Open Access Article
Open Access ArticleComparative study of antidiabetic, bactericidal, and antitumor activities of MEL@AgNPs, MEL@ZnONPs, and Ag–ZnO/MEL/GA nanocomposites prepared by using MEL and gum arabic
Abdelmoneim Bakur ab,
Tarig Elshaaranic,
Yongwu Niua and
Qihe Chen*a
ab,
Tarig Elshaaranic,
Yongwu Niua and
Qihe Chen*a
aDepartment of Food Science and Nutrition, Zhejiang University, Hangzhou 310058, China. E-mail: chenqh@zju.edu.cn
bDepartment of Food Sciences and Technology, University of Kordofan, El Obeid 51111, Sudan
cCollege of Chemical and Biological Engineering, Zhejiang University, Hangzhou 310027, China
First published on 27th March 2019
Abstract
In this study, a variety of nanocomposites, namely, MEL@AgNPs, MEL@ZnONPs, and Ag–ZnO/MEL/GA were biosynthesized using MEL and gum arabic to serve in biomedical applications. The synthesized nanocomposites were examined using X-ray diffraction (XRD), transmission electron microscopy (TEM), scanning electron microscopy (SEM), and FTIR spectroscopy. The physicochemical properties and biomedical activities of the synthesized nanocomposites were investigated. The Ag–ZnO/MEL/GA nanocomposites showed greater antidiabetic activity against α-amylase and α-glucosidase, and higher antibacterial activity compared to MEL@AgNPs and MEL@ZnONPs. Furthermore, HepG2 cells were exposed to MEL@AgNPs, MEL@ZnONPs, and Ag–ZnO/MEL/GA nanocomposites for 24 h and their IC50 values were 63.25, 26.91 and 28.97 μg mL−1 (P < 0.05), respectively. According to this comparative study, it is apparent that the Ag–ZnO/MEL/GA nanocomposites have a great potential to serve as antitumor agents against HepG2, and antidiabetic and antibacterial agents.
1. Introduction
Recently, the number of infections of pathogenic bacteria has increased and become a major health concern. The resistance of bacteria to antibiotics, and the appearance of new bacterial mutations, as well as a lack of drugs in underdeveloped countries, raise the demand for an effective and eco-friendly bactericidal agent.1,2 Resistance free bactericidal agents would be beneficial, particularly for foodborne diseases such as Escherichia coli, Staphylococcus aureus, Pseudomonas aeruginosa, Salmonella types, and Clostridium perfringens. Another major health concern is diabetes, which is a set of metabolic dysfunctions due to either defects in resistance or secretion of insulin. This disease causes the blood's glucose level to elevate; currently more than 387 million adults are affected in the worldwide.3 Nowadays, diabetes treatment is extensively studied using nanomaterials. They are small, biocompatible and have a high drug loading capacity and efficiency.4,5 Therefore, the development of new nanomedicine is increasingly required to inhibit carbohydrate-hydrolysing enzymes with minimum side effects.6 Another disease that can be treated using nanocomposites is cancer, which is a multifunctional disease which occurs when the proliferation of cells becomes uncontrolled. Cancer is one of the major causes of death throughout the world. Hepatocellular carcinoma (HCC) which is a kind of liver cancer has become one of the top four aggressive tumors around the globe.7,8 Therefore, new treatment of these diseases is highly demanded.Nanocomposites are materials that can be produced by incorporating two or more nano-fillers. In recent years they have attracted much attention due to their excellent multifunctional attributes and improved properties over a monometallic NPs.9 In this regard, composites containing both noble metal NPs and metal oxide are more attractive and have great potential in the biomedical domains. Among them, ZnONPs have gained more attention due to its functional properties. The photocatalytic and antimicrobial activities of ZnONPs can be enhanced through doping with Ag to produce Ag–ZnO.10 The interaction between ZnO and Ag in the Ag–ZnO composites provides an excellent inhibition of the bacterial growth.11,12 The properties of nanocomposites depend on their size, shape, interfacial characteristics, as well as the nature of nano-fillers.13,14 In line with this, biosurfactants have been emerged as an alternative green materials for the synthesis of NPs compared to the physiochemical and biological processes, biosurfactants are biodegradable, stable, less toxic, uniform in shape, and have improved biomedical activities of metallic NPs.15–19
In the current study, we used promising microbial surfactants, mannosylerythritol lipids (MEL), because of their various potential bioactivities, such as antimicrobial activity, antioxidant, interfacial, and antitumor properties. MEL has been used in many fields, e.g. pharmaceutical, environmental, and cosmetic.20–24 MEL has also gained particular attention due to their behavior of self-assembling properties could be utilized in gene transfection and drug delivery.25 As evident from previous reports, biosurfactants have ability to mediate the synthesis of NPs as both reducing and stabilizing agents, such as rhamnolipid, sophorolipids, and lipopeptide biosurfactant.18,26,27 Gum arabic (GA, E-414) is an edible and dried exudation obtained from the stems and branches of Acacia senegal. It is widely used as a thickening, emulsifier and stabilizer agent for food, pharmaceutical and cosmetics industries.28 GA also has been extended to use in the nanomedicine domains, because of its biocompatible for in vivo applications and excellent stabilization of nanomaterials.29
In the last decade, many methods and techniques are developed to fabricate nanocomposites using biosurfactants or biopolymer ingredients to obtain a unique multifunctional. Notable examples include sol-cast transformation method, which was used to synthesize GC/PEG/ZnO/Ag nanocomposites films.30 Also, the deposition–precipitation method was used to synthesize chitosan–Ag/ZnO nanocomposites.31 Solution casting is another method of preparing nanocomposites. Ag-doped ZnO as nanofiller with cashew gum and chitin was prepared through this method.32 Sol–gel technique was also used to prepare ZnONPs using Pseudomonas aeruginosa rhamnolipids.33 Also, the electrospinning process was employed to produce Ag–ZnO bimetallic nanoencapsulated in PVP/PCL nanofibres.34
Here, we focused on developing a novel, facile, and eco-friendly nanocomposites consisting of Ag and ZnO NPs coated by biopolymers (gum arabic) and biosurfactants (MEL) to serve as antidiabetic, antitumor, and bactericidal agents. To best of our knowledge, there was no study available on the synthesis of ZnO and Ag NPs using the mannosylerythritol lipids (MEL).
