Open Access Article
Open Access ArticlePreparation of lysozyme-imprinted nanoparticles on polydopamine-modified titanium dioxide using ionic liquid as a stabilizer
Zhongliang Zhao†
ab,
Caihong Zhu†a,
Qianping Guoa,
Yan Caia,
Xuesong Zhua and
Bin Li *ac
*ac
aCollege of Chemistry, Chemical Engineering and Materials Science, Orthopaedic Institute, Medical College, Soochow University, Suzhou, China
bDepartment of Orthopaedics, The First Affiliated Hospital, Medical College, Soochow University, Suzhou, China
cChina Orthopaedic Regenerative Medicine Group (CORMed), Hangzhou, China. E-mail: binli@suda.edu.cn; Fax: +86-512-6778-1163; Tel: +86-512-6778-1163
First published on 14th May 2019
Abstract
Molecular imprinting of proteins has evolved into an efficient approach for protein recognition and separation. However, maintaining the structural stability of proteins during the preparation process of molecularly imprinted polymers (MIPs) remains challenging. Ionic liquids (ILs), being capable of maintaining the stability of proteins, might enable effective imprinting and accurate recognition of proteins. In this study, lysozyme (Lyz)-imprinted titanium dioxide (TiO2) nanoparticles, TiO2@Lyz-MIPs, have been successfully prepared for selective recognition and separation of Lyz. This was achieved by the free radical polymerization of hydroxyethyl acrylate (HEA) and poly(ethylene glycol) dimethacrylate (PEGDMA) on polydopamine (PDA)-modified TiO2 nanoparticles using an IL, choline dihydrogen phosphate (chol dhp), as the stabilizer of Lyz. It was found that both PDA modification of TiO2 and the use of chol dhp as stabilizer improved the adsorption capacity of TiO2@Lyz-MIPs toward Lyz. When the concentration of HEA was 7 mg mL−1, the ratio of monomer to crosslinker was 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, and the concentration of chol dhp was 12.5 mg mL−1, the highest imprinting factor of 4.40 was achieved. TiO2@Lyz-MIPs exhibited relatively high adsorption capacity with the maximum adsorption capacity up to 120 mg g−1, which was more than four times higher than that of the non-imprinted polymers (NIPs) counterpart, TiO2@Lyz-NIPs. The adsorption rate of Lyz by TiO2@Lyz-MIPs was also much higher than that of TiO2@Lyz-NIPs. TiO2@Lyz-MIPs could successfully separate Lyz from diluted egg white, a complex mixture of proteins. Findings from this study indicate that effective recognition cavities toward Lyz were formed on the surface of Lyz-imprinted TiO2 nanoparticles prepared using IL as the template stabilizer. This approach may facilitate the development of MIPs for efficient protein recognition and separation.
1, and the concentration of chol dhp was 12.5 mg mL−1, the highest imprinting factor of 4.40 was achieved. TiO2@Lyz-MIPs exhibited relatively high adsorption capacity with the maximum adsorption capacity up to 120 mg g−1, which was more than four times higher than that of the non-imprinted polymers (NIPs) counterpart, TiO2@Lyz-NIPs. The adsorption rate of Lyz by TiO2@Lyz-MIPs was also much higher than that of TiO2@Lyz-NIPs. TiO2@Lyz-MIPs could successfully separate Lyz from diluted egg white, a complex mixture of proteins. Findings from this study indicate that effective recognition cavities toward Lyz were formed on the surface of Lyz-imprinted TiO2 nanoparticles prepared using IL as the template stabilizer. This approach may facilitate the development of MIPs for efficient protein recognition and separation.
1 Introduction
Molecular imprinting is a technology through which template-shaped cavities are created in polymer matrices during polymerization. It has been used in a variety of applications including sensors,1–3 chromatographic separation,4,5 and solid-phase extraction.6,7 However, the imprinting of macromolecules, such as proteins and enzymes, remains challenging due to the large molecular size and complexity of them. Especially, proteins are often denatured and unfolded and lose the conformational flexibility during the preparation of molecularly imprinted polymers (MIPs).8–10 Therefore, maintaining the structural stability of protein during MIP preparation is one of the greatest concerns in protein imprinting technology.11Ionic liquids (ILs), a class of organic salts that melt at low-temperatures, have been used as solvents, monomers, crosslinkers, and templates in MIP synthesis to accelerate the process and improve the selectivity and absorption of the template.12–14 In recent years, ILs have been found to stabilize enzymes and proteins,15–17 partially due to its ability to minimize the surface area of protein that is exposed to the solvent. For example, a biocompatible IL, choline dihydrogen phosphate (chol dhp), which has more “physical” effect than the “chemical” effect on protein structure, was able to dramatically retain the structure and activity of a number of proteins such as cytochrome c, Lyz, and ribonuclease A.18–20 As a result, chol dhp could help maintain template stability during MIP preparation and resulted in effective imprinting and recognition of proteins.21,22
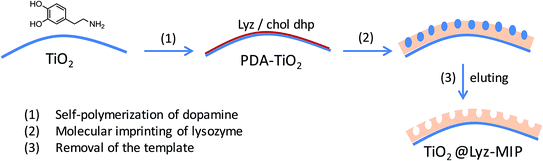
Recently, dopamine (DA), being able to self-polymerizes in aqueous conditions at room temperature, has been increasingly used as the monomer to prepare MIPs for imprinting fragile proteins and enzymes in a simple yet highly efficient way.23–25 Further, Li et al.26 found that MIPs formed by DA self-polymerization on polydopamine (PDA)-modified silica particles were able to prevent viral infections, while MIPs formed by DA self-polymerization on pure silica particles had no such effect. This may be due to the increased number of amino and catechol groups interacting with greater amounts of functional groups on the surface of virus to form three dimensional imprints, which in turn increased the binding capacity and selectivity of MIPs. However, there are no studies to use polydopamine surface-modified TiO2 nanoparticles as the matrix of protein molecular imprinting.
