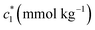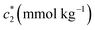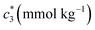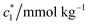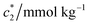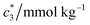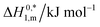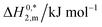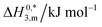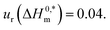Open Access Article
Open Access ArticleAggregation behavior of cetyldimethylethylammonium bromide under the influence of bovine serum albumin in aqueous/electrolyte solutions at various temperatures and compositions: conductivity and molecular dynamics study†
Md. Farid Ahmedab,
Mohammad Robel Molla ab,
Mousumi Sahac,
Imrul Shahriar
ab,
Mousumi Sahac,
Imrul Shahriar c,
Mohammad Saidur Rahman
c,
Mohammad Saidur Rahman ad,
Mohammad A. Halim
ad,
Mohammad A. Halim c,
Malik Abdul Rub
c,
Malik Abdul Rub ef,
Md. Anamul Hoque
ef,
Md. Anamul Hoque *a and
Abdullah M. Asiri
*a and
Abdullah M. Asiri ef
ef
aDepartment of Chemistry, Jahangirnagar University, Savar, Dhaka-1342, Bangladesh. E-mail: ahoque_ju@yahoo.com; ahoque_ju@juniv.edu; Fax: +880-2-7791052; Tel: PABX: +880-2-7791045-51 extn 1437
bBangladesh Council of Scientific and Industrial Research (BCSIR), Dhaka, Bangladesh
cDivision of Quantum Chemistry, The Red-Green Research Centre, BICCB, Dhaka, Bangladesh
dDepartment of Chemical Engineering, Faculty of Engineering, University of Malaya, 50603, Kuala Lumpur, Malaysia
eChemistry Department, Faculty of Science, King Abdulaziz University, Jeddah-21589, Saudi Arabia
fCenter of Excellence for Advanced Materials Research, King Abdulaziz University, Jeddah 21589, Saudi Arabia
First published on 25th February 2019
Abstract
Herein, we have investigated the interaction of bovine serum albumin (BSA), the most abundant globular protein, with a conventional cationic surfactant, cetyldimethylethylammonium bromide (CDMEAB), through a conductivity technique in the absence/presence of electrolyte solutions at various temperatures (298.15–323.15 K). The interaction of the protein with drugs/surfactants and other additives plays a crucial role in the body. Hence, the main concern of the study is to extract the impact of BSA on surfactant molecules and vice versa. From the specific conductivity versus concentration of surfactant plots, three different noticeable critical micelle concentration (c*) values were obtained for pure CDMEAB and its mixture with protein/protein + salts. The presence of BSA and electrolytes altered the c* values of CDMEAB revealing interactions among the studied constituents where the salt solutions reduced the c* values and created a convenient environment for favorable micellization. The negative magnitudes achieved for standard free energy changes (ΔG0m) suggest spontaneity of micellization while the values of ΔH0m and ΔS0m signified the existence of some electrostatic and hydrophobic interactions. The values of molar heat capacity (ΔC0m) were positive as well as small which was an indication of less structural deformation. Molecular Dynamics (MD) simulation for all atoms revealed that the salt ions promoted non-covalent interaction between BSA and CDMEAB, and such interactions were not observed in the absence of the salt. Protein structure remained nearly same in spite of strong interaction with CDMEAB as evident from the overall RMSD (root-mean-square deviation) values of the alpha carbons and backbone of the protein and RMSF (root-mean-square fluctuation) values of the amino acid residues present in BSA. In this work thermodynamic parameters of transfer (such as ΔG0m.tr., ΔH0m.tr., and ΔC0p.m.tr.) were also evaluated and the results are discussed in detail. Besides, contributions of enthalpy and entropy to free energy changes were also analyzed.
1. Introduction
Proteins are associated with various functions including transport, signaling, energy conversion, cell communication, catalysis, etc. in biological systems. Bovine serum albumin (BSA) along with other globular proteins has been stimulating a plethora of scientific investigations over the last few decades due to its typical architecture and better performance for carrier properties.1 This protein is one of the most familiar carrier proteins, which play a crucial role in transport as well as the release of various endogenous & exogenous materials in blood.1,2 The biological activities and benefits of proteins can be realized by means of insight from their performance to act as a hormone as well as transmitters in the central nervous system of living beings.3 BSA has the capability to bind with a diversified array of biologically active molecules (such as drugs, fatty acids, metabolites, amino acids, metals and various organic compounds). This affinity for binding of proteins (BSA) with drugs as well as amphiphilic substances opened a new area of research called “protein–surfactant interaction”, subsequently providing a wide range of applications in the field of drug delivery and drug formulations, biotechnology, detergents, foods and the cosmetics industry.4–7 Strong binding lessens the abundance of free drug in plasma while weak binding indicates short lifetime or less distribution. Spectroscopic elucidation of strong binding between BSA and drug such as ceftriaxone has been studied in the absence and presence of Zn by Yue et al.8 The protein-drug complex directs a model for studying drug–protein interactions which would later facilitate the research of metabolism and transportation. Li et al.9 studied the effect of chitosan on the microenvironment and the secondary structure of BSA which provides theoretical guidance for the design of new ingredients for the food industry.Assembly of amphiphilic molecules including surfactants/drugs etc. has a spontaneous propensity to form aggregates known as micelles due to the presence of weak non-covalent forces (i.e. van der Waals force) among the individual molecules. Micelles are formed at a particular concentration of that amphiphile known as “critical micelle concentration”.10–16 It is a unique feature of surfactant which helps to extract information about the interaction of drug and other biomolecules such as protein in the body. In the case of drug delivery, surfactants are important molecules to act as recipients/excipients. Hydrophobic nucleus/core of micelles is significant as it has the capability to enhance the solubility or bioavailability of hydrophobic drugs. Cationic surfactants are such surfactants that are utilized like anti-microbial substances in many types of antiseptics, disinfectants, perfumes as well as in cosmetics.11 Previous studies revealed that the presence of various additional substances exert their effects on the physiological feature of micelles including the extent of departure of opposite-ion binding, catalytic property, and reaction rate which are related to the phase-separation model.11,17
According to Deep and Ahluwalia, native globular protein's stability is noticeably influenced by pH, temperature, and insertion of small molecules like surfactants, co-enzymes, inhibitors and activators that bind specifically to the original state.18 The interaction between surfactant & globular proteins can help towards an understanding of the action of surfactant as a solubilizing agent for membrane proteins and lipids. Some excellent literatures are available on the study of the interaction between drugs/salts/polymers/polyols with surfactants; however, to the best of our knowledge, no study yet reported the interaction between BSA and CDMEAB.19–23 CDMEAB is a cationic surfactant which is used as disinfectant/laboratory purpose.24 Understanding the interaction between BSA and CDMEAB could be a key interest in the meadow study of protein-membrane chemistry model.
Therefore, in our present work, we have employed conductivity technique in order to investigate the interaction between BSA (Scheme 1(I)) and CDMEAB (Scheme 1(II)). Numerous studies revealed that when the aggregates are exposed to a variety of additives, the physiological properties (aggregate's stability, reaction rate, degree of ionization, and clouding phenomenon) could be affected due to the presence of additives.25–27 In a biological system, sodium ion plays a vital role to control the sodium pump and transmitting nerve systems.28 Salt ions have the unique property to decrease the critical micelle concentration (c*) by reducing the existing electrostatic repulsion force among the polar head groups of the surfactant, therefore, increasing the aggregation number (Nagg).29 Therefore, NaCl and Na2SO4 were utilized in this work to understand the effect of additives on different physicochemical parameters corresponding to BSA–CDMEAB interaction.
In recent years, MD simulation techniques are being implemented to understand the effect of surfactant on protein structure and on the protein folding phenomenon.30–32 Hoque et al. have studied the interaction of a cationic surfactant, cetylpyridinium chloride on BSA in presence and absence of electrolytes using MD simulation technique.33 MD simulation provided detailed information including non-covalent interactions between protein and surfactant, root-mean-square deviation of alpha carbon and backbone of the protein and root-mean-square fluctuation of each amino acid present in the protein. Delgado-Magnero and coworkers have utilized MD simulation method to understand the binding mechanism of nonionic surfactants with BSA and the results obtained suggest the potential use of polysorbates as excipients for minimizing the undesirable effects of protein adsorption and aggregation.34 In the present study, we investigate the nature and type of interactions present among the surfactant molecules and BSA employing MD simulation to support the experimental observations.