2. Experimental details
2.1. Materials
Gum arabic (GA, from acacia tree), a mannosylerythritol lipid (MEL) was provided by fermentation engineering lab, Dep. of Food Science and Nutrition, Zhejiang University, China. Zinc acetate dehydrate (Zn(CH3COO)2·2H2O), and silver nitrate (AgNO3) were purchased from Sinopharm Chemical Reagent Co., Ltd. China. All the other reagents were utilized as received at analytical reagent grade.2.2. Synthesis of nanocomposites
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 using magnetic stirring. After that, 0.1 M solution of KOH was added dropwise to the mixture under continuous stirring at ≥90 °C for 4 h. At last, the nanocomposites was thoroughly washed with distilled water and ethanol, and calcined at 500 °C for 2 h.
1 using magnetic stirring. After that, 0.1 M solution of KOH was added dropwise to the mixture under continuous stirring at ≥90 °C for 4 h. At last, the nanocomposites was thoroughly washed with distilled water and ethanol, and calcined at 500 °C for 2 h.2.3. Characterization of nanocomposites
FTIR spectra of the MEL@AgNPs, MEL@ZnONPs, and Ag–ZnO/MEL/GA nanocomposites were analyzed using FTIR spectrophotometer model (Vector 22, Bruker, Germany) in the range of 4000–500 cm−1. The crystallinity of the prepared nanocomposites was investigated via Siemens X-ray diffraction (XRD) analysis. Dried powder of nanocomposites was drop-coated onto glass slide, and the XRD pattern was recorded in a range of 20–80° at 2θ angle. The machine voltage was 45 keV, the current was 20 mA, and Cu-Kα radiation was used as an X-ray source. Furthermore, the structural features of the samples were investigated using transmission electron microscopy (TEM) (JEM-1230, JEOL, Akishima, Japan). The suspended samples were dropped into carbon-coated copper grids, and the excess samples were removed from the grid by using a cone of a blotting paper. The prepared thin film was reserved in a grid box sequentially. Moreover, the nanocomposites morphology was analyzed using scanning electron microscopic (SEM) (TM-1000, Hitachi, Japan). Thin films were prepared on a carbon-coated copper grid by dropping the samples onto the grid's surface, and the excess solution was removed using a blotting paper. Then, the films were dried under a mercury lamp for 5 min.2.4. Antidiabetic activity
2.5. Bactericidal efficacy test
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm, 8 min), and rinsed twice with PBS. About 2.5% of glutaraldehyde solution was utilized for cells fixation and dehydrated serially with an ethanol solution of 30%, 50%, 70%, 85%, 90%, and 100% concentration. After dehydration, the bacterial cells were dried with a critical point dryer, fixed on SEM support, sputtered with gold coating and observed in SEM (SEM, Model SU-8010, Hitachi, Japan). Furthermore, the TEM analysis was used to explore the bactericidal mechanism of the nanocomposites on E. coli cells. Thus, the fixation and dehydration steps were performed similarly to SEM steps. Then, a sequence of pre-treatment processes for the TEM analysis described by ref. 37 with slight medications were done, and the cells were observed under TEM (JEM-1230, JEOL, Akishima, Japan).
000 rpm, 8 min), and rinsed twice with PBS. About 2.5% of glutaraldehyde solution was utilized for cells fixation and dehydrated serially with an ethanol solution of 30%, 50%, 70%, 85%, 90%, and 100% concentration. After dehydration, the bacterial cells were dried with a critical point dryer, fixed on SEM support, sputtered with gold coating and observed in SEM (SEM, Model SU-8010, Hitachi, Japan). Furthermore, the TEM analysis was used to explore the bactericidal mechanism of the nanocomposites on E. coli cells. Thus, the fixation and dehydration steps were performed similarly to SEM steps. Then, a sequence of pre-treatment processes for the TEM analysis described by ref. 37 with slight medications were done, and the cells were observed under TEM (JEM-1230, JEOL, Akishima, Japan).2.6. Antitumor activity
The MTT assay was used to investigate the inhibition activity of the biosynthesized nanocomposites against HepG2 cells. HepG2 cells were obtained from Shanghai Cell Bank of China and maintained in a humidified incubator 5% CO2 at 37 °C, in DMEM (Dulbecco's Modification Eagle Medium) supplemented with 10% fetal bovine serum (FBS) and 1% antibiotics solution (100 U per mL penicillin and 100 μg per mL streptomycin). The HepG2 cells were placed into 96-well plates at a density of 1.25 × 105 cells per well and incubated for 24 h. After incubation, the cells were treated with the MEL@AgNPs, MEL@ZnONPs, and Ag–ZnO/MEL/GA nanocomposites at different concentrations (5–80 μg mL−1). The untreated cells were used as a control. The proliferation activity of the cells was determined by adding 5.0 mg per mL MTT reagent after 24 h of incubation. Finally, the absorbance was recorded at 570 nm in a microtitre plate reader (Thermo Electron Corp, Asheville, NC). The viability of HepG2 cells was expressed as a percentage of the control culture value, which was considered 100% viable.2.7. Statistical analysis
Statistical analysis was conducted using SPSS (version 19, USA). The data were expressed as a standard deviation (mean ± SD). The statistical significance of differences between groups was assessed using one-way analysis of variance (ANOVA). A significant difference was considered at (p < 0.05).3. Results and discussion