Over the past 40 years, titanium dioxide (TiO2) has been widely applied in photocatalysis for wastewater treatment, in solar cell and electronic devices, as a highly stable, low-cost, and non-toxic material. Recently, due to the appealing characteristics of molecular imprinting, TiO2-based MIPs have attracted increasing attention for its high selectivity and efficient separation potential.27–29 In recent, surface molecular imprinting based on nanoparticles has been thought as an effective method to overcome the protein imprinting difficulty in bulk approach, such as large mass transfer resistance and incomplete template removal. Compared to bulk MIPs, surface MIPs based on nanoparticles have extremely higher surface-to-volume ratio due to the small size, the increased surface area helps to provide more recognition sites and enough space for template diffusion. Second, in surface imprinting, the imprinting sites distributed close to the surface layer, resulting in lower mass transfer resistance and faster binding kinetics.30,31 So, in this study, TiO2 nanoparticles were used to prepare lysozyme (Lyz)-imprinted TiO2 nanoparticles by free radical polymerization of hydroxyethyl acrylate (HEA) and poly(ethylene glycol) dimethacrylate (PEGDMA) on PDA-modified surface of TiO2 using chol dhp to stabilize Lyz templates (Scheme 1). In this approach, PDA surface-modified TiO2 nanoparticles were prepared by DA self-polymerization to improve the interaction between TiO2 and Lyz, which might increase binding capacity and selectivity of TiO2@Lyz-MIPs. On the other hand, Lyz-imprinted nanoparticles were prepared using IL as a protein template stabilizer to ensure that the conformation of Lyz remained intact, which might also result in effective imprinting and recognition of TiO2@Lyz-MIPs. The molecular imprinting conditions were then optimized by adjusting the feed ratio of monomer, crosslinker and IL. The adsorption behaviors of imprinted TiO2 nanoparticles and their ability to isolate Lyz were evaluated.
2 Materials and methods
2.1 Materials
Titanium dioxide nanoparticles (TiO2, anatase, 60 nm) was obtained from Aladdin. Dopamine hydrochloride was from Sigma-Aldrich. Lysozyme (Lyz; chicken egg white, MW 14.4 kDa), bovine serum albumin (BSA; MW 66.4 kDa), and cytochrome c (cyt c; bovine heart, MW 12.4 kDa) were obtained from Solarbio. Acrylamide (AAm), methylene bisacrylamide (Bis), hydroxyethyl acrylate (HEA), poly(ethylene glycol) dimethacrylate (PEGDMA, MW 750 Da) were obtained from Alfa Aesar. Ammonium persulfate (APS) was obtained from Sinopharm Chemical Reagent Co., Ltd. N,N,N′,N′-Tetramethylethylenediamine (TEMED) was also obtained from Alfa Aesar. Choline dihydrogen phosphate (chol dhp) of 99.8% purity was obtained from Shanghai Cheng Jie Chemical Co., Ltd.2.2 Preparation of Lyz-imprinted TiO2 nanoparticles
Lyz-imprinted TiO2 nanoparticles (TiO2@Lyz-MIPs) were prepared in two steps, as shown in Scheme 1. First, polydopamine-modified TiO2 nanoparticles (PDA-TiO2) were made by mixing TiO2 nanoparticles (250 mg), dopamine hydrochloride (2 mg mL−1, 10.55 mmol L−1), and ammonium persulfate (1.2 mg mL−1, 5.26 mmol L−1) in 40 mL Tris–HCl (pH = 8.5, 10 mmol L−1) under shaking overnight at room temperature. After centrifuging (3000 rpm) and washing three times with deionized water, the PDA-TiO2 were collected for use. Second, the PDA-TiO2 were dispersed in 40 mL of PBS buffer (pH = 7.4, 10 mmol L−1), and the Lyz template (1 mg mL−1, 0.07 mmol L−1), the ionic liquid chol dhp stabilizer (12.5 mg mL−1, 62.14 mmol L−1), HEA (10 mg mL−1, 86.21 mmol L−1) and PEGDMA (0.5 mg mL−1, 0.67 mmol L−1) were added into the solution for preassembly. After 3 h, APS (1.25 mg mL−1, 5.48 mmol L−1) and 20 μL TEMED (0.39 mg mL−1, 3.33 mmol L−1) were then added under nitrogen protection to act as the initiator and accelerator, respectively, and the reaction was allowed to proceed for 24 h with shaking at room temperature. After polymerization, the resultant nanoparticles were washed with deionized water to remove unreacted monomers and oligomers, then washed repeatedly with 5% (v/v) acetic acid containing 5% (w/v) SDS solution to remove the embedded template. They were then re-washed several times with deionized water to remove any remaining acetic acid and SDS. Finally, TiO2@Lyz-MIPs was dried by lyophilization for further use. The Lyz-non-imprinted TiO2 nanoparticles (TiO2@Lyz-NIPs) control was prepared the same way and washed following the same procedure in the absence of the template. The detailed morphology of the imprinted nanoparticles was analyzed through scanning electron microscopy (FEI Co., USA). FT-IR spectra of TiO2 NPs, PDA-TiO2 NPs, and TiO2@Lyz-MIPs were performed on a Nicolet-6700 system (Thermo Nicolet Co., USA).2.3 Adsorption isotherms of TiO2@Lyz-MIPs
TiO2@Lyz-MIPs or TiO2@Lyz-NIPs (10 mg) were incubated in a solution of the protein (varying concentrations, 10 mL). After incubation, the solution was separated from TiO2@Lyz-MIPs or TiO2@Lyz-NIPs by centrifugation (6000 rpm), and the concentration of the protein in the solution was determined using a UV spectrophotometer. The amount of protein adsorbed on TiO2@Lyz-MIPs (or TiO2@Lyz-NIPs) was calculated according to the following equation| Q = (C0 − Ce)V/m |
Adsorption isotherm studies were conducted by incubating the TiO2@Lyz-MIPs or TiO2@Lyz-NIPs (10 mg) independently in solutions containing different concentrations of Lyz (initial concentration from 30 to 250 μg mL−1) at room temperature for 6 h. The amount of protein adsorbed by the nanoparticles was determined as described above.
