Open Access Article
Open Access ArticleAccelerated biodegradation of PLA/PHB-blended nonwovens by a microbial community†
Yalan Liua,
Zhicheng Zhanb,
Haixian Yeb,
Xiaoshan Lina,
Yurong Yan*b and
Yi Zhang *a
*a
aSchool of Biology and Biological Engineering, South China University of Technology, Guangzhou 510006, China. E-mail: btyzhang@scut.edu.cn; Fax: +86-20-39380601; Tel: +86-20-39380602
bSchool of Materials Science and Engineering, South China University of Technology, Guangzhou 510641, China. E-mail: yryan@scut.edu.cn; Fax: +86-20-22236883; Tel: +86-20-87113644
First published on 2nd April 2019
Abstract
In this study, the accelerated biodegradation of PLA/PHB (polylactic acid/polyhydroxybutyrate)-blended nonwovens was investigated in the presence of a microbial community. The PLA/PHB-blended nonwovens were buried in natural soil for 56 days, with soil samples collected for subsequent bacterial community domestication. The tensile strength and elongation at break of the PLA/PHB-blended nonwovens as well as the CO2 generated by the Gen III and natural soil communities were determined to assess the degradation rates of the polymer samples. After incubation for 15 days with the Gen III soil bacterial suspension, the surfaces and fibrous structure of nonwovens and the fibers within the nonwovens exhibited distinct changes. In addition, the amount of EvCO2 reached 566.79 mg, the tensile strength decreased from 10.95 ± 0.7 to 2.57 ± 0.31 MPa, a loss of 77%, and the elongation at break changed from 5.32 ± 0.45 to 7.07 ± 1.04%. The 16S rRNA pyrosequencing results showed that Proteobacteria and Firmicutes were the 2 most important bacterial phyla in the Gen III community, accounting for 80.4 and 19.4% of the total classified sequences, respectively. The results of this study demonstrate that compared to a natural soil microbial community, the domesticated strains in the Gen III community, especially members of the phyla Proteobacteria and Firmicutes, are useful in accelerating the degradation of PLA/PHB-blended nonwovens.
Introduction
Because of the worldwide use of plastics, a growing volume of these plastics end up as waste in numerous ecosystems.1 The production of and expenditure on plastic materials grow at an average rate of 9% per year, with the production of plastics having grown from 1.5 million tons in 1950 to 245 million tons in 2013.2 However, plastic product waste causes serious environmental pollution and is a threat to Earth's ecology as well as to human health.The generation of “white pollution” and its enormous threat to marine life has resulted in an increased awareness of the use of traditional nondegradable polymer materials. Industrial data show that the USA, Europe, and Asia account for 85% of plastic pollution in the oceans, with Asia accounting for 80% of the total volume.3 The amount of marine plastic pollution derived from land-based sources is from 4.8 to 12.7 million metric tons annually.4 Thus, the demand for biodegradable plastics has increased in recent years, with an increased awareness of the plastic waste problem and its impact on the environment eliciting new interest in the area of degradable polymers.5
Poly(L-lactic acid) (PLA) is a bio-based polymer that has attracted a great deal of attention in recent years due to its many useful properties, including its high transparency, high tensile strength and excellent biodegradability, with its final degradation products consisting of carbon dioxide, water and other small molecules under composting conditions.6 However, because of a number of drawbacks of PLA, such as its low impact strength and poor heat resistance, it needs to be modified in some circumstances. A number of polymers are used to increase the toughness of PLA, such as poly(butylene adipate-co-terephthalate) (PBAT),7 PCL, polyurethane, polyamide elastomer (PAE),8 starch,9 GMA-based copolymers, thermoplastic elastomers and GMA grafted copolymers.10
To improve the toughness and alter the biodegradation period, the use of biodegradable polyhydroxybutyrate (PHB) was evaluated in a previous study.11 In 1926, the French scientist Lemoigne first discovered and synthesized the most common monomer in Bacillus megaterium, polyhydroxybutyrate (PHB), the first PHA identified and studied. Some properties of PHB are the same as those of the conventional plastic polypropylene, such as tensile strength, impact strength, degree of crystallinity, and melting temperature, although PHB is more brittle than PP and is more susceptible to thermal degradation. Because of its biodegradability, biocompatibility and nontoxicity, PHB is a promising candidate in biomedical and agricultural applications.12