2. Experimental procedure
2.1. Materials and solutions
All the chemicals used in the study were of analytical grade and collected to use without further purification. CDMEAB (mass fraction purity, 0.99), BSA (mass fraction purity, >0.98), NaCl (mass fraction purity, 0.99), Na2SO4 (mass fraction purity, 0.99) were purchased from Across Organics (USA), Merck (Germany), Merck (India) and Merck (India) respectively.2.2. Conductivity measurement technique
CDMEAB and CDMEAB + BSA solutions were made in molal concentration (with/without a particular concentration of salts solution) by using distilled-deionized water having specific conductivity 1.7–2.0 μS cm−1 (over the temperature range 298.15–323.15 K). Weights of samples during solution preparation were taken by using an electric balance (Metler Toledo). In order to evaluate the magnitudes of specific conductivity of pure surfactant/(BSA + surfactant) system, we have used 4510 conductivity meter (manufacturer: Jenway, UK) including a glass cell electrode having cell constant 0.97 cm−1. Standard KCl solution having appropriate concentration was used to perform the calibration of the conductivity meter. Alternative current (AC) having frequency 60 Hz was employed to study the conductivity. At first, 25 mmol kg−1 solution of CDMEAB in water was prepared by utilizing an aqueous solution of BSA (at room temperature) and the surfactant solution was inserted into 20 mL BSA solution (with/without a fixed concentration of salts). These solutions were prepared by using salts (NaCl and Na2SO4) for examining the influence of salts in which the same concentration of salts was considered. Conductivity was measured after every addition of prepared solutions with thoroughly mixing and time was provided for the attainment of temperature equilibration; therefore, this process was repeated after every addition. The results obtained provide good matching with literature results.25–28,35 During the whole temperature range, RM6 Lauda circulating water bath was used to control the temperature and temperature error ±2 K was considered. Three different (c*) magnitudes were extracted from the abrupt alteration of specific conductivity with concentration of CDMEAB solution and (CDMEAB + BSA) mixtures.2.3. Molecular dynamics simulations
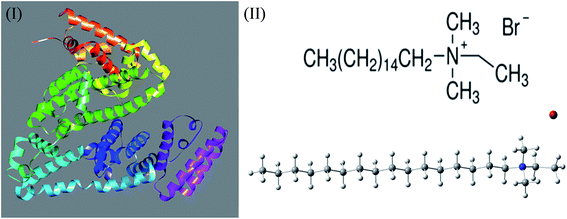
The crystal structure of serum albumin 4F5S from Bostaurus (Bovine) was collected from protein data bank (shown in Scheme 1(I)). This structure was chosen based on the resolved amino acid residues (25-607) and relatively high resolution of 2.47 Å compared to the other available structures in UniProt (http://www.uniprot.org/uniprot/P02769). The molecular dynamics simulation of one BSA and 40 CDMEAB molecules with water was carried out for 20 ns in the absence and presence of salt (NaCl) using YASARA Dynamics program.36,37 The Amber14 force field was used for all the simulations.38 A cut off radius of 8 Å was used for short-range van der Waals and Coulomb interactions. The long-range electrostatic interactions were calculated using the particle-mesh Ewald method.39 Periodic boundary condition (cell dimension of 105 Å × 105 Å × 105 Å) and temperature 298 K were considered for all simulations. Time step 1.25 fs was used and simulation snapshots were saved at every 100 ps. The solvent density of 0.998 g ml−1 for both systems was maintained by adding 34![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 699 molecules of water. Macro programmed protocol was used for steepest decent minimization and equilibration for salt and no salt environment. Molecular dynamics trajectory, root mean square deviation (RMSD) of alpha carbon and backbone, solvent accessible surface area (SASA), radius of gyration as well as the residue-specific root mean square fluctuations (RMSF) of protein were calculated by specific macros written in Yanaconda language. Collision cross section measurements of the protein structures were performed using the IMPACT program.40
699 molecules of water. Macro programmed protocol was used for steepest decent minimization and equilibration for salt and no salt environment. Molecular dynamics trajectory, root mean square deviation (RMSD) of alpha carbon and backbone, solvent accessible surface area (SASA), radius of gyration as well as the residue-specific root mean square fluctuations (RMSF) of protein were calculated by specific macros written in Yanaconda language. Collision cross section measurements of the protein structures were performed using the IMPACT program.40
3. Results and discussion
3.1. Determination of critical micelle concentration (c*) and counter-ion binding parameter (β) of pure and mixed systems
The concentration above which surfactant micellization takes place is called critical micelle concentration (c*). The c* values can be evaluated from the abrupt change of physico-chemical properties with the change of concentration of the surfactant solutions. In the current study, the specific conductivity (κ) of CDMEAB in H2O/H2O + BSA solutions having different concentrations is measured and the κ values are found to vary with the concentration of surfactant. Fig. 1 depicts the variation of specific conductivity associated with different concentration of CDMEAB in aqueous solution for pure CDMEAB and CDMEAB + BSA mixture. Every intersection points of the plots demonstrate the certain concentration of amphiphiles which is regarded as critical micelle concentration (c*). Here we have obtained three different noticeable break points for pure CDMEAB and (BSA + CDMEAB) mixtures which are designated as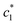 ,
, 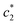 , and
, and 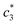 . The primary kink reflects the formation of BSA-surfactant complex (convey of the concentration of critical aggregation), second breakpoint indicates the CDMEAB micellization in presence of BSA whereas the tertiary breakpoint is acknowledged as the structural modification of micelles such as spherical shape to rod shape.41 More than one critical micelle concentration (c*) for various amphiphiles has been observed by many researchers as well as in our previous study.42,43
. The primary kink reflects the formation of BSA-surfactant complex (convey of the concentration of critical aggregation), second breakpoint indicates the CDMEAB micellization in presence of BSA whereas the tertiary breakpoint is acknowledged as the structural modification of micelles such as spherical shape to rod shape.41 More than one critical micelle concentration (c*) for various amphiphiles has been observed by many researchers as well as in our previous study.42,43
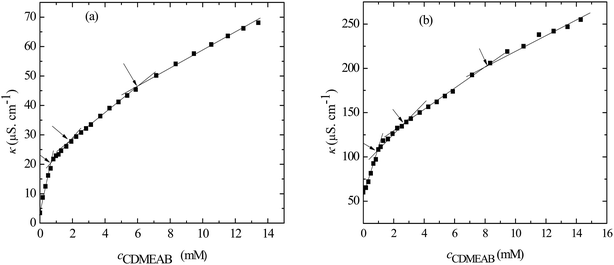 | ||
| Fig. 1 Specific conductivity (κ) versus concentration of CDMEAB for (a) pure CDMEAB and (b) (BSA + CDMEAB) mixed system containing 0.03 mmol kg−1 BSA in water at 303.15 K. | ||
The obtained magnitudes of 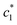 ,
, 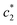 , and
, and 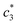 of CDMEAB are found 0.74, 1.49, and 6.57 mmol kg−1 respectively at 303.15 K. On the other hand, the values of
of CDMEAB are found 0.74, 1.49, and 6.57 mmol kg−1 respectively at 303.15 K. On the other hand, the values of 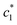 ,
, 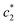 , and
, and 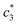 in aqueous BSA (0.005–0.100 mmol kg−1) +CDMEAB mixed systems are found (0.75–1.05 mmol kg−1), (2.22–2.49 mmol kg−1), and (6.35–7.67 mmol kg−1) respectively (Table 1). The values of CDMEAB are in good agreement with literature values.44 In presence of BSA,
in aqueous BSA (0.005–0.100 mmol kg−1) +CDMEAB mixed systems are found (0.75–1.05 mmol kg−1), (2.22–2.49 mmol kg−1), and (6.35–7.67 mmol kg−1) respectively (Table 1). The values of CDMEAB are in good agreement with literature values.44 In presence of BSA, 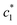 values are increased with increasing the concentration of BSA. The
values are increased with increasing the concentration of BSA. The 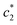 and
and 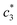 values are first increased, reach a maximum and then decreased. These variations in c* values signify the interaction between BSA and surfactant. The decrease in
values are first increased, reach a maximum and then decreased. These variations in c* values signify the interaction between BSA and surfactant. The decrease in 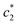 and
and 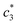 values at higher concentration of BSA possibly due to the electrostatic interaction between BSA and the positively charged surfactant. However, interactions sustained through the nonpolar interactions of the hydrophobic portion of BSA and CDMEAB. The values of the degree of micelle ionization (α) were extracted by means of taking the corresponding ratios of the slopes relating to the closest straight lines before & after the c*.21,22,45,46 Then α1, α2, and α3 can be obtained from the ratios of S2/S1, S3/S1 and S4/S1 respectively, where S1, S2, and S3 are the first, second and third straight lines. The fraction of counter ion binding (β) can be obtained from the relation: β = 1 − α.