2.4 Selective adsorption studies of TiO2@Lyz-MIPs
The specific recognition property of MIPs was evaluated using the imprinting factor (IF), defined as IF(α) = QMIP/QNIP where QMIP and QNIP are the adsorption capacity of the template of TiO2@Lyz-MIPs and TiO2@Lyz-NIPs, respectively.Selectivity of TiO2@Lyz-MIPs or TiO2@Lyz-NIPs toward Lyz was evaluated by comparing their ability to absorb the template protein with their ability to absorb the reference proteins. TiO2@Lyz-MIPs or TiO2@Lyz-NIPs (10 mg) was placed into a solution (10 mL) containing the template or reference protein(s) (250 μg mL−1). The solutions were shaken for 6 h at room temperature. The amount of protein adsorbed by the polymers was determined by monitoring the UV absorbance.
2.5 Competitive adsorption studies of TiO2@Lyz-MIPs
Competitive adsorption experiments for TiO2@Lyz-MIPs and TiO2@Lyz-NIPs were performed using a mixture of Lyz and BSA (Lyz 175 μg mL−1, BSA 175 μg mL−1) at 25 °C. TiO2@Lyz-MIPs or TiO2@Lyz-NIPs (6 mg) was placed into the mixed solution (6 mL) containing above two proteins (Lyz 175 μg mL−1, BSA 175 μg mL−1). The solutions were shaken for 6 h at 25 °C. After centrifugation, the supernatants were characterized using SDS polyacrylamide gel electrophoresis (SDS-PAGE).2.6 Separation of Lyz from diluted egg white solution
The imprinted TiO2@Lyz-MIPs were then applied in the separation of Lyz from diluted egg white solution. First, the Lyz source was prepared by separating the egg white from a fresh chicken egg and adding it to a PBS buffer to dilute it 32-fold. The mixture was centrifuged (12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm) at 4 °C for 15 min, after which the supernatant was collected as diluted egg white solution and stored at 4 °C. Second, the egg white solution was incubated with TiO2@Lyz-MIPs and TiO2@Lyz-NIPs in an ice bath for 6 h under gentle shaking, and the supernatants were collected by centrifugation (12
000 rpm) at 4 °C for 15 min, after which the supernatant was collected as diluted egg white solution and stored at 4 °C. Second, the egg white solution was incubated with TiO2@Lyz-MIPs and TiO2@Lyz-NIPs in an ice bath for 6 h under gentle shaking, and the supernatants were collected by centrifugation (12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm). Then, the diluted egg white solution and the supernatants were characterized using SDS-PAGE.
000 rpm). Then, the diluted egg white solution and the supernatants were characterized using SDS-PAGE.
3 Results and discussion
3.1 Preparation and characterizations of TiO2@Lyz-MIPs
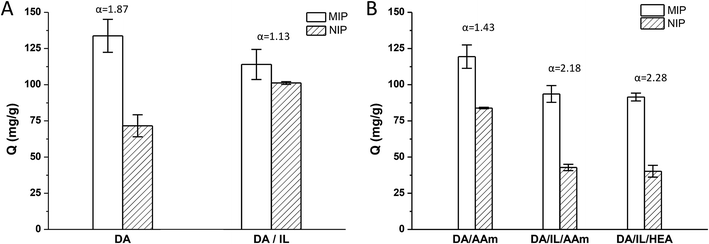
Recently, MIPs prepared by dopamine self-polymerization in aqueous conditions at room temperature were shown to be effective templates for imprinting fragile proteins and enzymes.22–24 In the present study, we used a similar method to prepare the Lyz-imprinted TiO2 nanoparticles. As shown in Fig. 1A, the imprinting factor was about 1.87. We then tried to use the chol dhp to stabilize the Lyz and hoped to improve the selectivity ability in polydopamine imprinting. However, contrary to expectations, the color of the reaction solution became much lighter, and the imprinting factor decreased to 1.13. This may be due to the formation of hydrogen bonds between phosphates of chol dhp and dopamine, which would inhibit the polymerization of dopamine.32It was reported that imprinted nanoparticles with high adsorption capacity and specific recognition ability could be obtained by radical-induced polymerization of low concentration monomers on vinyl-modified nanoparticles.33,34 So, in the following, we would like to combine the advantages of the two methods, using radical-polymerization of low concentration monomers with chol dhp as a stabilizer on polydopamine-modified TiO2 nanoparticles to prepare Lyz-imprinted TiO2 nanoparticles. On one hand, polydopamine-modified TiO2 would increase the interactions between TiO2 with the Lyz which might result in effective imprinting and selectivity of the achieved TiO2@Lyz-MIPs; on the other hand, the interactions between Lyz and functional monomer can be minimized by using chol dhp as stabilizer to ensure integrated conformation of Lyz to improve adsorption capacity and recognition ability.