Bearing in mind the need for greener bioprocess and novel enhancers for the synthesis nanomaterials using the microbial method. The advantages of using MEL over the surfactants due to their biocompatibility, excellent surface activity, diverse biochemical functions, and its wide range of applications.38 MEL can be utilized as a vehicle for the gene, and drug delivery is owing to their capability to form stable vesicles or nanostructures, which are employed as models for the cellular and molecular process. Also, the efficiency of gene transfection into mammalian cells has been increased considerably by the cationic liposome bearing MEL.20 Additionally, gums are emerging as biodegradable substances, which have been used as delivery systems.39 Based on this approach, we have prepared Ag, ZnO and Ag–ZnO nanocomposites by using biocompatible materials MEL and GA.3.1. Characterization of nanocomposites
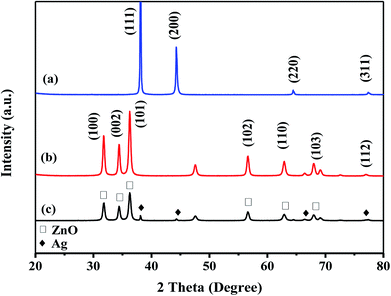
The crystalline structure of Ag, ZnO, and Ag–ZnO/MEL/GA nanocomposites was identified by X-ray diffraction technique (Fig. 1). The XRD analysis of MEL@AgNPs revealed four diffraction peaks at 2θ = 37.9°, 44.17°, 64.32° and 77.42° that associated to the (111), (200), (220) and (311) planes of the crystal lattice, respectively (Fig. 1a). Similar diffraction peaks were obtained by Sorbiun et al.40 XRD diffraction peaks of this nanocomposite emphasized the face-centered cubic (fcc) and the crystalline structure of AgNPs fabricated by MEL. Further, the diffraction peaks of MEL@ZnONPs at 2θ = 31.60°, 34.46°, 35.88°, 47.46°, 56.23°, 62.74° and 68.03° were corresponded to (100), (002), (101), (102), (110), (103) and (112), respectively. These peaks confirmed the hexagonal wurtzite structure of ZnO (Fig. 1b). The results are consistent with the previously reported studies.41 Eventually, Fig. 1c showed the XRD diffraction patterns of Ag–ZnO/MEL/GA nanocomposites. The diffraction peaks that designed with “♦” are attributed to AgNPs, while the diffraction peaks marked with “□” are related to the hexagonal wurtzite structure of ZnONPs. These diffraction peaks revealed that there is no notable shift in their position, and both the Ag and ZnO nanoparticles were successfully incorporated in the nanocomposites and attached to the surface of GA and MEL. The average crystallite size of Ag, ZnO and Ag–ZnO/MEL/GA nanocomposites was 43.14, 29.88, and 17.01 nm, respectively, obtained by the Scherrer formula.33,42,43 Generally, the NPs obtained by MEL are almost similar to those obtained by other biosurfactants and slightly larger than those obtained by chemical surfactants.17,44–46 The intensity of the diffraction peaks of ZnO in the nanocomposites became wider and weaker than that of pure ZnO. This was attributed to the decrement in the crystal size of ZnO when the ratio of Ag increased as the Ag could encapsulate ZnO and thus reduces the growth of ZnO crystal. A comparable phenomenon was stated in the earlier studies.42,47The FTIR spectra were recorded to verify the possible interactions of MEL in MEL@AgNPs, MEL@ZnONPs and Ag–ZnO/MEL/GA nanocomposites. Fig. 2 exhibited stretching vibrations of MEL at 3374, 2945, 1453, and 724 cm−1 which are correspond to (O–H), (C–H), (C–H) and (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) absorption bands, respectively. These bands are consistent with other's work.48 The absorption bands at 1453 and 2945 cm−1 confirmed the presence of the fatty acid chains. The bands of MEL in Ag and ZnO nanoparticles were shifting remarkably, which might be due to the reduction of Ag and ZnO.49 GA displayed notable stretching vibration bands at 3386, 2920 and 1420 cm−1 that correspond to O–H stretching vibration of polymer, C–H stretching of CH2 group and the deformations of the CH2 group, respectively. These results are in a good agreement with other reports.50–53 Finally, the absorption bands of Ag–ZnO/MEL/GA nanocomposites showed a minor shift and a low peak intensity compared to GA and MEL, which was ascribed to the strong intermolecular interactions between the functional groups in GA and MEL with Ag and ZnO NPs. Similar findings were also observed in the synthesized of ChNW/ZnO–Ag nanocomposites.42
O) absorption bands, respectively. These bands are consistent with other's work.48 The absorption bands at 1453 and 2945 cm−1 confirmed the presence of the fatty acid chains. The bands of MEL in Ag and ZnO nanoparticles were shifting remarkably, which might be due to the reduction of Ag and ZnO.49 GA displayed notable stretching vibration bands at 3386, 2920 and 1420 cm−1 that correspond to O–H stretching vibration of polymer, C–H stretching of CH2 group and the deformations of the CH2 group, respectively. These results are in a good agreement with other reports.50–53 Finally, the absorption bands of Ag–ZnO/MEL/GA nanocomposites showed a minor shift and a low peak intensity compared to GA and MEL, which was ascribed to the strong intermolecular interactions between the functional groups in GA and MEL with Ag and ZnO NPs. Similar findings were also observed in the synthesized of ChNW/ZnO–Ag nanocomposites.42
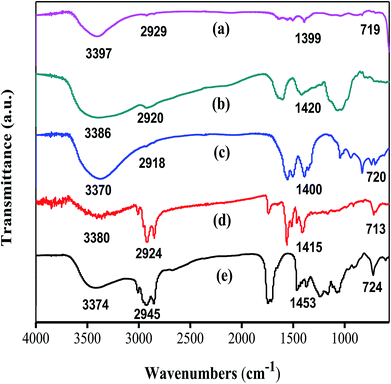 | ||
| Fig. 2 FTIR spectra of (a) MEL@ZnONPs (b) gum arabic (c) Ag–ZnO/MEL/GA nanocomposite (d) MEL@AgNPs (e) MEL. | ||
Fig. 3a–c showed the SEM images of the morphology of MEL@AgNPs, MEL@ZnONPs, and Ag–ZnO/MEL/GA nanocomposites. SEM micrographs illustrated that both the Ag and ZnO NPs samples are spherical in shape, uniform and dispersed with small size.40,54 Besides, the morphology of Ag–ZnO/MEL/GA nanocomposites exhibited a homogenous structure, smooth surface with slight agglomeration.55,56 The Ag and ZnO NPs are uniformly distributed on the surface of GA and MEL. This could be due to the role of the mannosylerythritol lipids (MEL) on the morphology of the nanoparticles.
