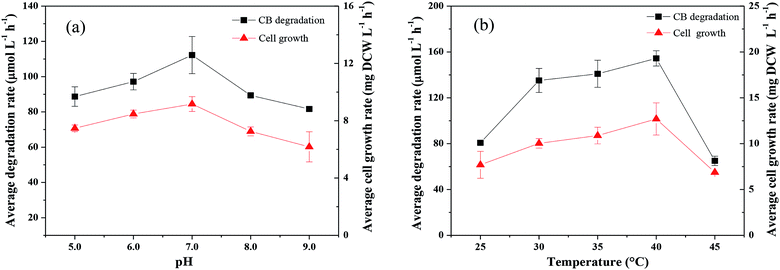
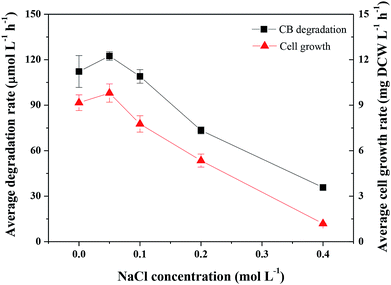
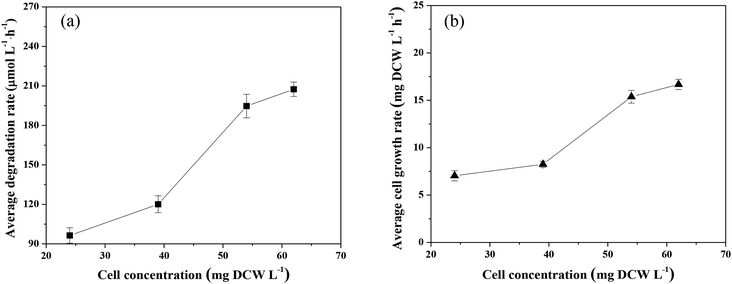
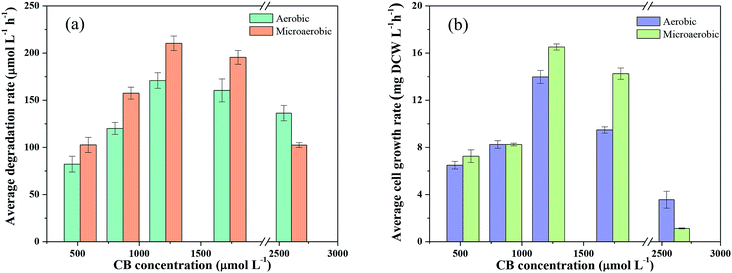
Factors affecting CB degradation
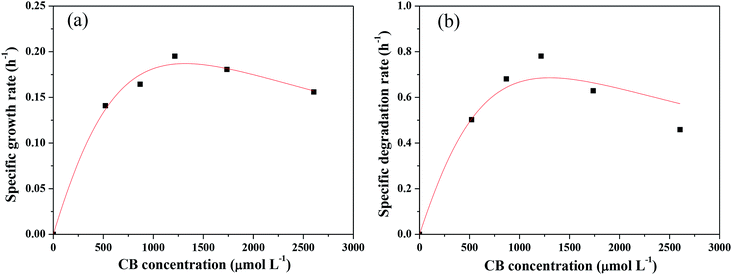
Kinetics of CB degradation by Ochrobactrum sp. ZJUTCB-1
The Monod kinetic model and the Haldane kinetic model are commonly used to predict substrate degradation.49,50 Considering the substrate inhibition occurred at high CB concentration, the Haldane kinetic model was used to predict the aerobic biodegradation of CB in this work.As shown in Fig. 7a, the Haldane kinetic model well fitted the experimental data (R2 > 0.95). The values of μmax, Ks, and Ki were 0.895 h−1, 2495 μmol L−1, and 697 μmol L−1, respectively. The specific growth rate of aerobic degradation initially increased and then decreased (Fig. 7a). This finding might indicate that the increasing available carbon source at concentrations below 1216 μmol L−1 was beneficial to bacterial growth. When the concentration was higher, the toxicity of CB inhibited cell growth. The maximum specific growth rate was more than two times higher than that of the other aerobic degradation bacteria reported in the literatures (0.15–0.36 h−1).24,25
Fig. 7b shows the change in the specific degradation rate of strain ZJUTCB-1 at different CB concentrations in aerobic condition. The experimental data also well fitted to the Haldane kinetic model (R2 > 0.90). Similar to the growth kinetic, the degradation kinetics also showed the same tendency, confirming that CB inhibited the activity of CB degradation. The values of vmax, Ks, Ki were 3.315 h−1, 2494.542 μmol L−1, and 697.149 μmol L−1, respectively. The maximum specific degradation rate with a cell concentration of 39 mg DCW L−1 was approximately 11 times of the reported data.25
CB degradation pathway of Ochrobactrum sp. ZJUTCB-1
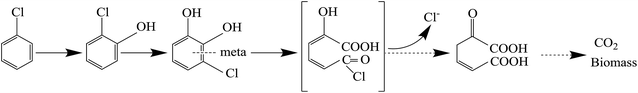
The pathway of CB degradation has been intensively investigated.15,51 In this work, the CB degradation pathway was also investigated. As shown in Fig. 8a, the characteristic peak of 2-chlorophenol could be detected under aerobic condition by GC-MS. The authenticity of the substance was confirmed by comparing the peak with the NIST library. Fig. 8b shows the main intermediate and cell growth under aerobic condition at different time intervals. 2-Chlorophenol accumulated at the initial stage of CB degradation, reaching the highest concentration of 2.34 mg L−1 (3 wt% initial CB concentration) at 4 h, and then gradually decreased. Meanwhile, noting that when the concentration of 2-chlorophenol decreased, the cell concentration still increased. These results confirmed that CB degradation proceeded with 2-chlorophenol as an intermediate. We inferred the degradation of CB was caused by monooxygenase, which initially produced 2-chlorophenol and then converted into 3-chlorocatechol. This step was similar to the result reported by Zhang et al.18 but was different from some previous works in which the degradation of CB was initiated by dioxygenase.43,52 The specific enzyme activity of 2,3-dioxygenase