values at higher concentration of BSA possibly due to the electrostatic interaction between BSA and the positively charged surfactant. However, interactions sustained through the nonpolar interactions of the hydrophobic portion of BSA and CDMEAB. The values of the degree of micelle ionization (α) were extracted by means of taking the corresponding ratios of the slopes relating to the closest straight lines before & after the c*.21,22,45,46 Then α1, α2, and α3 can be obtained from the ratios of S2/S1, S3/S1 and S4/S1 respectively, where S1, S2, and S3 are the first, second and third straight lines. The fraction of counter ion binding (β) can be obtained from the relation: β = 1 − α.
| cBSA (mmol kg−1) | |||
|---|---|---|---|
a Relative standard uncertainties (ur) is 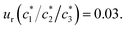 |
|||
| 0.000 | 0.74 | 1.49 | 6.57 |
| 0.005 | 0.75 | 2.27 | 6.79 |
| 0.010 | 0.77 | 2.32 | 7.67 |
| 0.030 | 0.91 | 2.49 | 6.84 |
| 0.050 | 0.93 | 2.31 | 6.52 |
| 0.100 | 1.05 | 2.22 | 6.35 |
At a particular concentration of BSA (0.03 mmol kg−1) in the c* values for BSA + CDMEAB mixed systems (ESI Fig. 1†) are decreased with temperature up to a fixed limit and then increased as well as provided a U-contoured architecture. The ESI Fig. 1† demonstrates that micellization became favored with increasing temperature due to the dehydration of hydrophobic moiety which was predominated over the dehydration of hydrophilic moiety; therefore, decline the propensity of hydrophilic hydration, which is highly significant than the propensity of hydrophobic dehydration at higher temperatures and lessen the micellization process.
3.2. Impact of salt solutions (NaCl and Na2SO4) on c* and β of (BSA + CDMEAB) mixed system at various temperatures
The obtained values of c* and β of pure CDMEAB & (BSA + CDMEAB) systems in the presence of two different sodium salts are viewed in Table 2. In cell membranes, the presence as well as the concentration of electrolytes is an important consideration as they may affect the interaction of surfactant. So, drug-surfactant interaction is an important approach where the coagulation process of surfactant molecules should be brought under the study of the impact of electrolytes, temperatures, pH, and the concentration of various additives. The values of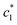 for (BSA + CDMEAB) in salt medium is noticeably decreased than pure CDMEAB and (BSA + CDMEAB) mixed system while the second
for (BSA + CDMEAB) in salt medium is noticeably decreased than pure CDMEAB and (BSA + CDMEAB) mixed system while the second 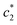 values show the opposite pattern. On the other hand, the third
values show the opposite pattern. On the other hand, the third 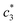 values of (BSA + CDMEAB) in presence of salts are comparatively lower than pure (BSA + CDMEAB) missed system. Overall, the electric double layer is compressed due to the presence of an aqueous solution of salts, and subsequently boosted the lessening of repulsion affinity among the head groups of CDMEAB and BSA. At a particular concentration of BSA and salts,
values of (BSA + CDMEAB) in presence of salts are comparatively lower than pure (BSA + CDMEAB) missed system. Overall, the electric double layer is compressed due to the presence of an aqueous solution of salts, and subsequently boosted the lessening of repulsion affinity among the head groups of CDMEAB and BSA. At a particular concentration of BSA and salts, 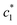 and
and 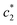 are first decreased and then increased. However, the
are first decreased and then increased. However, the 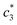 values did not exhibit any regular trend. In our study, we have chosen such salts that play a vital role in human body. Our principal interest is to understand the way how they interact with each other as well as providing additional information about thermodynamic conditions. These findings provoke better information about surfactant–protein interaction, drug–surfactant interaction in the presence of salts. Salts give a convenient environment for micellization. Table 2 demonstrate that the presence of salts decreases the c* values and enhances the aggregation process. The obtained values of c* are comparatively lower in case of Na2SO4 than NaCl which reveals that aggregation process is highly suitable with NaCl for (BSA + CDMEAB) mixture.
values did not exhibit any regular trend. In our study, we have chosen such salts that play a vital role in human body. Our principal interest is to understand the way how they interact with each other as well as providing additional information about thermodynamic conditions. These findings provoke better information about surfactant–protein interaction, drug–surfactant interaction in the presence of salts. Salts give a convenient environment for micellization. Table 2 demonstrate that the presence of salts decreases the c* values and enhances the aggregation process. The obtained values of c* are comparatively lower in case of Na2SO4 than NaCl which reveals that aggregation process is highly suitable with NaCl for (BSA + CDMEAB) mixture.
| Medium | cBSA/mmol kg−1 | ISalt/mmol kg−1 | T/K | β1 | β2 | β3 | |||
|---|---|---|---|---|---|---|---|---|---|
a Relative standard uncertainties (ur) are 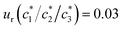 and ur(β1/β2/β3) = 0.04. and ur(β1/β2/β3) = 0.04. |
|||||||||
| CDMEAB | |||||||||
| H2O | 0.0 | 0.00 | 298.15 | 0.84 | 1.94 | 6.08 | 0.70 | 0.76 | 0.83 |
| 303.15 | 0.74 | 1.49 | 5.83 | 0.75 | 0.80 | 0.87 | |||
| 308.15 | 0.64 | 2.28 | 5.46 | 0.71 | 0.81 | 0.89 | |||
| 313.15 | 0.77 | 2.36 | 6.06 | 0.72 | 0.83 | 0.90 | |||
| 318.15 | 0.83 | 2.55 | 6.16 | 0.70 | 0.83 | 0.89 | |||
| 323.15 | 0.97 | 2.67 | 6.26 | 0.70 | 0.82 | 0.89 | |||
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||||||||
| BSA + CDMEAB | |||||||||
| H2O | 0.03 | 0.00 | 298.15 | 0.99 | 2.44 | 8.46 | 0.69 | 0.88 | 0.88 |
| 303.15 | 0.91 | 2.49 | 7.97 | 0.66 | 0.75 | 0.84 | |||
| 308.15 | 0.83 | 2.61 | 7.67 | 0.68 | 0.79 | 0.85 | |||
| 313.15 | 1.03 | 2.71 | 7.58 | 0.64 | 0.76 | 0.85 | |||
| 318.15 | 1.08 | 2.89 | 6.71 | 0.69 | 0.69 | 0.88 | |||
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||||||||
| BSA + CDMEAB | |||||||||
| H2O + NaCl | 0.03 | 0.50 | 303.15 | 0.88 | 3.26 | 7.24 | 0.28 | 0.79 | 0.84 |
| 1.00 | 303.15 | 0.78 | 3.08 | 7.31 | 0.69 | 0.76 | 0.82 | ||
| 1.50 | 303.15 | 0.71 | 2.86 | 7.52 | 0.54 | 0.67 | 0.77 | ||
| 2.00 | 303.15 | 0.85 | 2.67 | 7.41 | 0.58 | 0.69 | 0.84 | ||
| 3.00 | 303.15 | 0.89 | 2.63 | — | 0.58 | 0.69 | — | ||
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||||||||
| BSA + CDMEAB | |||||||||
| H2O + Na2SO4 | 0.03 | 0.50 | 303.15 | 0.79 | 1.96 | 5.87 | 0.57 | 0.72 | 0.81 |
| 1.00 | 303.15 | 0.71 | 2.67 | 6.56 | 0.57 | 0.67 | 0.77 | ||
| 1.50 | 303.15 | 0.65 | 3.09 | 6.68 | 0.60 | 0.69 | 0.77 | ||
| 2.00 | 303.15 | 0.67 | 3.78 | 7.91 | 0.60 | 0.62 | 0.72 | ||
| 3.00 | 303.15 | 0.79 | 4.82 | — | 0.57 | 0.56 | — | ||
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||||||||
| BSA + CDMEAB | |||||||||
| H2O + NaCl | 0.03 | 1.50 | 298.15 | 0.87 | 3.24 | 8.31 | 0.65 | 0.63 | 0.67 |
| 303.15 | 0.71 | 2.86 | 7.52 | 0.70 | 0.80 | 0.85 | |||
| 308.15 | 0.75 | 2.24 | 7.04 | 0.68 | 0.77 | 0.83 | |||
| 313.15 | 0.81 | 2.13 | 6.99 | 0.61 | 0.77 | 0.84 | |||
| 318.15 | 0.85 | 2.43 | 7.22 | 0.65 | 0.78 | 0.85 | |||
| 323.15 | 0.93 | 2.66 | 7.49 | 0.70 | 0.78 | 0.85 | |||
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||||||||
| BSA + CDMEAB | |||||||||
| H2O + Na2SO4 | 0.03 | 1.50 | 298.15 | 0.89 | 2.79 | 7.59 | 0.58 | 0.61 | 0.67 |
| 303.15 | 0.65 | 3.09 | 6.68 | 0.56 | 0.68 | 0.82 | |||
| 308.15 | 0.69 | 3.09 | 6.87 | 0.56 | 0.68 | 0.82 | |||
| 313.15 | 0.72 | 3.56 | 7.47 | 0.55 | 0.66 | 0.77 | |||
| 318.15 | 0.76 | 3.76 | 7.61 | 0.53 | 0.65 | 0.76 | |||
| 323.15 | 0.86 | 4.07 | 7.75 | 0.56 | 0.68 | 0.77 | |||
According to some previous study, chloride ion shows chaotropic nature along with reduction of the stability of hydrophobic moiety or associates of surfactants. Another anion is sulfate ion which carries high charge density and acts as water structure maker (a powerful kosmotrope). Therefore, raises the stability of hydrophobic aggregates of surfactants.47,48 Table 2 also indicates that Na2SO4 have better salting out effect than NaCl. The β magnitudes of CDMEAB & (BSA + CDMEAB) mixed systems in presence of electrolytes did not follow any hemolytic orientation while the aggregates stability is proved due to the high values of β (almost in all cases above 70% (±2), except 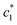 ).