Two polymerization systems were tested in this study including acrylamide/methylene bisacrylamide system (AAm/Bis) and hydroxyethyl acrylate/poly(ethylene glycol) dimethacrylate system (HEA/PEGDMA). All results were shown in Fig. 1B. The imprinting factor of the Lyz-imprinted TiO2 by AAm/Bis radical-polymerization on PDA-TiO2 (DA/AAm group) was 1.43, while the imprinting factor of the Lyz-imprinted TiO2 by AAm/Bis radical-polymerization with chol dhp as a stabilizer on PDA-TiO2 (DA/IL/AAm group) was increased to 2.18, which indicated that chol dhp could maintain Lyz stability and result in effective imprinting during AAm/Bis radical-polymerization of low concentration monomers. The Lyz-imprinted TiO2 by HEA/PEGDMA radical-polymerization were also prepared with chol dhp as a stabilizer on PDA-TiO2 (DA/IL/HEA group), and the imprinting factor was slightly increased to 2.28, perhaps due to the lower destabilization effect to the Lyz in HEA/PEGDMA system than that in AAm/Bis system.18
The imprinting capacity of the two groups were not much different, but considering the effect on the stability of the Lyz structure,21 HEA/PEGDMA system was selected as the optimal functional monomer and crosslinker to prepare the Lyz-imprinted TiO2 nanoparticles, as shown in Scheme 1. First, polydopamine-modified TiO2 nanoparticles were prepared through the self-polymerization of dopamine in Tris buffer (10 mmol L−1, pH = 8.5). Afterwards, the Lyz-imprinted TiO2 nanoparticles were prepared in PBS buffer by radical-polymerization of HEA/PEGDMA system with chol dhp as the stabilizer. Many factors during the imprinting procedure would affect the adsorption capacity, and in this study, the effects of different conditions including the feed amounts and the feed ratios of monomer, crosslinker, and the amounts of stabilizer “chol dhp” were investigated on the adsorption capacity of TiO2@Lyz-MIPs. The detailed preparation conditions and results were given in Table 1. As listed in Table 1, when the concentration of HEA, PEGDMA, and chol dhp was 10 mg mL−1 (86.21 mmol L−1), 0.5 mg mL−1 (0.67 mmol L−1), and 25 mg mL−1 (124.27 mmol L−1) respectively, the adsorption capacity of the TiO2@Lyz-MIPs (No. 1 in Table 1) was about 91.5 mg g−1, and the impacting factor was 2.28. Then referring to the reaction condition of No. 1 group, we found that the specific recognition properties of TiO2@Lyz-MIPs could be improved both by increasing the crosslinker concentration listed as No. 2, decreasing the concentration of ionic liquid listed as No. 3, and decreasing both the concentration of monomer and crosslinker without changing the feed ratio listed as No. 4, respectively. When the concentration of monomer was about 7 mg mL−1 (60.34 mmol L−1), the ratio of monomer to crosslinker was 20/1, the concentration of chol dhp was about 12.5 mg mL−1 (62.14 mmol L−1), the imprinting factor would be the highest at 4.40. The repeated preparation of TiO2@Lyz-MIPs (No. 5) was carried out for three times, and the adsorption amounts of TiO2@Lyz-MIPs were 91.5, 86.0, 87.3 mg g−1, the adsorption amounts of TiO2@Lyz-NIPs were 18.7, 15.8, 25.7 mg g−1, respectively. The results showed that the adsorption amount of Lyz on TiO2@Lyz-MIPs was stable, and the imprinting factor was also around 4, indicating that TiO2@Lyz-MIPs had good reproducibility.
| No. | HEA (mg mL−1)/(mmol L−1) | PEGDMA (mg mL−1)/(mmol L−1) | Chol dhp (mg mL−1)/(mmol L−1) | QMIP (mg g−1) | QNIP (mg g−1) | IF(α)c |
|---|---|---|---|---|---|---|
| a The Lyz concentration of all prepared samples is 1 mg mL−1. The results of each group were obtained from the average adsorption amounts of three parallel samples.b This group was imprinted by radical-polymerization of HEA/PEGDMA on the pure TiO2 nanoparticles.c IF(α): imprinting factor (α) = QMIP/QNIP. | ||||||
| 1 | 10/86.21 | 0.5/0.67 | 25/124.27 | 91.5 ± 2.7 | 40.2 ± 4.1 | 2.28 |
| 2 | 10/86.21 | 1.0/1.33 | 25/124.27 | 86.2 ± 11.4 | 29.5 ± 1.8 | 2.92 |
| 3 | 10/86.21 | 0.5/0.66 | 12.5/62.14 | 94.7 ± 1.6 | 23.4 ± 3.6 | 4.04 |
| 4 | 7/60.34 | 0.35/0.47 | 25/124.27 | 94.2 ± 15.2 | 30.5 ± 9.1 | 3.09 |
| 5 | 7/60.34 | 0.35/0.47 | 12.5/62.14 | 88.3 ± 2.9 | 20.0 ± 5.1 | 4.40 |
| 6b | 10/86.21 | 0.5/0.67 | 25/124.27 | 71.2 ± 7.3 | 36.6 ± 15.7 | 1.94 |
The adsorption capacity of TiO2@Lyz-MIPs (No. 6 in Table 1) imprinted by radical-polymerization of HEA/PEGDMA on pure TiO2 nanoparticles without polydopamine-modification was tested, and it was found that the imprinting factor was lower than that of the corresponding group (No. 1 in Table 1) prepared by radical-polymerization of HEA/PEGDMA on the PDA-TiO2 (1.94 vs. 2.28). This demonstrated that polydopamine-modification had a positive effect on the adsorption capacity as expected.