Objective evaluations of the biodegradability of biodegradable materials is an effective means of guiding the rational application of materials, with degradation in natural and specific accelerated degradation environments being commonly investigated. In natural environments, a long period of time is required, sometimes even years, whereas the use of specific environments to accelerate degradation by bacteria that break down organic matter allows key aspects of the degradation process to be investigated and evaluated. In 1977, microbial enzymes were identified that can attack synthetic, man-made polymers.13 However, the activity of these enzymes appeared to be limited to the biodegradation of aliphatic polyesters, which have few useful properties, excluding the use of biodegradable plastics from many applications. In 1995, Witt and Müller discovered aliphatic–aromatic copolyesters with a specific composition could be degraded by microorganisms, and they concluded that great improvements could be made in the characteristics of polymers.14 The biodegradation behavior of PLA and its blends has been investigated in a number of studies. Weng et al. studied the biodegradation of PLAs with different chemical structures under controlled composting conditions,15 while Rudnik and Briassoulis16 compared the biodegradation of PLA and PHA in soil environments and under composting conditions. Their results showed that these polymers could be biodegraded at a high rate in both real soil environments and under composting conditions. Thus, the microorganisms in soil and compost may play a major role in the biodegradation of renewable polymers.
In contrast, the degradation of PLA/PHB-blended nonwovens under natural conditions, including in soil environments and under composting conditions, was observed to occur at a very low rate,17 which is a major drawback limiting their usefulness. However, accelerating the biodegradation process would overcome this limitation and make these blends much more attractive. As the PLA/PHB = 80/20 is the most commonly used ratio in manufacturing applications, we applied the PLA/PHB-blended nonwovens with the same ratio in this study. In this study, we domesticated a soil bacterial community to enhance the efficiency and shorten the duration of the biodegradation process of PLA/PHB-blended nonwovens, the preliminary results of which showed that their biodegradation occurred much faster and more completely in domesticated bacterial suspensions than in a natural environment. These results indicated that the domesticated bacterial community was a strong driver promoting this biodegradation process.
Identifying the microbial communities involved in enhancing the biodegradation process and studying the microbial community composition changes during this process are necessary to reveal the mechanism promoting the rapid biodegradation of polymers. The results of previous studies demonstrated that advanced molecular techniques are provided powerful tools for detecting bacterial community structure and diversity in environmental samples, as well as for evaluating the biodegradation of PLA/PHB-blended nonwovens.18 In this study, high-throughput pyrosequencing was performed to quantify the abundance, diversity and structure of a domesticated bacterial community that promoted the rapid biodegradation of polymers from renewable biomass.
Experimental
Materials
The LB culture medium used in this study was produced by Guangdong Huankai Microbial Co., Ltd., while the chemicals (standard Q/HK3) K2HPO4·3H2O (Guangzhou Chemical Reagent Factory), MgSO4·7H2O (Guangzhou Chemical Reagent Factory), MnSO4·H2O (Shantou Guanghua Chemical Reagent Factory), NH4NO3 (Guangzhou Chemical Reagent Factory), and NaCl (Guangzhou Chemical Reagent Factory) were all analytical reagents and used without additional purification. Chloroform was purchased from Shantou Xilong Chemical Co. Ltd. The PLA (grade 6202D) and PHB (grade 18J45) used in this study were produced by Natureworks LLC (USA) and BASF SE (Germany), respectively.PLA/PHB-blended nonwovens
The PLA/PHB-blended nonwoven material was prepared on a traditional spunbonded nonwoven production line with a PLA/PHB weight ratio of 80/20 and was kindly provided by U.S. Pacific, China. The nonwoven samples had a basic weight of 40 g m−2 and were used as received other than being cut into 30 cm × 20 cm specimens for biodegradation assays.Biodegradation of PLA/PHB buried in a soil environment
The soil study site was located in Guangzhou University Town, China, at a longitude of 113.4 and a latitude of 23.0 according to GB/T 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 006. Nonwoven specimens were buried in ordinary orchard soil at a depth of 15–20 cm for 56 days, with 3 sample pieces excavated for testing every 14 days. After being washed with distilled water 3 times, the samples were dried at room temperature and then stored in a refrigerator for further testing.19 The average temperature and relativity humidity during the testing period are presented in Fig. S2.† During the experiment period (from March to May), the temperature in the Panyu District of Guangzhou City ranged from 16.6 to 30.5 °C, with an average temperature was 23.89 °C, while the humidity ranged from 51.5 to 96%, with an average humidity of 80.79% (Fig. S2†).