).
3.3. Temperature effects on the micellization of CDMEAB and (BSA + CDMEAB) mixtures
van der Waals interaction among the hydrophobic portion of surfactants are responsible for the aggregation phenomena. Effect of the variation of temperature of experimental system is one of the major considerable factors which affect the micellization process. Due to the propensity of hydration of polar portions, micellization may be delayed. Table 2 shows the values of c* at a particular temperature in the presence of electrolytes. In the case of pure CDMEAB, first and third c* values are initially decreased, attain the minimum position and then increased with rise of temperature whereas the second c* values are gradually increased with temperature. For the (BSA + CDMEAB) mixed system, the second and third c* values are gradually increased. After that, they decreased while first c* values are decreased, powered a nearest flat, and then increased with an accretion of temperatures. At a certain temperature (303.15 K), in salt medium, first c* values are increased initially and then decreased. On the other hand, the second & third c* values are gradually increased in Na2SO4 solution and gradually decreased in NaCl solution. Besides, with varying temperature in NaCl/Na2SO4 solution, c* values are first decreased and then increased (except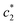 values in Na2SO4 solution).
values in Na2SO4 solution).
The effect of temperature on micellization of surfactant depends on the nature of surfactants whether it is ionic or non-ionic. Usually ionic amphiphiles exhibit U-shaped behavior,49,50 while non-ionic amphiphiles show regular trend;51–53 though it has been noted that non-ionic amphiphiles also provide the U-shaped curve.54 In our case, ESI Fig. 1† provides good harmony with literature. The alteration of critical micelle concentration due to the variation of temperatures may be analyzed by means of mode of hydration of the single surfactant and protein-surfactant mixtures. In the saturated solution of surfactant and its mixtures, both hydrophobic and hydrophilic hydration is possible while the aggregates of surfactant faced by hydrophilic hydration. However, reduction of both types of hydration is possible with an elevation of temperature and the de-solvation of polar head groups causes the favorable micellization. Many studies showed that elevation of temperature causes the disruption of H2O entities and rupturing of H-bonds around the hydrocarbon moiety of CDMEAB/BSA, stimulates the lessening of hydrophobic de-hydration, therefore, to decrease the micellization rate.11,17,50–52 Again, the β values of CDMEAB and (BSA + CDMEAB) mixed systems did not follow any regular bias with regard to the temperature in the presence and absence of sodium salts (Table 2).
3.4. Investigation of micellization process insight from thermodynamic states
The theoretical & practical investigation of drug–surfactant interaction, protein–surfactant interaction etc. significantly relies on the thermodynamic parameters. Collection of information of thermodynamic specifications helps to understand the consequences of a mode of collaboration among the studied constituents. Herein, we have evaluated different thermodynamic parameters of pure CDMEAB and (BSA + CDMEAB) systems in the presence as well as absence of salts through the following equations.32,55–58
ΔG0m = (1 + β)RT![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ln(Xc*) ln(Xc*)
| (1) |
ΔH0m = −(1 + β)RT2(∂![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ln ln![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Xc*/∂T) Xc*/∂T)
| (2) |
| ΔS0m=(ΔH0m − ΔG0m)/T | (3) |
Eqn (1) and (2) contains Xc* which denotes mole fraction of c*. ESI Fig. 1† depicts non-linear curve of ln(Xc*) versus T plot and these type of plots are used to calculate the magnitudes of ΔH0m. The values of tangents were measured as per temperature and then slopes are taken from the obtained tangents as the tantamount of ln(Xc*).59,60 The obtained magnitudes of different thermodynamic specifications for pure CDMEAB and (BSA + CDMEAB) mixed systems with and without salts (NaCl & Na2SO4) are summarized in Table 3.
| System | Medium | ISalta, mmol kg−1 | T/K | ΔG01,m, kJ mol−1 | ΔG02,m, kJ mol−1 | ΔG03,m, kJ mol−1 | ΔH01,m, kJ mol−1 | ΔH02,m, kJ mol−1 | ΔH03,m, kJ mol−1 | ΔS01,m, J mol−1 K−1 | ΔS02,m, J mol−1 K−1 | ΔS03,m, J mol−1 K−1 | ΔC01,m, kJ mol−1 K−1 | ΔC02,m, kJ mol−1 K−1 | ΔC03,m, kJ mol−1 K−1 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| a I = ionic strength of salts used.b Relative standard uncertainties (ur) limits are ur(ΔG0m), ur(ΔH0m), ur(ΔS0m), and (ΔmC0p) are 0.03, 0.03, 0.04 and 0.04 respectively. | |||||||||||||||
| CDMEAB | H2O | 0.00 | 298.15 | −46.77 | −44.75 | −41.46 | −41.67 | 20.50 | −11.56 | 17.11 | 218.83 | 100.28 | 3.75 | 0.52 | 1.12 |
| 303.15 | −49.60 | −47.66 | −43.13 | −23.21 | 23.11 | −6.09 | 87.05 | 233.44 | 122.18 | 4.07 | 0.53 | 1.21 | |||
| 308.15 | −49.80 | −46.76 | −44.75 | −0.89 | 25.65 | 0.34 | 158.72 | 234.98 | 146.33 | 4.39 | 0.53 | 1.30 | |||
| 313.15 | −50.02 | −47.95 | −45.04 | 21.48 | 28.50 | 7.08 | 228.33 | 244.15 | 166.45 | 4.72 | 0.53 | 1.38 | |||
| 318.15 | −50.31 | −48.14 | −45.54 | 45.74 | 31.03 | 14.36 | 301.91 | 248.84 | 188.29 | 5.04 | 0.53 | 1.47 | |||
| 323.15 | −51.22 | −48.76 | −46.07 | 71.66 | 33.73 | 21.60 | 380.25 | 255.27 | 209.41 | 5.34 | 0.54 | 1.55 | |||
| BSA + CDMEAB | H2O | 0.00 | 298.15 | −45.85 | −46.86 | −40.91 | −16.65 | 7.46 | −20.15 | 97.94 | 182.19 | 69.64 | 2.15 | 0.36 | 0.74 |
| 303.15 | −46.12 | −44.20 | −40.93 | −5.73 | 9.09 | −16.22 | 133.20 | 175.77 | 81.52 | 2.20 | 0.40 | 0.78 | |||
| 308.15 | −47.70 | −45.74 | −42.06 | 4.81 | 11.71 | −12.19 | 170.40 | 186.42 | 96.93 | 2.26 | 0.45 | 0.81 | |||
| 313.15 | −46.48 | −45.55 | −42.97 | 16.32 | 14.01 | −8.27 | 200.40 | 190.20 | 110.81 | 2.32 | 0.49 | 0.85 | |||
| 318.15 | −48.36 | −43.97 | −44.81 | 29.69 | 15.83 | −3.84 | 245.30 | 187.95 | 128.78 | 2.38 | 0.53 | 0.89 | |||
| 323.15 | −45.92 | −46.95 | −45.41 | 40.01 | 19.35 | 0.78 | 265.90 | 205.16 | 142.92 | 2.44 | 0.58 | 0.92 | |||
| BSA + CDMEAB | H2O–NaCl | 1.50 | 298.15 | −43.71 | −39.32 | −36.52 | −78.09 | −65.10 | −27.83 | −115.29 | −86.48 | 29.17 | 5.16 | 3.59 | 1.52 |
| 303.15 | −48.40 | −44.68 | −41.48 | −52.95 | −49.20 | −21.50 | −14.99 | −14.91 | 65.91 | 5.59 | 4.13 | 1.76 | |||
| 308.15 | −48.25 | −45.84 | −42.06 | −22.62 | −25.84 | −10.86 | 83.17 | 64.91 | 101.25 | 6.05 | 4.69 | 2.00 | |||
| 313.15 | −46.82 | −46.95 | −42.93 | 9.56 | 1.41 | 0.28 | 180.05 | 154.43 | 137.98 | 6.52 | 5.24 | 2.26 | |||
| 318.15 | −48.31 | −47.30 | −43.82 | 42.28 | 28.05 | 11.93 | 284.72 | 236.83 | 175.23 | 6.98 | 5.80 | 2.50 | |||
| 323.15 | −50.17 | −47.69 | −44.33 | 78.35 | 57.03 | 24.38 | 397.70 | 324.06 | 212.62 | 7.41 | 6.35 | 2.74 | |||
| BSA + CDMEAB | H2O–Na2SO4 | 1.50 | 298.15 | −41.42 | −39.62 | −36.76 | −92.92 | 15.58 | −13.08 | −172.71 | 185.16 | 79.42 | 5.81 | 0.60 | 1.14 |
| 303.15 | −43.80 | −41.38 | −39.94 | −64.48 | 18.77 | −7.69 | −68.21 | 198.40 | 106.39 | 6.25 | 0.63 | 1.26 | |||
| 308.15 | −45.38 | −42.13 | −41.98 | −31.23 | 22.21 | −0.80 | 45.93 | 208.81 | 133.64 | 6.68 | 0.66 | 1.37 | |||
| 313.15 | −45.53 | −41.72 | −41.03 | 4.40 | 25.22 | 6.66 | 159.45 | 213.74 | 152.30 | 7.14 | 0.69 | 1.52 | |||
| 318.15 | −45.46 | −41.84 | −41.52 | 39.90 | 28.52 | 14.42 | 268.32 | 221.16 | 175.81 | 7.58 | 0.72 | 1.62 | |||
| 323.15 | −46.57 | −43.01 | −42.30 | 79.05 | 32.66 | 22.48 | 388.75 | 234.17 | 200.47 | 8.00 | 0.75 | 1.73 | |||