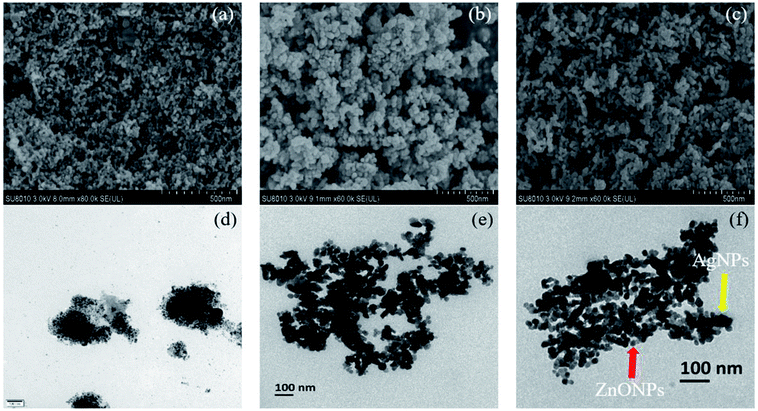 | ||
| Fig. 3 SEM (a–c) and TEM (d–f) micrographs of MEL@AgNPs, MEL@ZnONPs, and Ag–ZnO/MEL/GA nanocomposite, respectively. | ||
Additionally, the TEM images of MEL@AgNPs, MEL@ZnONPs, and Ag–ZnO/MEL/GA nanocomposites are shown in Fig. 3d–f. The images confirmed the spherical shape of nanocomposites. The average size the pure ZnO was 29.88 nm, while that of Ag–ZnO/MEL/GA nanocomposites was 17.01 nm. The small size of Ag–ZnO/MEL/GA nanocomposites was due to the increase in the Ag ratio.
3.2. Inhibition of α-amylase and α-glucosidase
Diabetes mellitus (DM) is multifunctional metabolic disorder characterized by inadequate secretion or weakened action of insulin resulting in high-level of blood sugar (hyperglycemia). The digestive enzymes (α-amylase and α-glucosidase) play an essential role in the digestion of carbohydrate.57 In this study, the digestive enzymes α-amylase and α-glucosidase were used to assess the antidiabetic efficacy of the prepared MEL@AgNPs, MEL@ZnONPs, and Ag–ZnO/MEL/GA nanocomposites. As shown in Fig. 4, all three samples demonstrated significant inhibition in a concentration-dependent reduction for both digestive enzymes.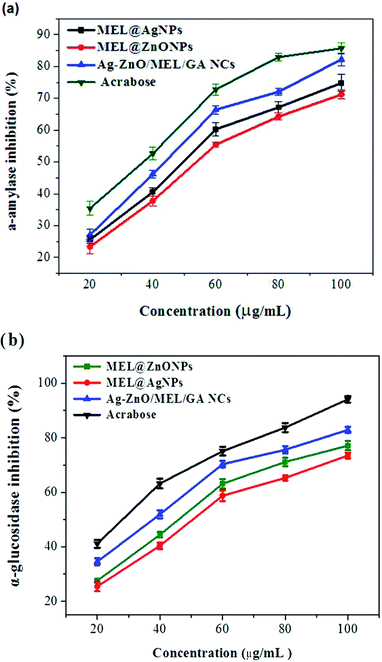 | ||
| Fig. 4 Antidiabetic efficacy of the prepared MEL@AgNPs, MEL@ZnONPs and Ag–ZnO/MEL/GA nanocomposite based on inhibition of α-amylase and α-glucosidase. | ||
In the case of α-amylase inhibitory activity, Fig. 4a illustrates the IC50 values of Ag, ZnO, and Ag–ZnO/MEL/GA nanocomposites, which were found to be 54, 49, and 43 μg mL−1, respectively. While in α-glucosidase inhibitory activity, these were found to be 51, 46, and 37, respectively (Fig. 4b). These results are consistent with other studies.58–60 It has been found that the Ag–ZnO/MEL/GA nanocomposites show a potent antidiabetic activity in comparison with Ag and ZnO NPs, whereas ZnONPs revealed marginally higher activity compared with AgNPs in inhibition of α-amylase and α-glucosidase. Several publications have documented that the SPB1 biosurfactant could be a promising candidate for the treatment of diabetes in addition to delaying the later complications from it. The SPB1 biosurfactant has the ability to reduce the blood glucose level due to the efficiency of modulating the immune system leading to the decrease of β-cells damages. In diabetic group, lipid droplets were observed in the cytoplasm of hepatic and renal tissues, due to the lack of insulin responsible for metabolizing fats.61 The chemical surfactants are usually used to enhance the solubility and delivery of antidiabetic drugs such as SDS, CTAB and Tween-80.62 Also, Edwards and his colleagues63 suggested that using different kind of gums could be effective in reducing postprandial hyperglycemia in human. Furthermore, zinc is one of the essential trace elements acting as insulin in the human body and enhances glucose consumption.64,65 From the prior results, it can be concluded that the use of Ag–ZnO/MEL/GA nanocomposites, biosynthesized using the MEL and gum arabic, will be considerably helpful to inhibit the carbohydrate-digesting enzymes and could demonstrate an efficient way to control diabetes.