006. Nonwoven specimens were buried in ordinary orchard soil at a depth of 15–20 cm for 56 days, with 3 sample pieces excavated for testing every 14 days. After being washed with distilled water 3 times, the samples were dried at room temperature and then stored in a refrigerator for further testing.19 The average temperature and relativity humidity during the testing period are presented in Fig. S2.† During the experiment period (from March to May), the temperature in the Panyu District of Guangzhou City ranged from 16.6 to 30.5 °C, with an average temperature was 23.89 °C, while the humidity ranged from 51.5 to 96%, with an average humidity of 80.79% (Fig. S2†).
Soil samples collection and bacterial community domestication
Soil samples were collected from an orchard at the same longitude and latitude at a depth of 15–20 cm and were sieved through a 2 mm-sized sieve to remove particles larger than 2 mm.20 The soil was prepared according to ISO-11274 1998, and the pH of the suspensions was tested according to ISO 10390 2005. LB culture medium was added to the suspensions to form uniform suspensions and then was sterilized at 115 °C for 20 min.Sterile LB culture medium (2.1 g) and 100 mL of distilled water was added to a 500 mL flask, sterilized at 115 °C for 20 min and then cooled down for further use. Next, 10 g of screened soil samples were added into the sterilized LB liquid medium and cultured for 24 hours with shaking at 190 rpm at 37 °C, after which the first generation of the soil bacterial suspension (‘Gen I’) was collected. The second generation of bacteria ‘Gen II’ was derived from a suspension culture of liquid medium seeded with 1 mL of ‘Gen I’ inoculated into 100 mL of sterilized LB liquid medium using the same conditions described above. The third and the fourth generations of bacteria (‘Gen III’ and ‘Gen IV’, respectively) were prepared in the same way and so on until the composition of the domesticated bacterial community stabilized (Fig. S1, Table S1†). The final resulting bacterial suspensions were stored at 4 °C. Because the ‘Gen III’ community exhibited a stable composition, it was used as an inoculant in further tests.
Biodegradation tests of polymer samples in the soil bacterial suspensions
Jars with a 500 mL working volume were used as reactors. The jars were covered and sealed with 8 layers of gauze, sterilized for 20 min at 115 °C, and then allowed to cool. Each reactor was filled with 7.35 g of LB culture medium, 350 mL of distilled water, and 5 pieces of nonwoven samples, which were all completely immersed into the liquid medium. One milliliter of the ‘Gen III’ suspensions was inoculated into the jar and was cultured for 24 hours with shaking at 150 rpm at room temperature. All of the reactors were cultured at room temperature for 24 hours in a shaker and incubated at room temperature for 15 days. Samples were collected from the reactor every 3 days and were washed carefully with distilled water 4 times, dried at room temperature and then stored in a refrigerator for further testing.21Assessment of accelerated biodegradation
Evaluation of the biodegradability of the nonwoven blends
According to ISO 14852 (ref. 24), determination of the CO2 generated during aerobic polymer biodegradation is the most commonly used method to assess the degree of biodegradation. The detection device shown in Fig. 1 was designed according to ISO 14852 and consisted of an air pump, air CO2 absorption bottles, a bioreactor and evolved CO2 absorption bottles. The air was passed through the absorption bottles containing NaOH solution at a rate of 50–100 mL min−1 to eliminate CO2, after which the CO2-free air entered the bioreactor. The degradation of the PLA/PHB-blended nonwovens was carried out using a thermostatic magnetic stirrer to control the temperature and rotation speed. Evolved CO2 from the degraded PLA/PHB-blended nonwovens was absorbed by Ba(OH)2 in the absorption bottles, and the resulting BaCO3 was collected to quantity the evolved CO2.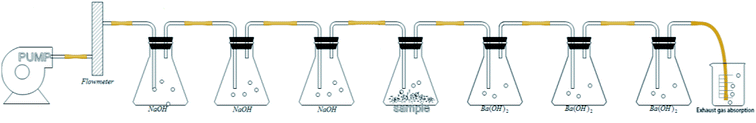 | ||
| Fig. 1 Device used to detect the amount of CO2 evolved by PLA/PHB-blended nonwovens during the degradation process. | ||
Five grams of the PLA/PHB-blended samples was cut into pieces and placed into the bioreactors containing 200 mL of detection solution. Next, 1.0 mL of the mixed bacterial suspension was used to inoculate the bioreactor and was cultured at 30 °C for a specific length of time. Following the biodegradation experiments, the weight of the collected BaCO3 was quantified to calculate the amount of evolved CO2 (EvCO2) generated during the biodegradation assay. The theoretical evolved CO2 (ThCO2) can be calculated from the initial input amount of the blended nonwoven sample and the total organic carbon (TOC) of that material. A control, untreated sample was also run at the same time as the treatments.