Table 2 compares the adsorption capacities of MIPs prepared on nano/micro-particles with reported different methods. Compared with other MIPs prepared from the same constituent monomers, TiO2@Lyz-MIPs by this new method showed relatively higher adsorption amounts and higher imprinting factor. This may be due to the immobilization of Lyz by PDA layer and the protection of ionic liquids on protein structure.
| Matrix | Template | Monomer | QMIP (mg g−1) | QNIP (mg g−1) | IF(α) | Ref. |
|---|---|---|---|---|---|---|
| a This work. | ||||||
| TiO2@PDA | Lyz | HEA/PEGDMA | 88.3 ± 2.9 | 20.0 ± 5.1 | 4.40 | a |
| PVP | Lyz | HEA/EGDMA | 43.2 ± 3.8 | 18.3 ± 3.4 | 2.36 | 21 |
| GO | AA | HEA | 6.2 | 1.4 | 4.43 | 35 |
| Fe3O4@SiO2 | Lyz | MBA/AAm/MAA | 17.1 ± 0.4 | 5.8 ± 0.3 | 2.95 | 36 |
| PCL | Lyz | MAA/DMAEMA | 835 ± 106 | 557 ± 19.6 | 1.50 | 37 |
FT-IR was then used to confirm the successful synthesis of Lyz-imprinted TiO2 nanoparticles. As shown in Fig. 2A, the presence of PDA-TiO2 was confirmed by the characteristic peaks of 2850–2950 cm−1 assigned to the asymmetric stretching vibration of O–H bonds on benzene rings, and the characteristics peaks of 1200–1600 cm−1 were assigned to the bending and shearing vibrations of N–H in polydopamine. Furthermore, the observed differences between FT-IR spectra of PDA-TiO2 and TiO2@Lyz-MIPs verified the existence of HEA chains with emergence of peaks at 1534 cm−1. All above results confirmed that TiO2@Lyz-MIPs were modified onto the titanium dioxide nanoparticles successfully. The morphological studies of TiO2, PDA-TiO2, TiO2@Lyz-MIPs and TiO2@Lyz-NIPs was examined by scanning electron microscopy (SEM) (Fig. 2B). Compared to the original TiO2, the PDA-TiO2, TiO2@Lyz-MIPs and TiO2@Lyz-NIPs showed no apparent increase in size.
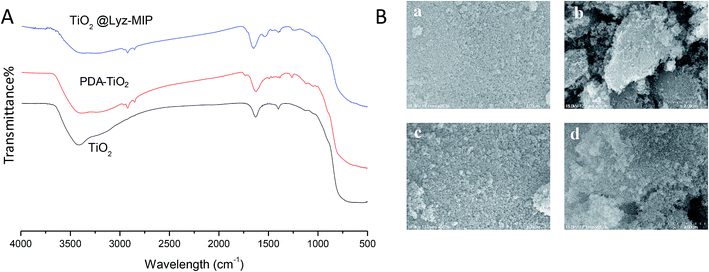 | ||
| Fig. 2 Characterizations of the imprinted nanoparticles. (A) FT-IR spectra; (B) SEM observations of TiO2 (a), PDA-TiO2 (b), TiO2@Lyz-MIPs (c) and TiO2@Lyz-NIPs (d). | ||
3.2 Adsorption properties of the Lyz-imprinted TiO2 nanoparticles
During the imprinting process, the template protein molecules were embedded in the polymerization layer, and then after the removal of these embedded protein molecules, the protein imprinted sites would be created. These sites would have a physical shape complementary to the template protein, and consequently can specifically recognize the template protein.38–40In this work, the adsorption kinetics and isotherms of TiO2@Lyz-MIPs (No. 5 in Table 1) and the corresponding TiO2@Lyz-NIPs were investigated to evaluate the adsorption capacity. As an important consideration in practical application for fast separation, the adsorption kinetics experiment was carried out using 250 μg mL−1 Lyz solutions on TiO2@Lyz-MIPs and TiO2@Lyz-NIPs at different time intervals. As presented in Fig. 3, the adsorption capacity of both imprinted TiO2@Lyz-MIPs and non-imprinted TiO2@Lyz-NIPs rapidly increased in the first 30 min, while the adsorption rate of TiO2@Lyz-MIPs is much higher than that of TiO2@Lyz-NIPs. This indicated that the interaction force between TiO2@Lyz-MIPs and Lyz was higher than that between TiO2@Lyz-NIPs and Lyz, which may be due to the existence of Lyz-shaped imprinted holes on the surface of titanium dioxide. After a period of adsorption, the adsorption rate of both imprinted TiO2@Lyz-MIPs and non-imprinted TiO2@Lyz-NIPs was decreased. However, increasing the extraction time still had a small-scale positive trend with adsorption capacity and almost reached equilibrium after 180 min. Adsorption capacity is a key factor to evaluate the selective binding and recognition ability of the imprinting effect on prepared materials. The adsorption isotherms on the TiO2@Lyz-MIPs and TiO2@Lyz-NIPs were then tested. As shown in Fig. 4, TiO2@Lyz-MIPs exhibited relatively higher adsorption capacity than that of TiO2@Lyz-NIPs within the concentration range covered, suggesting that TiO2@Lyz-MIPs had specific adsorption. When the initial concentration of Lyz was lower than 30 μg mL−1, the adsorption capacity gap was very small between TiO2@Lyz-MIPs and TiO2@Lyz-NIPs. And the special adsorption became nearly saturated when the concentration was 150 μg mL−1. The maximum adsorption capacity attained from TiO2@Lyz-MIPs reached the value of about 120 mg g−1, while the maximum adsorption capacity of TiO2@Lyz-NIPs was about 30 mg g−1. All of this indicated that TiO2@Lyz-MIPs had relatively high adsorption and the presence of a template in synthesis played a vital role in the formation of imprinted sites on the surface of TiO2@Lyz-MIPs.