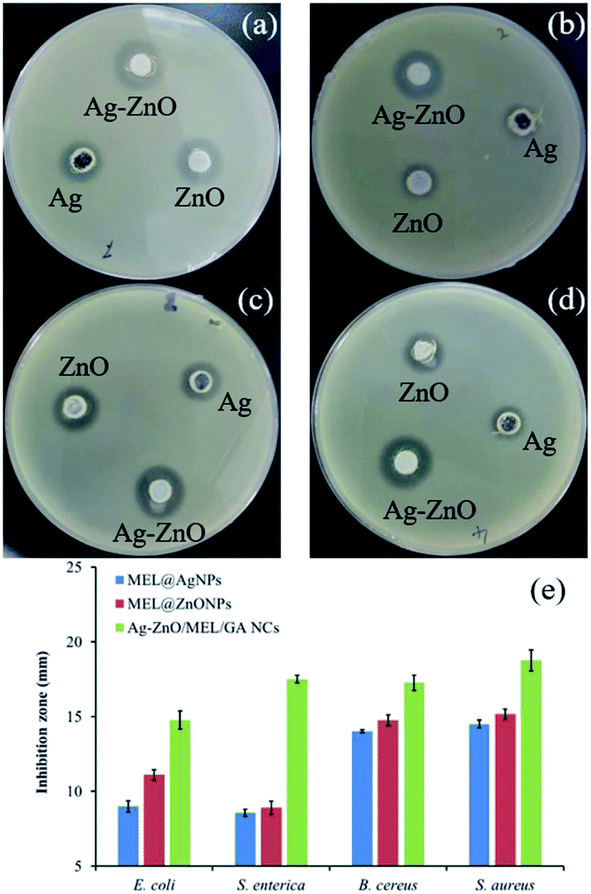
3.3. Bactericidal potential studies
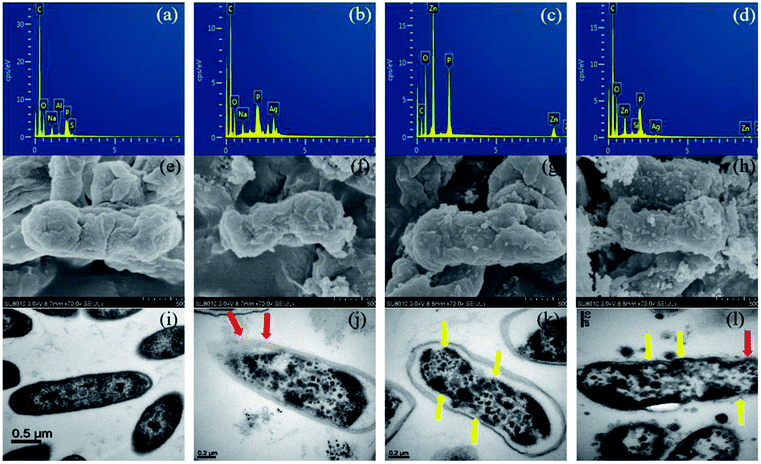 | ||
| Fig. 6 EDX spectra (a–d), SEM (e–h) and TEM (i–l) analysis of untreated E. coli, treated with MEL@AgNPs, MEL@ZnONPs and Ag–ZnO/MEL/GA nanocomposites, respectively for each analysis. | ||
Several mechanisms have been proposed for the antibacterial activity of silver NPs. For instance, silver NPs may adhere to the surface of the cell membrane disturbing permeability and respiration functions of the cell17 but can also penetrate inside the bacteria.77 Other hypothesis states that the deadly effect of Ag, resulting from the interaction of ionic silver with thiol groups, inhibits vital enzymes.78,79 Moreover, other studies formulated hypothesis that ZnONPs can deform the wall and membrane of E. coli cells due to roughness and toxic oxygen radicals generated from the surface of ZnONPs, in addition, penetration of ZnO inside the cells of E. coli leads to membrane destroy, inhibition growth and finally cell death.80,81 Furthermore, the gum arabic shows antibacterial activity against E. coli due to the phytochemical materials.82 In addition to these, MEL has been demonstrated antimicrobial activity especially against Gram-positive bacteria.83 Interestingly, other studies have verified the improved antibacterial actions of NPs synthesized by using surfactants, e.g., Tween 80, CTAB, SDS, and polymers, e.g., PVP 360.16,84 So far, the exact mechanism behind the bactericidal activity is still not well known.
3.4. Cell viability assay
MTT assay was performed to evaluate the cytotoxicity effect of biosynthesized MEL@AgNPs, MEL@ZnONPs and Ag–ZnO/MEL/GA nanocomposites towards HepG2 cancer cells using increased concentration levels (5–80 μg mL−1). Generally, the cell viability of HepG2 was decreased significantly with increasing the concentrations of the synthesized samples. The IC50 values of biosynthesized MEL@AgNPs, MEL@ZnONPs and Ag–ZnO/MEL/GA nanocomposites for inhibition HepG2 cell were found to be 63.25, 26.91, and 28.97ΜG/ML, respectively (Fig. 7). It was observed that the HepG2 cells treated with the lower doses of 5 μg mL−1 do not show any significant cytotoxicity. Other researchers have reported similar results.85 Interestingly, there was no significant difference between MEL@ZnONPs and Ag–ZnO/MEL/GA nanocomposites. In contrast, previous studies indicated that Ag–ZnO nanocomposites' cytotoxicity is lower than a MEL@ZnONPs in the same dose range.86 Arooj et al.87 confirmed that the antitumor efficiency of Ag–ZnO NCs (with 10–30% Ag amount) were more than ZnONPs to malignant melanoma HT144 cells under daylight exposure. In our study, we did not determine the Ag content as well as did not use a light source to improve the cytotoxicity of Ag–ZnO NCs.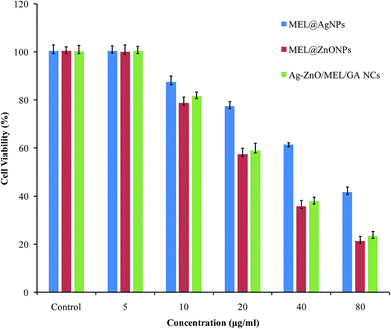 | ||
| Fig. 7 Antitumor activity of various concentrations of MEL@AgNPs, MEL@ZnONPs and Ag–ZnO/MEL/GA nanocomposites against the liver cancer cell line HepG2. | ||
The reason for the potential antitumor activity of Ag–ZnO/MEL/GA nanocomposites might is due to the availability of biomolecules existing in both MEL and GA, which possessed antitumor efficacy against some cancer cells line. Besides the role of surfactants in NPs decoration, they play an important role in improving the solubility and cytotoxicity of anticancer drugs.88 MEL has also induces tyrosinase activity and promotes the production of melanin.23 Many studies reported the antitumor activity was enhanced with the presences of the surfactants.71,88 Nevertheless, this is the first comparative study in the antitumor activity of Ag, ZnO and Ag–ZnO nanocomposites using MEL and GA against the HepG2 cell line. More studies are needed to figure out the biological mechanism of Ag–ZnO/MEL/GA nanocomposites.