Total DNA extraction and PCR amplification
Microbes in ‘Gen III’ were collected for DNA extraction, which was performed using a soil total DNA isolation kit (OMEGA Bio-Tek, USA) following the manufacturer's instructions. The specific primers used to amplify the V3 region (161 bp) of the bacterial 16S rRNA gene were F341 (5′-CCTACGGGAGGCAGCAG-3′) and R534 (5′-ATTACCGCGGCTGCTGG-3′).25 The PCR amplification system included the following: 10× PCR buffer, 0.2 mmol L−1 dNTPs, upstream and downstream primers (0.4 μmol L−1 each), 1 U Ex Taq polymerase (TaKaRa), 10 ng of the sample DNA as template, and water to a total volume of 25 μL.26 The amplification steps were as follows: a 95 °C predenaturation for 5 min followed by 25 cycles of 94 °C for 45 s, 65–55 °C for 30 s (the annealing temperature was progressively reduced from 65 to 55 °C at a rate of 1 °C every 2 cycles, with 5 cycles at 55 °C), and 72 °C for 1 min, with a final extension at 72 °C for 8 min.High-throughput pyrosequencing analysis
The DNA samples were sent to Sangon Biotech Co. Ltd. (Shanghai, China) for pyrosequencing of the V3 region of the 16S rRNA gene following the 454 GS-FLX Titanium protocol (454 Life Sciences/Roche Diagnostics, CT, USA), which yields reads lengths of ∼400 bp.27,28 The paired sequences were combined using FLASH (http://ccb.jhu.edu/software/FLASH) with the minimum overlap value set to 10 bp and the default settings used for the other parameters. The PCR artifacts were denoised by eliminating the low-quality sequences using Trimmomatic.29 In UPARSE, the quality-filtered sequences were clustered into de novo OTUs at a 97% similarity threshold, with chimeric sequences removed using Uchime.30 USEARCH was used to map the reads to the OTUs to generate an OTU abundance table. We removed one outlier with a very low sequence count and microbial diversity. MOTHUR was used to construct rarefaction curves and to calculate ecological indices of the Chao1 estimator and Shannon diversity/richness. The sequences were classified using RDP classifier at a 80% confidence threshold for sequences longer than 200 bp. The genera obtained from the RDP classifier were clustered via the complete linkage hierarchical clustering technique using the R package Hclust CLUST (http://sekhon.berkeley.edu/stats/html/hclust.htmL). The goal of the analysis was to distinguish between the bacterial species at genus, family, order, class or phylum level rather than to generate complete profiles of bacterial genes.Results and discussion
Changes in the mechanical properties and physical appearance of the PLA/PHB-blended nonwovens
During degradation, the primary polymer chains break down or become crosslinked, which depends on the mechanism of the degradation, and all degradation processes impact the mechanical properties of products. The tensile strength and the elongation at break values of the tested PLA/PHB-blended nonwoven samples are reported in Table 1.| Test type | Incubation time (days) | Tensile strength (MPa) | Elongation at break (%) |
|---|---|---|---|
| Degradation in soil bacterial suspensions Gen III | 0 | 10.95 ± 0.77 | 5.32 ± 0.45 |
| 3 | 6.82 ± 0.44 | 5.33 ± 0.32 | |
| 6 | 5.69 ± 0.70 | 7.94 ± 1.09 | |
| 9 | 4.96 ± 0.54 | 7.60 ± 1.70 | |
| 12 | 2.83 ± 1.00 | 4.67 ± 1.14 | |
| 15 | 2.57 ± 0.31 | 7.07 ± 1.04 | |
| Degradation in the soil burial environment | 0 | 10.95 ± 0.77 | 5.32 ± 0.45 |
| 14 | 9.40 ± 0.89 | 7.26 ± 0.71 | |
| 28 | 8.48 ± 1.41 | 5.82 ± 0.71 | |
| 42 | 8.50 ± 1.00 | 4.80 ± 1.00 | |
| 56 | 7.73 ± 0.54 | 4.45 ± 0.54 |