The obtained magnitudes of ΔG0m (free energy changes) are conceived to be negative which provides strong evidence that the aggregation process is spontaneous in nature. The negative values of free energy changes are elevated with an elevation of temperatures. Such negative values of ΔG0m for the micellization of surfactants were also reported.19–22,55–58 These values are comparatively lower in presence of salts suggesting the slight reduction of dynamic forces for perfect interaction.
In the case of pure CDMEAB and (BSA + CDMEAB), the ΔH01,m, and ΔH03,m values are negative at the initial temperatures which suggest the process is exothermic in nature. These negative values are decreased with rising temperatures and revealed the endothermic process by altering the negative values to positive sign. The ΔH02,m values are positive (i.e. endothermic process) which are raised by means of rising temperatures. Again, all the values of entropy changes (ΔS01,m, ΔS02,m, and ΔS03,m) of pure CDMEAB and (BSA + CDMEAB) are positive and these values are raised by means of rising temperatures. Changes of entropy values point out that the aggregation process is entirely entropy controlled, especially at elevated temperatures. However, at the initial temperatures, the systems are both entropy and enthalpy controlled. These values of enthalpy and entropy also indicate the existence of hydrophobic (at the higher temperatures) and electrostatic (at the lower temperatures) interactions.
In presence of salts, the values of enthalpy (ΔH01,m, ΔH02,m, and ΔH03,m) for pure CDMEAB and (BSA + CDMEAB) are negative at the initial temperatures (except ΔH02,m values in presence of Na2SO4), which suggests the process is exothermic in nature. These negative values are decreased with rising temperatures and revealed the endothermic process by altering the negative values to positive sign. On the other hand, the values of entropy changes are positive except ΔS01,m and ΔS02,m (in presence of NaCl) & ΔS01,m (in presence of Na2SO4) values at 298.15 K and 303.15 K temperatures.
The negative values of ΔH0m demonstrate the existence of electrostatic interaction between the head groups, while the positive ΔH0m magnitudes demonstrate the disruption of water structure as well as the presence of hydrophobic interaction. Such negative and positive values of ΔH0m also reported by some groups previously.61–64
The positive magnitude of entropy changes (ΔS0m) of pure CDMEAB and (BSA + CDMEAB) mixed system can be analyzed by means of taking two considerable factors. Firstly, the disruption of iceberg structures as well as shifting of the hydrophobic moiety from H2O environment to hydrophobic environment of micelle cores. Secondly, the enhancement of free rotation of hydrophobic moiety inside the micelle core compared to H2O environment.65 On the other hand, a negative magnitude of entropy change (ΔS0m) may be possible only when the molecules of BSA and CDMEAB are affected due to the construction of iceberg structure around them.
ESI Fig. 2† shows the results of free energy changes (ΔG0m) which is form the enthalpic contribution (H01,m) together with entropic augmentation (−TΔS01,m). ESI Fig. 2† also depicts that enthalpy augmentation is gradually enhanced with rising temperatures while entropy augmentation showed opposite style in case of (BSA + CDMEAB) system in water. On the other hand, the supplementation of enthalpy along with entropy showed an interesting pattern in the salt medium. With an elevation of temperature, enthalpy contribution is increased and entropy contribution is decreased & showed a cross point. It is also clear from ESI Fig. 2† that both contributions are comparatively higher in salts medium than aqueous medium and provided the following pattern:
| Na2SO4> NaCl > H2O |
Molar heat capacity is an important thermodynamic aspect through which we may noticeably understand the way by which drug–surfactant or protein–surfactant interaction get motion restriction along with structural rearrangement/response to binding of ligands. The value of changes of molar heat capacity (ΔC0m) for the micellization of pure CDMEAB and (BSA + CDMEAB) mixed system are summarized in Table 3. These values were achieved by taking the corresponding slopes from the plots of enthalpy changes (ΔH0m) vs. temperature (T) by utilizing the below-mentioned relation (4).66,67
| ΔC0m = ((∂H0m)/∂T)P | (4) |
The achieved magnitudes of ΔC0m are positive in all cases and these values are clearly higher in the presence of salts than aqueous medium & followed the following trend (Table 3);
| (BSA + CDMEAB + NaCl) > (BSA + CDMEAB + Na2SO4) > (BSA + CDMEAB) > CDMEAB |
The alteration of molar heat capacity in case of BSA & CDMEAB mixtures can be considered to associate with motion restriction event. It is also nearly related to the molecular surface area as well as staying in the similar horizon of change in the solvent accessible surface area.68 The little positive molar heat capacity values, as well as entropy values, indicate the less structural changes in studied surfactant's micelle over the period of binding interaction.43
3.5. Transfer attributes of thermodynamics for (BSA + CDMEAB) mixed systems
The following eqn (5)–(7) were utilized in order to manipulate the transfer attributes of thermodynamic (such as free energy of transfer = ΔG0m.tr. enthalpy of transfer = ΔH0m.tr. & molar heat capacity of transfer = ΔC0p.m.tr.) for (BSA + CDMEAB) mixed systems.26,69,70| ΔG0m.tr. = ΔG0m(aq. additive) − ΔG0m(aq.) | (5) |
| ΔH0m.tr. = ΔH0m(aq. additive) − ΔH0m(aq.) | (6) |
| ΔC0p.m.tr. = ΔC0p.m.(aq. additive) − ΔC0p.m.(aq.) | (7) |
The evaluated values of free energy of transfer (ΔG0m.tr.), enthalpy of transfer (ΔH0m.tr.), & molar heat capacity of transfer (ΔC0p.m.tr.) for (BSA + CDMEAB) mixed systems in pure water and in salts solution are shown in ESI Table 1†. The calculated values of free energy of transfer (ΔG0m.tr.) are positive in the cases of (BSA + CDMEAB) mixed system in water and salts (except ΔG02.m.tr. at 298.15 K in water). The first transfer enthalpy values for (BSA + CDMEAB) system in water are positives at (298.15–308.15) K, while other values including second & third are negative. On the other hand, in the presence of electrolytes, the obtained values of transfer enthalpy are negative and the negative values are gradually lessened with rising temperatures and at the high temperature, it changed to positive magnitude. Including our previous study, some researchers also reported the negative as well as positive values of transfer enthalpy.3,26,71 Jha et al.72 reported that negative values of transfer enthalpy are possible about the driving of organic (amino acids) and inorganic (various salts) molecules to urea solution from the water. The transfer of amphiphilic molecules from water to (BSA + CDMAEB) and (BSA + CDMEAB + salts) medium creates exothermic as well as endothermic environment respectively. The molar heat capacity of transfer (ΔC0p.m.tr.) is negative (except at 318.15 & 323.15) of (BSA + CDMEAB) mixed system in water while the others values of (BSA + CDMEAB) mixed system in salts are positive. These positive values of ΔC0p.m.tr. signify that the micelles faced high hydration tendency which is determinable to achieve high H-bonding. On the contrary, negative magnitudes of ΔC0p.m.tr. signify that the hydration of the polar head group is more significant than structured water molecules about a non-polar group of monomeric surfactants.73 Apparently, the ΔC02,m.tr. value at 318.15 K is 0.00 kJ K−1 mol−1 which is not actually 0.00 kJ K−1 mol−1 except very low value. All the molar heat capacity of transfer (ΔC0p.m.tr.) are approximately constant which relates the noticeable structural changeover.