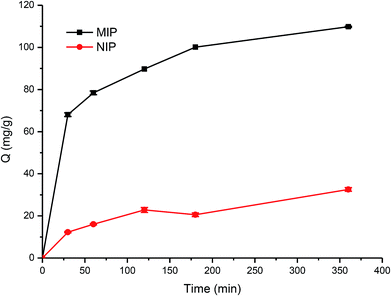 | ||
| Fig. 3 Absorption kinetic curve of Lyz onto TiO2@Lyz-MIPs and TiO2@Lyz-NIPs. Adsorption condition: V = 10 mL, mMIP = mNIP = 10 mg, CLyz = 250 μg mL−1, in water. | ||
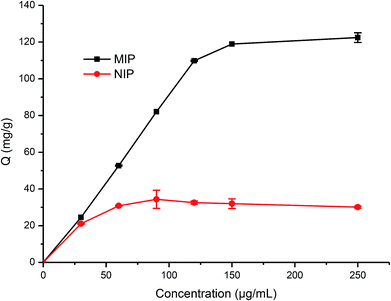 | ||
| Fig. 4 Absorption isotherm curve of Lyz onto TiO2@Lyz-MIPs and TiO2@Lyz-NIPs. Adsorption condition: V = 10 mL, mMIP = mNIP = 10 mg, CLyz = 30–250 μg mL−1, in water. | ||
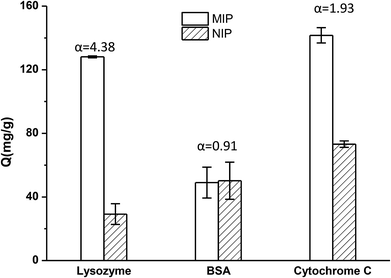
The selectivity tests of TiO2@Lyz-MIPs and TiO2@Lyz-NIPs were carried out by using BSA and cyt c as the competitor proteins. The initial protein concentration used for binding was 250 μg mL−1. The experimental result was shown in Fig. 5. From this, we found that the TiO2@Lyz-MIPs exhibited a much higher imprinting factor of Lyz (αLyz = 4.38) with respect to the control TiO2@Lyz-NIPs. The former also exhibited a higher imprinting factor than that of BSA (αBSA = 0.91) and cyt c (αcyt c = 1.93). In addition, the separation factor (β), calculated as αLyz/αBSA and αLyz/αcyt c, was 4.81 of BSA and 2.27 of cyt c. All of these results indicated that the imprinted TiO2@Lyz-MIPs had recognition sites on the surface of TiO2 nanoparticles, which were capable of selectively binding Lyz.
To further compare the selectivity of TiO2@Lyz-MIPs and TiO2@Lyz-NIPs, separation of Lyz from protein mixture (containing 175 μg mL−1 Lyz and 175 μg mL−1 BSA) was performed. The protein mixture after treatment with TiO2@Lyz-MIPs and TiO2@Lyz-NIPs was analysed by SDS-PAGE (Fig. 6). The two bands in lane 2 belong to Lyz and BSA, respectively. After treatment with TiO2@Lyz-MIPs, the band of Lyz disappeared obviously and the band of BSA had little change (lane 3), suggesting most of Lyz was captured by TiO2@Lyz-MIPs. Different from lane 3, the band of Lyz still could observed in lane 4, indicating TiO2@Lyz-NIPs had no selectivity toward Lyz.