The results presented in Table 1 show that a decrease in tensile strength occurred under both degradation conditions, but opposing trends and fluctuations in the changes were observed for the elongation at breaks. When the samples were degraded in Gen III soil bacterial suspension, the tensile strength decreased from 10.95 ± 0.77 to 2.57 ± 0.31 MPa, a loss of 77%, while the elongation at break fluctuated but increased from 5.32 ± 0.45 to 7.07 ± 1.04%. For samples buried in the soil, slower decreasing trends were observed for both the tensile strength (from 10.95 ± 0.77 to 7.73 ± 0.54 MPa, a loss of 30%) and in the elongation at break values (from 5.32 ± 0.45 to 4.45 ± 0.54%). When the tested samples were immersed in liquid (Gen III), the bonding points in the nonwovens exhibited a debonding behavior that decreased the tensile strength of nonwovens and increased the sliding between fibers inside the nonwovens, which increased the elongation at break values of the tested samples. However, because the swelling and soaking behavior of liquid is not appreciable in soil, biodegradation is the only mechanism through which the fibers can be degraded. Thus, the observed breakage of nonwovens was only related to the biodegradation of the fibers, and the variation showed changes in both tensile strengths and elongation at break values.
Comparing with the degradation observed in Gen III soil bacterial suspension and in the buried soil environment, we observed that the tensile strength of the PLA/PHB-blended nonwovens in the buried soil environment for 45 days was the same as that of the samples incubated with the Gen III soil bacterial suspension for 3 days. The variation in the nonwoven samples incubated in the bacterial suspension compared to that observed in the samples buried in natural soil is further evidence that the degradation of the PLA/PHB-blended nonwovens can be accelerated in soil bacterial suspensions.
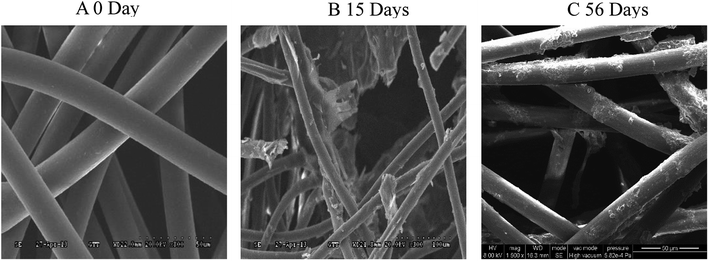
Morphological analysis of the degradation of PLA/PHB-blended nonwovens
The morphology of the PLA/PHB (80/20) blends showed a sea-island pattern with of island phase of PHB and sea phase of PLA.31 The SEM image in Fig. 2 shows the morphology of the PLA/PHB-blended nonwovens. The morphologies of test samples that were buried in the buried soil environment for 56 days and those of samples that were exposed to the Gen III soil bacterial suspensions for 15 days in the laboratory are shown in Fig. 2A and B, respectively. Morphological changes can be clearly observed for all of the samples. At the start of biodegradation (0 day), obviously regular, homogenous fibers with smooth surfaces inside the PLA/PHB-blended nonwovens were observed. After 56 days of being buried in the soil, slight scratches appeared in the fiber surfaces, and the fibrous structure of the test samples had undergone minimal damage. These changes indicated that the biodegradation processes had begun, albeit slowly. However, after incubation for 15 days with the Gen III soil bacterial suspension, the surfaces and fibrous structure of the nonwovens and fibers within the nonwovens exhibited distinct changes. The scanning electron microscopy results showed that pores had appeared on the surface of the samples, and microcracks were also visible within the fibrous reticular structure. The presence of these phenomena indicated that the PLA/PHB-blended nonwovens had rapidly degraded in the domesticated bacterial suspension.For the dispersion of PHB (island) in the PLA (sea) matrix, the crystal structure of PLA could be improved. The possibility of small molecules such as water to penetrate the surface of both crystal structure and the amorphous region would be also increased. They could also have contributed the acceleration of the bio-degradation of the PLA/PHB-blended nonwovens. The relatively high concentrations of the bacterial inoculum and the high inorganic salt concentration in our suspension were the primary reasons for the rapid biodegradation of the PLA/PHB-blended nonwovens. The domesticated bacterial community may have increased the degradation of the nonwovens by secreting enzymes that catalyzed the hydrolysis of the ester linkages in the samples. When the macromolecular chains of the PLA/PHB-blended fibers within the nonwovens break down into smaller pieces, bacteria can use the small-molecule substances as nutrient resources during the biodegradation process.32 Compared to the samples degrading naturally in soil, the degradation of the polymer samples in the domesticated bacterial suspension was accelerated.