3.6. Investigation of enthalpy-entropy compensation for (BSA + CDMEAB) mixed systems
The following eqn (8) was utilized to evaluate the enthalpy-entropy compensation phenomena among the studied components. A linear relationship was also studied between ΔH0m and ΔS0m (ESI Fig. 3†). The straight lines between ΔH0m and ΔS0m provided excellent R2 values with a range of 0.950–0.999 which are more or less approximate to unity.26,74,75
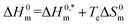 | (8) |
In the eqn (8), 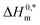 and Tc signs are denoted as intrinsic enthalpy gain and compensation temperature respectively. The values of
and Tc signs are denoted as intrinsic enthalpy gain and compensation temperature respectively. The values of 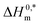 and Tc for (BSA + CDMEAB) mixtures in water and salts solution are summarized in Table 4. The intrinsic enthalpy
and Tc for (BSA + CDMEAB) mixtures in water and salts solution are summarized in Table 4. The intrinsic enthalpy 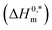 values are found negative, suggesting the convenient and stable micelle formation even at ΔS0m = 020, therefore stable micelle formation in water compared to the salt solution is also noticed by means of reduction of
values are found negative, suggesting the convenient and stable micelle formation even at ΔS0m = 020, therefore stable micelle formation in water compared to the salt solution is also noticed by means of reduction of 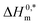 values in salts medium.
values in salts medium.
The 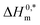 values demonstrate the solute–solute interaction along with an increase in hydrophobicity during micelle growth. The compensation temperature (Tc) values lie in the range of 278–363 K in the current study which shows good agreement with the literature values.10,20,48 Lumry et al.76 reported compensation temperature (Tc) from 270 to 300 K and suggested that it is the indication of H2O contributions in the protein solution as well as comparable to a biological fluid. Their finding supports our result with a good harmony. Ionic surfactants in water solution also showed same phenomena about compensation temperature.77
values demonstrate the solute–solute interaction along with an increase in hydrophobicity during micelle growth. The compensation temperature (Tc) values lie in the range of 278–363 K in the current study which shows good agreement with the literature values.10,20,48 Lumry et al.76 reported compensation temperature (Tc) from 270 to 300 K and suggested that it is the indication of H2O contributions in the protein solution as well as comparable to a biological fluid. Their finding supports our result with a good harmony. Ionic surfactants in water solution also showed same phenomena about compensation temperature.77
3.7. Insights from molecular dynamics
Molecular dynamics (MD) simulation can provide microscopic details about the interactions present between a protein and drug or surfactant molecules. Simulation results can provide information about the stability of a system and the structural changes that a protein may undergo in presence of other small molecules. To gain atomistic level understanding of the aggregation behavior of CDMEAB surfactant molecules under the influence of BSA, we have performed 20 ns all-atom simulation in the presence and absence of NaCl (Fig. 2). Salt ions are found to have significant contribution in stabilizing the protein-surfactant system as the positively charged sodium ions bind to the negatively charged amino acid residues of BSA. For instance, one sodium ion interacts with four amino acid residues Asp 13, Asp 254, Asp 255 and Asp 258 through three electrostatic interactions with the COO− moiety of the corresponding amino acids and a metal-acceptor bond with the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O group of the peptide linkage between Asp 254 and Asp 255 (ESI Fig. 4†). BSA contains 99 negatively charged residues including aspartic acid and glutamic acid. An interesting fact is that BSA being a plasma protein has significant ability to bind metal ions including Na+. Fig. 3(A) shows the change in overall Gibbs free energy of the system with simulation timescale and it is observed that the presence of 5% NaCl causes a reduction in the overall Gibbs free energy of the system. Therefore, it may be inferred that the presence of salt ion causes the protein-surfactant system to stabilize to a greater extent. The 20 ns simulation snapshot in ESI Fig. 5† depicts that in presence of NaCl, the surfactant molecules form larger aggregates and come in close contact with BSA compared to when salt ions are not present. BSA has three distinct albumin domains: 19–209; 210–402; 403–600. When salt ions are present, the surfactant molecules are found to be equally distributed in these three domains but in absence of salt, no surfactant molecules interact with the 210–402 domain. The surfactant molecules are also found to be strongly interacting with the unstructured regions of BSA.
O group of the peptide linkage between Asp 254 and Asp 255 (ESI Fig. 4†). BSA contains 99 negatively charged residues including aspartic acid and glutamic acid. An interesting fact is that BSA being a plasma protein has significant ability to bind metal ions including Na+. Fig. 3(A) shows the change in overall Gibbs free energy of the system with simulation timescale and it is observed that the presence of 5% NaCl causes a reduction in the overall Gibbs free energy of the system. Therefore, it may be inferred that the presence of salt ion causes the protein-surfactant system to stabilize to a greater extent. The 20 ns simulation snapshot in ESI Fig. 5† depicts that in presence of NaCl, the surfactant molecules form larger aggregates and come in close contact with BSA compared to when salt ions are not present. BSA has three distinct albumin domains: 19–209; 210–402; 403–600. When salt ions are present, the surfactant molecules are found to be equally distributed in these three domains but in absence of salt, no surfactant molecules interact with the 210–402 domain. The surfactant molecules are also found to be strongly interacting with the unstructured regions of BSA.
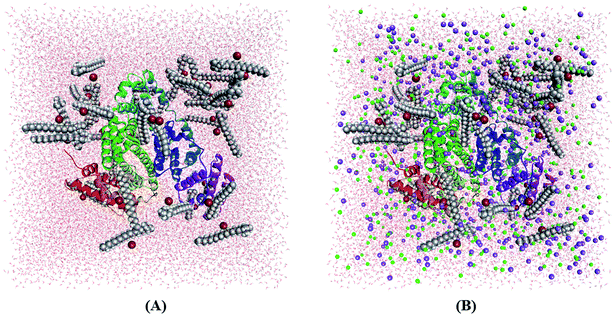 | ||
| Fig. 2 Simulation box containing BSA + 40 CDMEAB in (A) no-salt and (B) 5% NaCl environments (sodium in purple and chloride in green). | ||
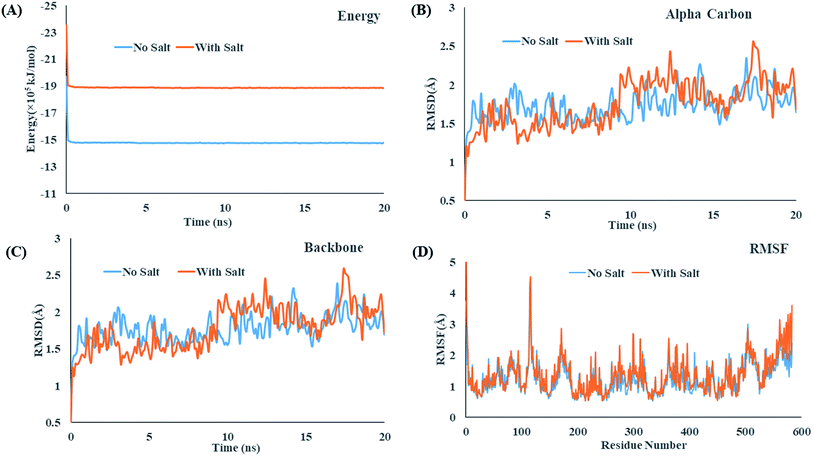 | ||
| Fig. 3 (A) Gibbs free energy change over 5 ns molecular dynamics, comparative RMSD trajectory of (B) alpha carbon, (C) backbone of BSA, (D) RMSF trajectory of BSA. | ||
Fig. 3(B and C) shows that the root means square deviation (RMSD) values of the alpha carbons and backbones of BSA remains almost similar throughout the simulation timescale and fluctuates around an average value. Presence of salt causes no significant change in the RMSD values which infers that the salt ions cause no significant structural changes of the protein. On the other hand, Fig. 4(A) depicts that the Solvent Accessible Surface Area (SASA) of the protein is much smaller in presence of NaCl. An explanation for the phenomenon can be inferred for the 20 ns simulation snapshot shown in ESI Fig. 5†. It reveals that in presence of salt, the larger aggregates of surfactant molecules stay in close contact with the protein surface that makes the surface less accessible to solvent molecules.