was measured to be 1.03 μmol min−1 mg−1 protein, while that of 1,2-dioxygenase was not detected, suggesting that 2,3-dioxygenase played a major role in CB degradation. Furthermore, chlorine ion concentration increased in the aqueous phase, and 93% of CB was dechlorinated at the end of experiments (Fig. 8c). Therefore, 3-chlorocatechol was deduced to be oxidized via meta-cleavage pathway to acylchloride. The resulting acylchloride was dechlorinated and converted into 4-oxalocrotonate, which entered the Kreb's cycle subsequently.15,52 A possible pathway for CB degradation by Ochrobactrum sp. ZJUTCB-1 under aerobic condition was proposed in Fig. 9. Furthermore, the existence of 2,3-dioxygenase also confirmed that the great degradation performance of the strain ZJUTCB-1 under microaerobic conditions might be attributed to the high oxygen affinity of 2,3-dioxygenase.The variation of carbon-based products, including the C-biomass, C–CO2, and C–2-chlorophenol, during the aerobic degradation within 5 h was determined. As shown in Fig. 8d, the total carbon determined with C–CB, C-biomass, C–CO2, and C–2-chlorophenol decreased by 9.24%, which suggested that several intermediates were undetectable. Under the cultivation time of 5 h, 39.3% of carbon from CB was converted into CO2, and a higher fraction (43.4%) of carbon from CB was converted into biomass. These results further certified that the biomass assimilation and CO2 production were the two major fates of the carbon in the removed CB.
Conclusions
The present work screened a new CB-degrading bacterium, and it was identified as Ochrobactrum sp. ZJUTCB-1. The isolate exhibited superior performance in aerobic CB-degrading activity with a maximum average degradation rate of 170.9 μmol L−1 h−1. The strain can efficiently degrade CB under a range of temperatures (30–40 °C) and pH (6.0–7.0) with optimum at 40 °C and pH 7.0. Salt concentrations higher than 0.05 mol L−1 remarkably reduced the biodegradation capability. CB degradation patterns were well described by Haldane model. Moreover, the truly oxic condition was not an essential element for the CB degradation by Ochrobactrum sp. ZJUTCB-1, because the degradation rate of CB did not decrease under oxygen limitation. 2-Chlorophenol was determined as the main intermediate and Cl−, and CO2 as the final products. CB was aerobically degraded via 2-chlorophenol into 3-chlorocatechol and subsequently subjected to the meta-cleavage pathway for further metabolism.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 21876157, 21606204) and the Program for Changjiang Scholars and Innovative Research Team in University (IRT13096).References
- R. A. Pandey, P. R. Joshi, S. N. Mudliar and S. C. Deshmukh, Bioresour. Technol., 2010, 101, 5168–5174 CrossRef CAS PubMed.
- O. Lhotský, E. Krákorová, L. Linhartová, Z. Křesinová, J. Steinová, L. Dvořák, T. Rodsand, A. Filipová, K. Kroupová, L. Wimmerová, J. Kukačka and T. Cajthaml, Sci. Total Environ., 2017, 607–608, 1451–1465 CrossRef PubMed.
- B. B. Feng, Y. X. Wei, Y. N. Qiu, S. F. Zuo and N. Ye, J. Rare Earths, 2018, 36, 1169–1174 CrossRef CAS.