Detection of biodegradation using the domesticated bacterial suspension
To verify the biodegradability of the PLA/PHB-blended nonwovens in the Gen III soil bacterial suspension, the standard ISO 14852 method was used to determine the amount of evolved CO2. For this experiment, the ratio of EvCO2 to ThCO2 was used to indicate the degree of biodegradation of the PLA/PHB-blended nonwovens. The biodegradation of the PLA/PHB-blended nonwovens was assessed in the presence of 0.5% of the Gen III soil bacterial suspension using a ratio of nonwovens to liquid of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 40. As shown in Fig. 3, the percentage of evolved CO2 from the nonwoven samples increased in the presence of the bacterial suspension. The biodegradation of the PLA/PHB-blended nonwovens began 3 days after inoculation, and after degrading for 15 days, the amount of EvCO2 reached 566.79 mg, indicating a biodegradation yield of 9.93%. In contrast, the percentage of evolved CO2 increased slowly and remained at a low level in the control, where the domesticated bacterial suspension was substituted with sterilized water. The results were similar to those of a previous study24 and showed that the renewable PLA/PHB-blended nonwovens could be rapidly degraded and utilized by the domesticated bacterial suspension.
40. As shown in Fig. 3, the percentage of evolved CO2 from the nonwoven samples increased in the presence of the bacterial suspension. The biodegradation of the PLA/PHB-blended nonwovens began 3 days after inoculation, and after degrading for 15 days, the amount of EvCO2 reached 566.79 mg, indicating a biodegradation yield of 9.93%. In contrast, the percentage of evolved CO2 increased slowly and remained at a low level in the control, where the domesticated bacterial suspension was substituted with sterilized water. The results were similar to those of a previous study24 and showed that the renewable PLA/PHB-blended nonwovens could be rapidly degraded and utilized by the domesticated bacterial suspension.
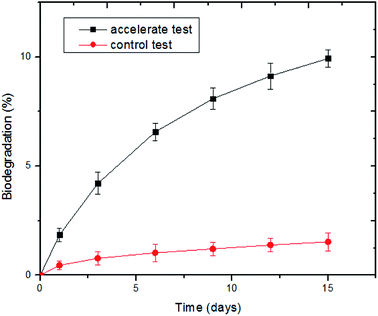 | ||
| Fig. 3 Percentage of evolved CO2 from the PLA/PHB-blended nonwovens with different degradation time in the presence of the domesticated bacterial suspension. | ||
Analysis of OTU abundance and species identification
The biodegradability of mixed PLA/PHB-blended nonwovens depends on the composition of the degrading microorganisms in the domesticated bacterial suspension. To quantify the abundance, diversity and structure of the domesticated bacterial community, a high-throughput pyrosequencing analysis was performed. The amplicon libraries were constructed using primers to amplify the V3 region of the 16S rRNA gene, and high-throughput sequencing was performed on a 454 GS-FLX Titanium protocol to distinguish the bacteria at different taxonomic levels.After sequencing, 2.5 M raw sequence reads were obtained from the samples, and the corresponding number of bases in the raw sequences was 0.38 Gb. After the connector sequences, low-complexity, and low-quality sequences were removed, 98.1% of the remaining reads were retained. As shown in Table 2, 9913 sequences were obtained after assembly, and after removing the primers and redundant sequences, 8441 sequences remained. The number of bacterial species was estimated by the number of observed operational taxonomic units (OTUs). In the Gen III samples, 546 OTUs were identified that covered 96.3% of the remaining sequences. To further study the bacterial community structure, taxonomies of the microbes were analyzed at phylum and genus levels depending on the sequencing data.