3.3 Separation of Lyz from egg white using TiO2@Lyz-MIPs
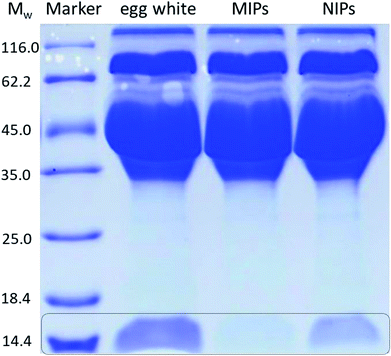
In order to evaluate the potential use of TiO2@Lyz-MIPs, the separation of Lyz directly from diluted chicken egg white, a sample of complex protein mixture, was tested. The results of the SDS-PAGE were shown in Fig. 7. Compared with the 32-fold diluted egg white samples (lane 2), the intensity of Lyz bands at Mw of 14.4 kDa faded after being treated with TiO2@Lyz-MIPs (lane 3) and TiO2@Lyz-NIPs (lane 4). However, the intensity of Lyz bands was much lighter after being treated with TiO2@Lyz-MIPs compared with that treated with TiO2@Lyz-NIPs, meaning that markedly more Lyz was adsorbed by TiO2@Lyz-MIPs. This implies that Lyz could be successfully separated from a protein mixture as complex as egg white by TiO2@Lyz-MIPs. Therefore, TiO2@Lyz-MIPs may have good imprinting effect and considerable selectivity for actual protein separation applications.4 Conclusions
In this work, an effective surface Lyz imprinted TiO2@Lyz-MIPs for selective separation of Lyz have been successfully developed by radical-polymerization of HEA/PEGDMA on the polydopamine-modified surface of TiO2 with ionic liquid “chol dhp” as a stabilizer to Lyz. The combination of ionic liquid and surface molecular imprinting technology could overcome the difficulty caused by protein structural variability, and achieve efficient recognition in protein separation process. The molecular imprinting conditions were optimized. It was found that polydopamine-modification would have a positive effect on the adsorption capacity and that using of chol dhp as stabilizer could also result in effective imprinting. When the concentration of HEA was 7 mg mL−1 (60.34 mmol L−1), the ratio of monomer to crosslinker was 20/1, the concentration of chol dhp was 12.5 mg mL−1 (62.14 mmol L−1), the imprinting factor was the highest at 4.40. FT-IR measurement confirmed the successful synthesis of TiO2@Lyz-MIPs. The adsorption capacity of TiO2@Lyz-MIPs rapidly increased in the first 30 min, meanwhile, the adsorption rate of TiO2@Lyz-MIPs was much higher than that of TiO2@Lyz-NIPs. TiO2@Lyz-MIPs exhibited relatively high adsorption capacity within the concentration range from 30 μg mL−1 to 250 μg mL−1. The maximum adsorption capacity of TiO2@Lyz-MIPs could reach to 120 mg g−1, which was more than four times higher than that of TiO2@Lyz-NIPs. The TiO2@Lyz-MIPs was also able to separate Lyz from the diluted egg white. All these results demonstrated that recognition cavities have been formed on the surface of TiO2@Lyz-MIPs, and this method would propose a good opportunity to promote the development of molecular imprinting technique for protein.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors are grateful to the funding support from the National Natural Science Foundation of China (81471790, 81871805), Jiangsu Provincial Special Program of Medical Science (BL2012004), Jiangsu Provincial Clinical Orthopedic Center, the Priority Academic Program Development (PAPD) of Jiangsu Higher Education Institutions, and the Key Laboratory of Stem Cells and Biomedical Materials of Jiangsu Province and Chinese Ministry of Science and Technology.Notes and references
- Q. Yang, J. H. Li, X. Y. Wang, H. L. Peng, H. Xiong and L. X. Chen, Biosens. Bioelectron., 2018, 112, 54–71 CrossRef CAS PubMed.
- Y. Saylan, F. Yilmaz, E. Ozgur, A. Derazshamshir, H. Yavuz and A. Denizli, Sensors, 2017, 17, e898 CrossRef PubMed.
- M. L. Yola and N. Atar, Curr. Anal. Chem., 2017, 13, 13–17 CrossRef CAS.
- D. Bitas and V. Samanidou, Molecules, 2018, 23, e316 CrossRef PubMed.
- M. Rutkowska, J. Plotka-Wasylka, C. Morrison, P. P. Wieczorek, J. Namiesnik and M. Marc, TrAC, Trends Anal. Chem., 2018, 102, 91–102 CrossRef CAS.
- C. L. Wang, X. L. Hu, P. Guan, D. F. Wu, L. W. Qian, J. Li and R. Y. Song, J. Pharm. Biomed. Anal., 2015, 102, 137–143 CrossRef CAS.
- X. C. Guo, Z. Y. Xia, H. H. Wang, W. Y. Kang, L. M. Lin, W. Q. Cao, H. W. Zhang and W. H. Zhou, Talanta, 2017, 166, 101–108 CrossRef CAS.
- K. G. Yang, L. H. Zhang, Z. Liang and Y. K. Zhang, Anal. Bioanal. Chem., 2012, 403, 2173–2183 CrossRef CAS.
- G. Q. Pan, Q. P. Guo, C. B. Cao, H. L. Yang and B. Li, Soft Matter, 2013, 9, 3840–3850 RSC.
- G. Q. Pan, Q. P. Guo, Y. Ma, H. L. Yang and B. Li, Angew. Chem., Int. Ed., 2013, 52, 6907–6911 CrossRef CAS PubMed.
- L. W. Qian, X. L. Hu, P. Guan, D. Wang, J. Li, C. B. Du, R. Y. Song, C. L. Wang and W. Q. Song, Anal. Chim. Acta, 2015, 884, 97–105 CrossRef CAS PubMed.