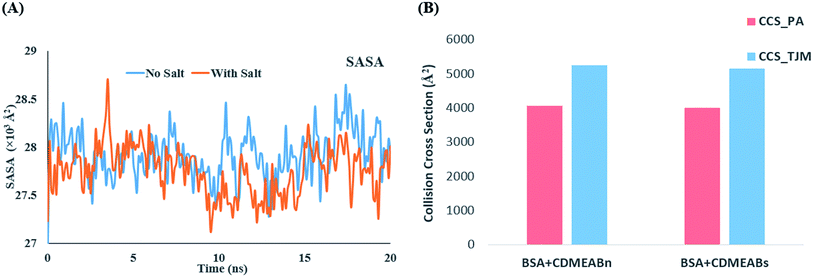 | ||
| Fig. 4 (A) Change in SASA over the simulation timescale (B) collision cross sections of BSA in salt (CDMEABs) and no salt (CDMEABn) environment. | ||
Fig. 3(D) shows that the root means square fluctuation (RMSF) of the amino acid residues (1–560) remains almost similar in the both aqueous and salt environment but the RMSF values of the residues 561–583 are significantly larger when salt ions are present. As proteins may undergo significant structural changes while interacting with other molecules, in recent years, the calculation of collision cross section (CCS) has received great attention for providing precise information about the conformation and size of a protein in different environments. Both the projection approximation (PA) and trajectory method (TJM) has been utilized to measure perform the CCS analysis of BSA in the presence and absence of salt.78,79 The CCS_PA and CCS_TJM values of BSA in presence of salt are 4000.59 Å2 and 5157.86 Å2 respectively which are slightly lower that when no salt ions are present (Fig. 4(B)). In spite of strong interaction between the protein and surfactant molecules, the overall collision cross section of BSA + CDMEAB system remains almost similar to the CCS value of native BSA.
4. Conclusion
Micellization and thermodynamic properties of cationic surfactant CDMEAB with the globular protein bovine serum albumin (BSA) have been investigated in aqueous solution together with the presence of two different salts at various temperatures through conductometric method. The values of critical micelle concentration (c*) enhanced due to the presence of BSA while the presence of salts comparatively declined the c* values. Different parameters of thermodynamics (ΔH0m and ΔS0m) demonstrated the existence of hydrophobic, exothermic and electrostatic interactions which are acting as binding forces among the protein and surfactant. Negative free energy changes (ΔG0m) are a strong indication of spontaneous micellization. The compensation temperature (Tc) of the studied systems suggest that the behavior of BSA + surfactant system along with additives are much comparable to a biological fluid. The negative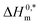 values demonstrate the excellent stability of BSA + surfactant aggregate. Molecular dynamics (MD) simulation results unveiled some interesting features of the molecular interaction between BSA and CDMEAB in the presence and absence of salt environment. Firstly, strong and homogenous interaction was predicated between BSA and CDMEAB in the presence of NaCl. Secondly, RMSD and RMSF values of alpha carbon and amino acid residues and collision cross section (CCS) of the protein remained same in aqueous and salt environments that showed that there is no significant structural change occurred to the protein structure.
values demonstrate the excellent stability of BSA + surfactant aggregate. Molecular dynamics (MD) simulation results unveiled some interesting features of the molecular interaction between BSA and CDMEAB in the presence and absence of salt environment. Firstly, strong and homogenous interaction was predicated between BSA and CDMEAB in the presence of NaCl. Secondly, RMSD and RMSF values of alpha carbon and amino acid residues and collision cross section (CCS) of the protein remained same in aqueous and salt environments that showed that there is no significant structural change occurred to the protein structure.
Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
Dr Mohammad A. Halim likes to acknowledge The World Academy of Science (TWAS) for providing funding (17-479 RG/CHE/AS_I) to purchase high performance computer for performing molecular dynamics simulation.References
- K. A. Majorek, P. J. Porebski, A. Dayal, M. D. Zimmerman, K. Jablonska, A. J. Stewartd, M. Chruszcz and W. Minor, Mol. Immunol., 2012, 52, 174 CrossRef CAS PubMed.
- T. Kosa, T. Maruyama and M. Otagiri, Pharmaceutical Research, 1997, 14, 1607 CrossRef CAS PubMed.
- Y. Matsui and A. Okimoto, Bull. Chem. Soc. Jpn., 1978, 51, 3030 CrossRef CAS.
- D. Bhatt, K. C. Maheria and J. Parikh, J. Surfactants Deterg., 2013, 16, 547 CrossRef CAS.
- K. S. Sharma, S. R. Patiland and A. K. Rakshit, Colloids Surf., A, 2003, 219, 67 CrossRef CAS.
- D. Kumar and M. A. Rub, J. Mol. Liq., 2018, 269, 1 CrossRef CAS.
- T. Randolf and L. Jones, Pharm. Biotechnol., 2002, 13, 159 Search PubMed.
- Q. Yue, T. Shen, C. Wang, C. Gao and J. Liu, Int. J. Spectrosc., 2012, 2012, 1678 Search PubMed.
- G. Li, J. Huang, T. Chen, X. Wang, H. Zhang and Q. Chen, Carbohydr. Polym., 2017, 176, 75 CrossRef CAS PubMed.
- S. M. A. Ahsan, M. R. Molla, M. S. Rahman, M. A. Hoque, M. A. Rub and M. A. Khan, J. Mol. Liq., 2019, 274, 484 CrossRef CAS.
- D. Attwood and A. Florence, Surfactant Systems–Their Chemistry, Pharmacy and Biology, Chapman and Hall, New York, 1983 Search PubMed.
- N. Azum, A. Z. Naqvi, M. A. Rub and A. M. Asiri, J. Mol. Liq., 2017, 240, 189 CrossRef CAS.
- U. Ashraf, O. A. Chat, M. Maswal, S. Jabeen and A. A. Dar, RSC Adv., 2015, 5, 83608–83618 RSC.
- N. Azum, M. A. Rub and A. M. Asiri, J. Dispersion Sci. Technol., 2017, 38, 96 CrossRef CAS.
- D. Kumar, K. E. Neo and M. A. Rub, J. Surfactants Deterg., 2016, 19, 101 CrossRef CAS.
- Y. K. Shrestha and F. Yan, RSC Adv., 2014, 4, 37274 RSC.
- M. J. Rosen, Surfactants and interfacial phenomena, Wiley, New York, 1959 Search PubMed.
- S. Deep and J. C. Ahluwalia, Phys. Chem. Chem. Phys., 2001, 3, 4583 RSC.
- M. R. Amin, S. Mahbub, S. Hidayathulla, M. M. Alam, M. A. Hoque and M. A. Rub, J. Mol. Liq., 2018, 269, 417 CrossRef CAS.
- S. Aktar, R. M. Molla, S. Mahbub, A. M. Rub, M. A. Hoque and D. S. Islam, J. Dispersion Sci. Technol., 2018 DOI:10.1080/01932691.2018.1472005.
- M. A. Hoque, M. M. Alam, M. R. Molla, S. Rana, M. A. Rub, M. A. Halim, M. A. Khan and A. Ahmed, J. Mol. Liq., 2017, 244, 512 CrossRef CAS.
- M. A. Hoque, M. M. Alam, M. R. Molla, S. Rana, M. A. Rub, M. A. Halim, M. A. Khan and F. Akhtar, Chin. J. Chem. Eng., 2018, 26, 159 CrossRef.
- H. Hamishehkar, S. Hosseini, A. H. Naseri, A. Safarnejad and F. Rasoulzadeh, BioImpacts, 2016, 6, 125 CrossRef CAS PubMed.
- D. Otzen, Biochim. Biophys. Acta, Proteins Proteomics, 2011, 1814, 562 CrossRef CAS PubMed.