4. Conclusions
In conclusion, we have successfully biosynthesized MEL@AgNPs, MEL@ZnONPs and Ag–ZnO/MEL/GA nanocomposites using MEL and gum arabic. XRD, TEM, SEM, and FTIR spectroscopy confirmed the formation of these materials. Moreover, the biosynthesized materials demonstrated significant antidiabetic efficacy in a concentration-dependent reduction for the carbohydrate-digesting enzymes (α-amylase and α-glucosidase). The bactericidal efficacy was investigated against pathogenic food bacteria, Gram-negative (E. coli and S. enterica) and Gram-positive (B. cereus and S. aureus) using the well-diffusion technique, and the results of the zone of inhibition (ZOI) were measured. Also, the cytotoxicity test was performed against the human HepG2 cell line. However, further investigations are desired to explore the biological effects in vivo for the efficacy of Ag–ZnO/MEL/GA nanocomposites.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We acknowledge the Public Projects of Zhejiang Province (grant number LGF18C200003) and Nature Science Foundation of Zhejiang Province (grant number LR13C200002) China, for financially supported this study.References
- K. L. Kotloff, J. P. Winickoff, B. Ivanoff, J. D. Clemens, D. L. Swerdlow, P. J. Sansonetti, G. K. Adak and M. M. Levine, Bull. World Health Organ., 1999, 77, 651–666 CAS.
- Z.-C. Xiong, Z.-Y. Yang, Y.-J. Zhu, F.-F. Chen, Y.-G. Zhang and R.-L. Yang, ACS Appl. Mater. Interfaces, 2017, 9, 22212–22222 CrossRef CAS PubMed.
- L. Guariguata, D. R. Whiting, I. Hambleton, J. Beagley, U. Linnenkamp and J. E. Shaw, Diabetes Res. Clin. Pract., 2014, 103, 137–149 CrossRef CAS PubMed.
- V. Karthick, V. G. Kumar, T. S. Dhas, G. Singaravelu, A. M. Sadiq and K. Govindaraju, Colloids Surf., B, 2014, 122, 505–511 CrossRef CAS PubMed.
- R. Sharma, U. Gupta, N. K. Garg, R. K. Tyagi and N. K. Jain, Colloids Surf., B, 2015, 127, 172–181 CrossRef CAS PubMed.
- U. Etxeberria, A. L. De La Garza, J. Campin, J. A. Martnez and F. I. Milagro, Expert Opin. Ther. Targets, 2012, 16, 269–297 CrossRef CAS PubMed.
- D. S. Shewach and R. D. Kuchta, Chem. Rev., 2009, 109, 2859–2861 CrossRef CAS PubMed.
- Q. H. Ye, L. X. Qin, M. Forgues, P. He, J. W. Kim, A. C. Peng, R. Simon, Y. Li, A. I. Robles, Y. Chen, Z. C. Ma, Z. Q. Wu, S. L. Ye, Y. K. Liu, Z. Y. Tang and X. W. Wang, Nat. Med., 2003, 9, 416–423 CrossRef CAS PubMed.
- R. Sahay, V. J. Reddy and S. Ramakrishna, Int. J. Mech. Mater. Eng., 2014, 9, 25 CrossRef.
- R. Aladpoosh and M. Montazer, Carbohydr. Polym., 2016, 141, 116–125 CrossRef CAS PubMed.
- S. Agnihotri, G. Bajaj, S. Mukherji and S. Mukherji, Nanoscale, 2015, 7, 7415–7429 RSC.
- I. Matai, A. Sachdev, P. Dubey, S. Uday Kumar, B. Bhushan and P. Gopinath, Colloids Surf., B, 2014, 115, 359–367 CrossRef CAS PubMed.
- X. Xu, F. Liu, L. Jiang, J. Y. Zhu, D. Haagenson and D. P. Wiesenborn, ACS Appl. Mater. Interfaces, 2013, 5, 2999–3009 CrossRef CAS PubMed.
- L. Zhang, J. Gu, L. Song, L. Chen, Y. Huang, J. Zhang and T. Chen, J. Mater. Chem. A, 2016, 4, 10810–10815 RSC.
- G. S. Kiran, J. Selvin, A. Manilal and S. Sujith, Crit. Rev. Biotechnol., 2011, 31, 354–364 CrossRef CAS PubMed.
- A. M. Alkilany, P. K. Nagaria, C. R. Hexel, T. J. Shaw, C. J. Murphy and M. D. Wyatt, Small, 2009, 5, 701–708 CrossRef CAS PubMed.
- L. Kvítek, A. Panáček, J. Soukupová, M. Kolář, R. Večeřová, R. Prucek, M. Holecová and R. Zbořil, J. Phys. Chem. C, 2008, 112, 5825–5834 CrossRef.
- A. S. Reddy, C.-Y. Chen, S. C. Baker, C.-C. Chen, J.-S. Jean, C.-W. Fan, H.-R. Chen and J.-C. Wang, Mater. Lett., 2009, 63, 1227–1230 CrossRef CAS.
- Y. Xie, R. Ye and H. Liu, Colloids Surf., A, 2006, 279, 175–178 CrossRef CAS.
- D. Kitamoto, T. Morita, T. Fukuoka, M.-a. Konishi and T. Imura, Curr. Opin. Colloid Interface Sci., 2009, 14, 315–328 CrossRef CAS.
- L. Fan, H. Li, Y. Niu and Q. Chen, PLoS One, 2016, 11, e0148198 CrossRef PubMed.
- Y. Wakamatsu, X. Zhao, C. Jin, N. Day, M. Shibahara, N. Nomura, T. Nakahara, T. Murata and K. K. Yokoyama, Eur. J. Biochem., 2001, 268, 374–383 CrossRef CAS PubMed.
- X. Zhao, Y. Wakamatsu, M. Shibahara, N. Nomura, C. Geltinger, T. Nakahara, T. Murata and K. K. Yokoyama, Cancer Res., 1999, 59, 482–486 CAS.
- M. Takahashi, T. Morita, T. Fukuoka, T. Imura and D. Kitamoto, J. Oleo Sci., 2012, 61, 457–464 CrossRef CAS PubMed.
- Y. Inoh, D. Kitamoto, N. Hirashima and M. Nakanishi, J. Controlled Release, 2004, 94, 423–431 CrossRef CAS PubMed.
- M. B. Kasture, P. Patel, A. A. Prabhune, C. V. Ramana, A. A. Kulkarni and B. L. V. Prasad, J. Chem. Sci., 2008, 120, 515–520 CrossRef CAS.