- X. M. Zhang, W. M. Yang, N. W. Wang, X. N. Ni, H. J. Wang, Z. Y. Gu, X. Y. Wu and W. Z. Xu, Adv. Polym. Technol., 2017, 36, 220–229 CrossRef CAS.
- M. Jia, J. Yang, Y. K. Sun, X. Bai, T. Wu, Z. S. Liu and H. A. Aisa, Anal. Bioanal. Chem., 2018, 410, 595–604 CrossRef CAS PubMed.
- R. Viveiros, S. Rebocho and T. Casimiro, Polymers, 2018, 10, e306 CrossRef PubMed.
- Y. Akdogan, M. J. N. Junk and D. Hinderberger, Biomacromolecules, 2011, 12, 1072–1079 CrossRef CAS PubMed.
- T. Vasantha, A. Kumar, P. Attri, P. Venkatesu and R. S. R. Devi, Protein Pept. Lett., 2014, 21, 15–24 CrossRef CAS.
- M. Bisht and P. Venkatesu, New J. Chem., 2017, 41, 13902–13911 RSC.
- K. Fujita, D. R. MacFarlane and M. Forsyth, Chem. Commun., 2005, 4804–4806 RSC.
- K. Fujita and H. Ohno, Biopolymers, 2010, 93, 1093–1099 CrossRef CAS PubMed.
- K. Fujita, K. Murata, M. Masuda, N. Nakamura and H. Ohno, RSC Adv., 2012, 2, 4018–4030 RSC.
- L. W. Qian, X. L. Hu, P. Guan, B. Gao, D. Wang, C. L. Wang, J. Li, C. B. Du and W. Q. Song, Anal. Bioanal. Chem., 2014, 406, 7221–7231 CrossRef CAS PubMed.
- N. Zhang, X. Hu, P. Guan, C. Du, J. Li, L. Qian, X. Zhang, S. Ding and B. Li, Chem. Eng. J., 2017, 317, 356–367 CrossRef CAS.
- Z. W. Xia, Z. A. Lin, Y. Xiao, L. Wang, J. N. Zheng, H. H. Yang and G. N. Chen, Biosens. Bioelectron., 2013, 47, 120–126 CrossRef CAS PubMed.
- G. D. Riveros, K. Cordova, C. Michiels, H. Verachtert and G. Derdelinckx, Talanta, 2016, 160, 761–767 CrossRef CAS PubMed.
- H. J. Li, X. N. Wang, Z. R. Wang, Y. Wang, J. D. Dai, L. Gao, M. B. Wei, Y. S. Yan and C. X. Li, Microchim. Acta, 2018, 185, e193 CrossRef PubMed.
- N. Li, Y. J. Liu, F. Liu, M. F. Luo, Y. C. Wan, Z. Huang, Q. Liao, F. S. Mei, Z. C. Wang, A. Y. Jin, Y. Shi and B. Lu, Acta Biomater., 2017, 51, 175–183 CrossRef CAS PubMed.
- Y. Liu, J. L. Zhu, X. Liu and H. X. Li, RSC Adv., 2016, 6, 69326–69333 RSC.
- J. M. George, A. Antony and B. Mathew, Microchim. Acta, 2018, 185, e358 CrossRef PubMed.
- C. Lai, X. Zhou, D. Huang, G. Zeng, M. Cheng, L. Qin, H. Yi, C. Zhang, P. Xu, C. Zhou, R. Z. Wang and C. Huang, J. Taiwan Inst. Chem. Eng., 2018, 91, 517–531 CrossRef CAS.
- J. Luo, Q. Ma, W. Wei, Y. Zhu, R. Liu and X. Y. Liu, ACS Appl. Mater. Interfaces, 2016, 8, 21028–21038 CrossRef CAS PubMed.
- J. Wackerlig and R. Schirhagl, Anal. Chem., 2016, 88, 250–261 CrossRef CAS PubMed.
- F. Ponzio, P. Bertani and V. Ball, J. Colloid Interface Sci., 2014, 431, 176–179 CrossRef CAS PubMed.
- Y. Liu, Y. Gu, M. Li and Y. Wei, New J. Chem., 2014, 38, 6064–6072 RSC.
- X. M. Deng, C. Y. Chen, J. F. Xie, C. Q. Cai and X. M. Chen, RSC Adv., 2016, 6, 43223–43227 RSC.
- J. Ji, X. L. Sun, X. M. Tian, Z. J. Li and Y. Z. Zhang, Anal. Lett., 2013, 46, 969–981 CrossRef CAS.
- Y. X. Wang, Z. H. Chai, Y. J. Sun, M. Gao and G. Q. Fu, J. Biomater. Sci., Polym. Ed., 2015, 26, 644–656 CrossRef CAS PubMed.
- H. R. Culver, S. D. Steichen and N. A. Peppas, Biomacromolecules, 2016, 17, 4045–4053 CrossRef CAS PubMed.
- A. Bossi, F. Bonini, A. P. F. Turner and S. A. Piletsky, Biosens. Bioelectron., 2007, 22, 1131–1137 CrossRef CAS PubMed.
- N. M. Bergmann and N. A. Peppas, Prog. Polym. Sci., 2008, 33, 271–288 CrossRef CAS.
- L. X. Chen, X. Y. Wang, W. H. Lu, X. Q. Wu and J. H. Li, Chem. Soc. Rev., 2016, 45, 2137–2211 RSC.
Footnote |
| † These authors contributed equally to this study. |
| This journal is © The Royal Society of Chemistry 2019 |