- Y. Y. Guo, Y. R. Li, J. Wang, T. Y. Zhu and M. Ye, Chem. Eng. J., 2014, 236, 506–512 CrossRef CAS.
- P. T. Hoa, K. Suto, C. Inoue and J. Hara, AIP Conf. Proc., 2007, 898, 12–16 CrossRef CAS.
- L. Y. Jiang, G. F. Nie, R. Y. Zhu, J. D. Wang, J. M. Chen, Y. B. Mao, Z. W. Cheng and W. A. Anderson, J. Environ. Sci., 2017, 55, 266–273 CrossRef CAS PubMed.
- C. Luo, Z. Chen, D. L. Wu and L. M. Ma, Chem. Eng. J., 2014, 241, 376–383 CrossRef CAS.
- D. D. Pang, J. Gao, F. Ouyang, R. S. Zhu and C. Xie, Catalysts, 2017, 7, 126 CrossRef.
- X. L. Weng, P. F. Sun, Y. Long, Q. J. Meng and Z. B. Wu, Environ. Sci. Technol., 2017, 51, 8057–8066 CrossRef CAS PubMed.
- W. Zhao and B. L. Shi, Sep. Sci. Technol., 2009, 44, 1737–1752 CrossRef CAS.
- M. A. Surhio, F. N. Talpur, S. M. Nizamani, F. Amin, C. W. Bong, C. W. Lee, M. A. Ashraf and M. R. Shah, RSC Adv., 2014, 4, 55960–55966 RSC.
- Z. W. Cheng, X. M. Zhang, C. Kennes, J. M. Chen, D. Z. Chen, J. X. Ye, S. H. Zhang and D. D. Dionysiou, Sci. Total Environ., 2019, 650, 2095–2106 CrossRef CAS PubMed.
- J. P. You, M. Du, H. Chen, X. Y. Zhang, S. H. Zhang, J. M. Chen, Z. W. Cheng, D. Z. Chen and J. X. Ye, Int. Biodeterior. Biodegrad., 2018, 129, 202–208 CrossRef CAS.
- S. H. Zhang, J. P. You, N. An, J. K. Zhao, L. D. Wang, Z. W. Cheng, J. X. Ye, D. Z. Chen and J. M. Chen, Chem. Eng. J., 2018, 351, 515–522 CrossRef CAS.
- J. A. Field and R. Sierra-Alvarez, Biodegradation, 2008, 19, 463–480 CrossRef CAS PubMed.
- M. Rehfuss and J. Urban, Syst. Appl. Microbiol., 2005, 28, 695–701 CrossRef CAS PubMed.
- R. Manikandan, H. J. Prabhu and P. Sivashanmugam, J. Biotechnol., 2007, 6, 1338–1342 CAS.
- L. L. Zhang, S. Q. Leng, R. Y. Zhu and J. M. Chen, Appl. Microbiol. Biotechnol., 2011, 91, 407–415 CrossRef CAS PubMed.
- M. V. Monferrán, J. R. Echenique and D. A. Wunderlin, Chemosphere, 2005, 61, 98–106 CrossRef PubMed.
- J. Kiernicka, C. Seignez and P. Peringer, Lett. Appl. Microbiol., 1999, 28, 27–30 CrossRef CAS PubMed.
- H. Li, Y. H. Liu, N. Luo, X. Y. Zhang, T. G. Luan, J. M. Hu, Z. Y. Wang, P. C. Wu, M. J. Chen and J. Q. Lu, Res. Microbiol., 2006, 157, 629–636 CrossRef CAS PubMed.
- T. K. Vyas and S. R. Murthy, J. Basic Microbiol., 2015, 55, 382–388 CrossRef CAS PubMed.
- B. R. Yang, Z. Q. Sun, L. P. Wang, Z. X. Li and C. Ding, Bioresour. Technol., 2017, 231, 19–25 CrossRef CAS PubMed.
- I. I. R. Baptista, N. Y. Zhou, E. A. C. Emanuelsson, L. G. Peeva, D. J. Leak, A. Mantalaris and A. G. Livingston, Biotechnol. Bioeng., 2008, 99, 68–74 CrossRef CAS PubMed.
- Z. W. Cheng, C. Li, C. Kennes, J. X. Ye, D. Z. Chen, S. H. Zhang, J. M. Chen and J. M. Yu, Int. Biodeterior. Biodegrad., 2017, 123, 276–285 CrossRef CAS.
- A. Kaschl, C. Vogt, S. Uhlig, I. Nuenhuis, H. Weiss, M. Kästner and H. H. Richnow, Environ. Toxicol. Chem., 2005, 24, 1315–1324 CrossRef CAS PubMed.