- E. Priyadarshini, N. Pradhan, A. K. Pradhan and P. Pradhan, Spectrochim. Acta, Part A, 2016, 163, 127–133 CrossRef CAS PubMed.
- B. H. Ali, A. Ziada and G. Blunden, Food Chem. Toxicol., 2009, 47, 1–8 CrossRef CAS PubMed.
- A. C. A. Roque, A. Bicho, I. L. Batalha, A. S. Cardoso and A. Hussain, J. Biotechnol., 2009, 144, 313–320 CrossRef CAS PubMed.
- Y. Liu and H.-I. Kim, Carbohydr. Polym., 2012, 89, 111–116 CrossRef CAS PubMed.
- Z. Lu, J. Gao, Q. He, J. Wu, D. Liang, H. Yang and R. Chen, Carbohydr. Polym., 2017, 156, 460–469 CrossRef CAS PubMed.
- M. T. Ramesan, C. Siji, G. Kalaprasad, B. K. Bahuleyan and M. A. Al-Maghrabi, J. Polym. Environ., 2018, 26, 2983–2991 CrossRef CAS.
- B. N. Singh, A. K. S. Rawat, W. Khan, A. H. Naqvi and B. R. Singh, PLoS One, 2014, 9, e106937 CrossRef PubMed.
- M. Hu, C. Li, X. Li, M. Zhou, J. Sun, F. Sheng, S. Shi and L. Lu, Artif. Cells, Nanomed., Biotechnol., 2018, 46, 1248–1257 CrossRef CAS PubMed.
- K. Balan, W. Qing, Y. Wang, X. Liu, T. Palvannan, Y. Wang, F. Ma and Y. Zhang, RSC Adv., 2016, 6, 40162–40168 RSC.
- Y. Zi, M. Zhu, X. Li, Y. Xu, H. Wei, D. Li and C. Mu, Carbohydr. Polym., 2018, 192, 118–125 CrossRef CAS PubMed.
- J. Yang, H. Lu, M. Li, J. Liu, S. Zhang, L. Xiong and Q. Sun, Carbohydr. Polym., 2017, 178, 311–321 CrossRef CAS PubMed.
- J. I. Arutchelvi, S. Bhaduri, P. V. Uppara and M. Doble, J. Ind. Microbiol. Biotechnol., 2008, 35, 1559–1570 CrossRef CAS PubMed.
- A. J. Ribeiro, F. R. L. de Souza, J. M. N. A. Bezerra, C. Oliveira, D. Nadvorny, M. F. de La Roca Soares, L. C. C. Nunes, E. C. Silva-Filho, F. Veiga and J. L. Soares Sobrinho, Carbohydr. Polym., 2016, 147, 188–200 CrossRef CAS PubMed.
- M. Sorbiun, E. Shayegan Mehr, A. Ramazani and S. Taghavi Fardood, J. Mater. Sci.: Mater. Electron., 2018, 29, 2806–2814 CrossRef CAS.
- S. B. Rana, V. K. Bhardwaj, S. Singh, A. Singh and N. Kaur, J. Exp. Nanosci., 2014, 9, 877–891 CrossRef CAS.
- A. A. Oun and J.-W. Rhim, Carbohydr. Polym., 2017, 169, 467–479 CrossRef CAS PubMed.
- S. Azizi, M. B. Ahmad, M. Z. Hussein and N. A. Ibrahim, Molecules, 2013, 18, 6269–6280 CrossRef CAS PubMed.
- X. Lu, A. Dandapat, Y. Huang, L. Zhang, Y. Rong, L. Dai, Y. Sasson, J. Zhang and T. Chen, RSC Adv., 2016, 6, 60916–60921 RSC.
- X. Lu, Y. Huang, B. Liu, L. Zhang, L. Song, J. Zhang, A. Zhang and T. Chen, Chem. Mater., 2018, 30, 1989–1997 CrossRef CAS.
- Y. Huang, L. Dai, L. Song, L. Zhang, Y. Rong, J. Zhang, Z. Nie and T. Chen, ACS Appl. Mater. Interfaces, 2016, 8, 27949–27955 CrossRef CAS PubMed.
- S. Azizi, R. Mohamad, R. A. Rahim, A. B. Moghaddam, M. Moniri, A. Ariff, W. Z. Saad and F. Namvab, Appl. Surf. Sci., 2016, 384, 517–524 CrossRef CAS.
- T. Fukuoka, T. Morita, M. Konishi, T. Imura and D. Kitamoto, J. Oleo Sci., 2007, 56, 435–442 CrossRef CAS PubMed.
- S. Gomez-Grana, M. Perez-Ameneiro, X. Vecino, I. Pastoriza-Santos, J. Perez-Juste, J. M. Cruz and A. B. Moldes, Nanomaterials, 2017, 7(6), 139 CrossRef PubMed.
- M. M. Solomon, H. Gerengi, S. A. Umoren, N. B. Essien, U. B. Essien and E. Kaya, Carbohydr. Polym., 2018, 181, 43–55 CrossRef CAS PubMed.
- S. I. Alhassan, P. A. P. Mamza, A. M. Ja'o and D. E. Arthur, SDRP Journal of Computational Chemistry & Molecular Modelling, 2015, 1, 1–8 Search PubMed.
- H. Adam, M. A. Siddig, A. A. Siddig and N. A. Eltahir, Sudan Med. Monit., 2013, 8, 174 CrossRef.
- D. Mudgil, S. Barak and B. S. Khatkar, Int. J. Biol. Macromol., 2012, 50, 1035–1039 CrossRef CAS PubMed.
- L. Dai, L. Song, Y. Huang, L. Zhang, X. Lu, J. Zhang and T. Chen, Langmuir, 2017, 33, 5378–5384 CrossRef CAS PubMed.
- Y. Huang, A. Dandapat and D.-H. Kim, Nanoscale, 2014, 6, 6478–6481 RSC.
- Y. Huang and D.-H. Kim, Nanoscale, 2012, 4, 6312–6317 RSC.
- V. Malapermal, I. Botha, S. B. N. Krishna and J. N. Mbatha, Saudi J. Biol. Sci., 2017, 24, 1294–1305 CrossRef CAS PubMed.