- D. Kumar and M. A. Rub, J. Mol. Liq., 2018, 250, 329 CrossRef CAS.
- A. Z. Naqvi, M. A. Rub and Kabir-ud-Din, Colloid J., 2015, 77, 525 CrossRef CAS.
- M. A. Rub, N. Azum and A. M. Asiri, Russ. J. Phys. Chem. B, 2016, 10, 1007 CrossRef.
- I. Restrepo-Angulo, A. D. Vizcaya-Ruiz and J. Camacho, J. Appl. Toxicol., 2010, 30, 497 CrossRef CAS PubMed.
- W. Caetano and M. Tabak, J. Colloid Interface Sci., 2000, 225, 69 CrossRef CAS PubMed.
- M. Heshmat, V. P. Nicu and E. J. Baerends, J. Phys. Chem. A, 2012, 116, 3454 CrossRef CAS PubMed.
- D. Lu, Z. Liu and J. Wu, J. Chem. Phys., 2007, 126, 064906 CrossRef PubMed.
- S. Jana, S. Ghosh, S. Dalapati and N. Guchhait, Photochem. Photobiol. Sci., 2012, 11, 323 RSC.
- M. A. Hoque, F. Ahmed, M. A. Halim, M. R. Molla, S. Rana, M. A. Rahman and M. A. Rub, J. Mol. Liq., 2018, 260, 121 CrossRef CAS.
- K. H. Delgado-Magnero, P. A. Valiente, M. Ruiz-Peña, A. Pérez-Gramatges and T. Pons, Colloids Surf., B, 2014, 116, 720–726 CrossRef CAS PubMed.
- R. Amin, M. R. Molla, S. Rana, M. A. Hoque, M. A. Rub, M. Kabir and A. M. Asiri, Phys. Chem. Liq., 2018, 1–17, DOI:10.1080/00319104.2018.1519565.
- E. Krieger, T. Darden, S. B. Nabuurs, A. Finkelstein and G. Vriend, Proteins: Struct., Funct., Bioinf., 2004, 57, 678 CrossRef CAS PubMed.
- F. Krieger, B. Fierz, O. Bieri, M. Drewello and T. Kiefhaber, J. Mol. Biol., 2003, 332, 265 CrossRef CAS PubMed.
- J. A. Maier, C. Martinez, K. Kasavajhala, L. Wickstrom, K. E. Hauser and C. Simmerling, J. Chem. Theory Comput., 2015, 11, 3696 CrossRef CAS PubMed.
- T. Darden, D. York and L. Pedersen, J. Chem. Phys., 1993, 98, 10089 CrossRef CAS.
- E. G. Marklund, M. T. Degiacomi, C. V. Robinson, A. J. Baldwin and J. L. Benesch, Structure, 2015, 23, 791 CrossRef CAS PubMed.
- I. García-Mateos, S. Pérez and M. M. Velázquez, J. Colloid Interface Sci., 1997, 194, 356 CrossRef.
- M. A. Rub, A. M. Asiri, J. M. Khan, F. Khan and R. H. Khan, J. Taiwan Inst. Chem. Eng., 2014, 45, 2068 CrossRef CAS.
- M. A. Hoque, M.-O.-F. Patoary, M. R. Molla, M. A. Halim, M. A. Khan and M. A. Rub, J. Dispersion Sci. Technol., 2017, 38, 1578 CrossRef CAS.
- Sk. M. A. Ahsan, M. D. Hossain, M. A. Hoque and M. A. Khan, Indian J. Chem., Sect. A: Inorg., Bio-inorg., Phys., Theor. Anal. Chem., 2016, 55, 160 Search PubMed.
- M. Rub, A. Asiri, D. Kumar, N. Azum and F. Khan, Acta Phys.-Chim. Sin., 2014, 30, 699 CAS.
- F. Khan, M. S. Sheikh, M. A. Rub, N. Azum and A. M. Asiri, J. Mol. Liq., 2016, 222, 1020 CrossRef CAS.
- M. A. Hoque, M. D. Hossain and M. A. Khan, J. Chem. Thermodyn., 2013, 63, 135 CrossRef CAS.
- M. Rahman, M. A. Khan, M. A. Rub and M. A. Hoque, J. Mol. Liq., 2016, 223, 716 CrossRef CAS.
- M. Schick, J. Phys. Chem., 1963, 67, 1796 CrossRef CAS.
- G. Nemethy and A. Ray, J. Phys. Chem., 1971, 75, 809 CrossRef CAS.
- B. Sharma and A. K. Rakshit, J. Colloid Interface Sci., 1989, 129, 139 CrossRef CAS.
- C. La Mesa, Phys. Chem. Liq., 1990, 94, 323 CrossRef CAS.
- E. Crook, G. Trebbi and D. Fordyce, J. Phys. Chem., 1964, 68, 3592 CrossRef CAS.
- S. Chauhan, S. Kumari and K. Singh, J. Chem. Thermodyn., 2017, 105, 337 CrossRef CAS.
- J. Mata, D. Varade and P. Bahadur, Thermochim. Acta, 2005, 428, 147 CrossRef CAS.
- M. R. Molla, S. Rana, M. A. Rub, A. Ahmed and M. A. Hoque, J. Surfactants Deterg., 2018, 21, 231 CrossRef CAS.
- S. Mahbub, M. Rahman, S. Rana, M. A. Rub, M. A. Hoque and M. A. Khan, J. Surfactants Deterg., 2019, 22, 137–152 CrossRef CAS.
- N. Azum, M. A. Rub and A. M. Asiri, J. Solution Chem., 2016, 45, 791 CrossRef CAS.
- M. A. Rub, N. Azum, F. Khan and A. M. Asiri, J. Chem. Thermodyn., 2018, 121, 199 CrossRef.
- G. McDonnell and A. D. Russell, Clin. Microbiol. Rev., 1999, 12, 147 CrossRef CAS PubMed.
- F. Khan, M. A. Rub, N. Azum and A. M. Asiri, J. Phys. Org. Chem., 2018, 31, e3812 CrossRef.
- D. Kumar, S. Hidayathulla and M. A. Rub, J. Mol. Liq., 2018, 271, 254 CrossRef CAS.
- D. Kumar and M. A. Rub, J. Mol. Liq., 2017, 238, 389 CrossRef CAS.
- M. K. Al-Muhanna, M. A. Rub, N. Azum, S. B. Khan and A. M. Asiri, J. Dispersion Sci. Technol., 2016, 37, 450 CrossRef CAS.
- G. Stainsby and A. Alexander, Trans. Faraday Soc., 1950, 46, 587 RSC.
- M. A. Hoque, M. M. Rashid, M. R. Molla and M. A. Rub, J. Solution Chem., 2017, 46, 682 CrossRef CAS.
- O. Owoyomi, J. Ige and O. Soriyan, Chem. Sci. J., 2011, 25, 1 Search PubMed.
- I. Jelesarov and H. R. Bosshard, J. Mol. Recognit., 1999, 12, 3 CrossRef CAS PubMed.
- K. S. Sharma and A. K. Rakshit, J. Surfactants Deterg., 2004, 7, 305 CrossRef CAS.
- M. Rahman, M. A. Khan, M. A. Rub, M. A. Hoque and A. M. Asiri, J. Chem. Eng. Data, 2017, 62, 1464 CrossRef CAS.
- A. Rakshit and B. Sharma, Colloid Polym. Sci., 2003, 281, 45 CrossRef CAS.
- R. Jha and J. C. Ahluwalia, J. Chem. Soc., Faraday Trans., 1993, 89, 3465 RSC.
- H. S. Nalwa, Handbook of surfaces and interfaces of materials, five-volume set, Elsevier, 2001 Search PubMed.
- L.-J. Chen, S.-Y. Lin and C.-C. Huang, J. Phys. Chem. B, 1998, 102, 4350 CrossRef CAS.
- M. Rahman, M. A. Hoque, M. A. Khan, M. A. Rub and A. M. Asiri, Chin. J. Chem. Eng., 2018, 26, 1110 CrossRef.
- R. Lumry and S. Rajender, Biopolymers, 1970, 9, 1125 CrossRef CAS PubMed.
- G. Sugihara and M. Hisatomi, J. Colloid Interface Sci., 1999, 219, 31 CrossRef CAS PubMed.
- E. Mack Jr, J. Am. Chem. Soc., 1925, 47, 2468 CrossRef.
- M. Mesleh, J. Hunter, A. Shvartsburg, G. C. Schatz and M. Jarrold, J. Phys. Chem., 1996, 100, 16082 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9ra00070d |
| This journal is © The Royal Society of Chemistry 2019 |