- I. S. Moreira, C. L. Amorim, M. F. Carvalho and P. M. L. Castro, Int. Biodeterior. Biodegrad., 2012, 72, 76–81 CrossRef CAS.
- N. Kimura, W. Kitagawa, T. Mori, N. Nakashima, T. Tamura and Y. Kamagata, Appl. Microbiol. Biotechnol., 2006, 73, 474–484 CrossRef CAS PubMed.
- K. Pollmann, S. Beil and D. H. Pieper, Appl. Environ. Microbiol., 2001, 67, 4057–4063 CrossRef CAS PubMed.
- X. W. Jiang, H. Liu, Y. Xu, S. J. Wang, D. J. Leak and N. Y. Zhou, Arch. Microbiol., 2009, 191, 485–492 CrossRef CAS PubMed.
- A. Alfreider, C. Vogt and W. Babel, Appl. Environ. Microbiol., 2003, 69, 1372–1376 CrossRef CAS PubMed.
- J. F. Andrews, Biotechnol. Bioeng., 1968, 10, 707–723 CrossRef CAS.
- E. Schmidt, G. Remberg and H. J. Knackmuss, Biochem. J., 2005, 192, 331–337 CrossRef PubMed.
- J. A. Asturias and K. N. Timmis, Microbiology, 1993, 175, 4631–4640 CAS.
- A. Farrell and B. Quilty, Biodegradation, 1999, 10, 353–362 CrossRef CAS PubMed.
- M. M. Bradford, Anal. Biochem., 1976, 72, 248–254 CrossRef CAS.
- Y. R. Wu, T. T. He, M. Q. Zhong, Y. L. Zhang, E. M. Li, T. W. Huang and Z. Hu, J. Environ. Sci., 2009, 21, 1446–1451 CrossRef CAS.
- X. L. Wu, Y. Y. Wang, R. X. Liang, Q. Y. Dai and W. L. Chao, Bull. Environ. Contam. Toxicol., 2010, 85, 235–237 CrossRef CAS PubMed.
- D. Ghosal, J. Chakraborty, P. Khara and T. K. Dutta, FEMS Microbiol. Lett., 2010, 313, 103–110 CrossRef CAS PubMed.
- W. S. El-Sayed, M. K. Ibrahim, M. Abu-Shady, F. El-Beih, N. Ohmura, H. Saiki and A. Ando, J. Biosci. Bioeng., 2003, 96, 310–312 CrossRef CAS PubMed.
- G. Y. Li, J. K. Xiong, P. K. Wong and T. C. An, Environ. Pollut., 2016, 208, 796–802 CrossRef CAS PubMed.
- J. L. Nelson, J. M. Fung, H. Cadillo-Quiroz, X. Cheng and S. H. Zinder, Environ. Sci. Technol., 2011, 45, 6806–6813 CrossRef CAS PubMed.
- X. M. Liang, M. R. Howlett, J. L. Nelson, G. Grant, S. Dworatzek, G. Lacrampe-Couloume, S. H. Zinder, E. A. Edwards and B. Sherwood Lollar, Environ. Sci. Technol., 2011, 45, 8321–8327 CrossRef CAS PubMed.
- M. H. El-Naas, S. A. Al-Muhtaseb and S. Makhlouf, J. Hazard. Mater., 2009, 164, 720–725 CrossRef CAS PubMed.
- B. Bhunia, B. Basak, T. Mandal and P. Bhattacharya, Int. J. Biol. Macromol., 2013, 54, 1–8 CrossRef CAS PubMed.
- Z. G. He, F. L. Gao, T. Sha, Y. H. Hu and C. He, J. Hazard. Mater., 2009, 163, 869–873 CrossRef CAS PubMed.
- T. Deng, H. Y. Wang and K. Yang, Water Sci. Technol., 2018, 77, 504–510 CrossRef CAS PubMed.
- Ö. Çinar, K. Demiröz, G. Kanat, Y. Uysal and C. Yaman, Clean: Soil, Air, Water, 2009, 37, 657–662 Search PubMed.
- J. L. Wang and W. Wan, Int. J. Hydrogen Energy, 2009, 34, 3313–3323 CrossRef CAS.
- A. Nuhoglu and B. Yalcin, Process Biochem., 2005, 40, 1233–1239 CrossRef CAS.
- M. Lapertot, C. Seignez, S. Ebrahimi and P. Peringer, Ind. Eng. Chem. Res., 2006, 45, 6778–6784 CrossRef CAS.
- A. E. Mars, T. Kasberg, S. R. Kaschabek, M. H. Van Agteren, D. B. Janssen and W. Reineke, J. Bacteriol., 1997, 179, 4530–4537 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2019 |