- G. D. Saratale, R. G. Saratale, G. Benelli, G. Kumar, A. Pugazhendhi, D.-S. Kim and H.-S. Shin, J. Cluster Sci., 2017, 28, 1709–1727 CrossRef CAS.
- D. Rehana, D. Mahendiran, R. S. Kumar and A. K. Rahiman, Bioprocess Biosyst. Eng., 2017, 40, 943–957 CrossRef CAS PubMed.
- A. Bayrami, S. Parvinroo, A. Habibi-Yangjeh and S. Rahim Pouran, Artif. Cells, Nanomed., Biotechnol., 2018, 46, 730–739 CrossRef CAS PubMed.
- R. Zouari, R. B. Abdallah-Kolsi, K. Hamden, A. E. Feki, K. Chaabouni, F. Makni-Ayadi, F. Sallemi, S. Ellouze-Chaabouni and D. Ghribi-Aydi, Pept. Sci., 2015, 104, 764–774 CrossRef CAS PubMed.
- N. Seedher and M. Kanojia, AAPS PharmSciTech, 2008, 9, 431–436 CrossRef CAS PubMed.
- C. A. Edwards, N. A. Blackburn, L. Craigen, P. Davison, J. Tomlin, K. Sugden, I. T. Johnson and N. W. Read, Am. J. Clin. Nutr., 1987, 46, 72–77 CrossRef CAS PubMed.
- H. Haase, S. Overbeck and L. Rink, Exp. Gerontol., 2008, 43, 394–408 CrossRef CAS PubMed.
- J. Jansen, W. Karges and L. Rink, J. Nutr. Biochem., 2009, 20, 399–417 CrossRef CAS PubMed.
- W. Salem, D. R. Leitner, F. G. Zingl, G. Schratter, R. Prassl, W. Goessler, J. Reidl and S. Schild, Int. J. Med. Microbiol., 2015, 305, 85–95 CrossRef CAS.
- N. Papo and Y. Shai, J. Biol. Chem., 2005, 280, 10378–10387 CrossRef CAS PubMed.
- K. Y. Yoon, J. H. Byeon, C. W. Park and J. Hwang, Environ. Sci. Technol., 2008, 42, 1251–1255 CrossRef CAS PubMed.
- J. Nashida, N. Nishi, Y. Takahashi, C. Hayashi, M. Igarashi, D. Takahashi and K. Toshima, J. Org. Chem., 2018, 83, 7281–7289 CrossRef CAS PubMed.
- D. Kitamoto, H. Isoda and T. Nakahara, J. Biosci. Bioeng., 2002, 94, 187–201 CrossRef CAS PubMed.
- G. Kaur, S. Kumar, R. Kant, G. Bhanjana, N. Dilbaghi, S. K. Guru, S. Bhushan and S. Jaglan, RSC Adv., 2016, 6, 57084–57097 RSC.
- V. K. Sharma, R. A. Yngard and Y. Lin, Adv. Colloid Interface Sci., 2009, 145, 83–96 CrossRef CAS PubMed.
- S. K. Basha, K. V. Lakshmi and V. S. Kumari, Sens Biosensing Res., 2016, 10, 34–40 CrossRef.
- M. Arakha, M. Saleem, B. C. Mallick and S. Jha, Sci. Rep., 2015, 5, 9578 CrossRef CAS PubMed.
- Y.-H. Hsueh, K.-S. Lin, W.-J. Ke, C.-T. Hsieh, C.-L. Chiang, D.-Y. Tzou and S.-T. Liu, PLoS One, 2015, 10, e0144306 CrossRef PubMed.
- A. A. Tayel, W. F. El-Tras, S. Moussa, A. F. El-Baz, H. Mahrous, M. F. Salem and L. Brimer, J. Food Saf., 2011, 31, 211–218 CrossRef CAS.
- M. Jose Ruben, E. Jose Luis, C. Alejandra, H. Katherine, B. K. Juan, R. Jose Tapia and Y. Miguel Jose, Nanotechnology, 2005, 16, 2346 CrossRef.
- Y. Matsumura, K. Yoshikata, S. Kunisaki and T. Tsuchido, Appl. Environ. Microbiol., 2003, 69, 4278–4281 CrossRef CAS.
- A. Gupta, M. Maynes and S. Silver, Appl. Environ. Microbiol., 1998, 64, 5042–5045 CAS.
- G. Applerot, A. Lipovsky, R. Dror, N. Perkas, Y. Nitzan, R. Lubart and A. Gedanken, Adv. Funct. Mater., 2009, 19, 842–852 CrossRef CAS.
- R. Brayner, R. Ferrari-Iliou, N. Brivois, S. Djediat, M. F. Benedetti and F. Fievet, Nano Lett., 2006, 6, 866–870 CrossRef CAS.
- S. M. A. Alawi, M. A. Hossain and A. A. Abusham, Beni-Suef University Journal of Basic and Applied Sciences, 2018, 7, 22–26 CrossRef.
- D. Kitamoto, H. Yanagishita, T. Shinbo, T. Nakane, C. Kamisawa and T. Nakahara, J. Biotechnol., 1993, 29, 91–96 CrossRef CAS.
- L. Kvitek, A. Panáček, J. Soukupova, M. Kolář, R. Večeřová, R. Prucek, M. Holecova and R. Zbořil, J. Phys. Chem. C, 2008, 112, 5825–5834 CrossRef CAS.
- R. Wahab, M. A. Siddiqui, Q. Saquib, S. Dwivedi, J. Ahmad, J. Musarrat, A. A. Al-Khedhairy and H.-S. Shin, Colloids Surf., B, 2014, 117, 267–276 CrossRef CAS PubMed.
- M. M. Rad, N. Najafzadeh, N. Tata and A. Jafari, Pharmaceut. Chem. J., 2018, 52, 112–116 CrossRef CAS.
- S. Arooj, S. Nazir, A. Nadhman, N. Ahmad, B. Muhammad, I. Ahmad, K. Mazhar and R. Abbasi, Beilstein J. Nanotechnol., 2015, 6, 570–582 CrossRef CAS.
- A. M. Alkilany and C. J. Murphy, Langmuir, 2009, 25, 13874–13879 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2019 |