Bacterial diversity and community structure analysis
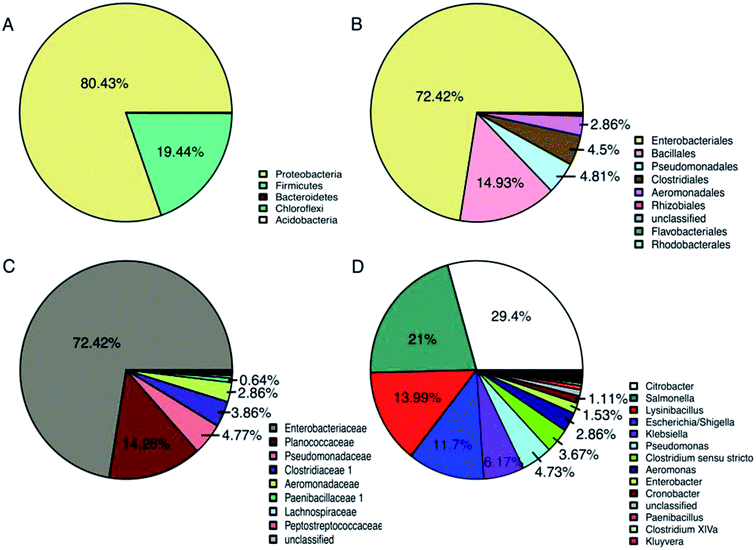
The distribution of partial bacterial 16S rRNA gene sequences at different levels from the domesticated Gen III sample are shown in Fig. 4. From a total of 8441 sequences obtained from the pyrosequencing analysis, only 2 bacterial phyla were identified, with Proteobacteria and Firmicutes accounting for 80.43 and 19.44%, respectively, representing over 99% of total classified sequences (Fig. 4A). In the most abundant phylum, Proteobacteria, the unique tag sequences could be classified into 3 orders and 8 genera (>1% of all sequences). At the order level (Fig. 4B), these taxa included Enterobacteriales (70.91%), Pseudomonadales (4.73%), and Aeromonadales (2.86%). At the genus level (Fig. 4D), the primary genera were Citrobacter (29.4%), Salmonella (21.00%), Escherichia (11.70%), Klebsiella (6.17%), Pseudomonas (4.73%), Aeromonas (2.86%), Enterobacter (1.53%), and Cronobacter (1.11%). In the second most abundant phylum Firmicutes, the unique tag sequences could be classified into 2 orders and 2 genera (>1% of all sequences). At the genus level, these taxa included Lysinibacillus (13.99%) and Clostridium sensu stricto (3.67%), which belong to orders Bacillales and Clostridiales, respectively (Fig. 4D).The above results suggested that due to the abundance of the tag sequences, members of the phyla Proteobacteria (especially those belonging to the genera Citrobacter, Salmonella, Escherichia, and Pseudomonas) and Firmicutes (especially those belonging to the genera Lysinibacillus and Clostridium) may play significant roles in the biodegradation of PLA/PHB-blended nonwovens. Previously, bacterial communities capable of biodegrading mixed PLA and PHB materials were assessed and identified using clone libraries and DNA sequencing to the phylum and class levels. In these studies, the majority of cloned gene sequences belonged to different bacterial phyla, namely, Actinobacteria, Bacteroidetes, Chloroflexi, Firmicutes, Gemmatimonadetes, Planctomycetes, Proteobacteria, and TM7.33,34 However, in the present study, only 2 bacterial phyla (Proteobacteria and Firmicutes) were detected. Although the phylum Actinobacteria has often been reported to be capable of biodegrading PLA and other biopolymers,34,35it was not observed in this study. Interestingly, the composition of bacterial phyla obtained from the domesticated cultivation in this study was different from previous results reported from natural soil and composts.
In some cases, the bacterial composition results obtained from cultivation and gene sequencing data were not identical. The microbial diversities identified from the cultivation and gene sequencing methods were different. It is possible that Actinobacteria had a slower growth rate than did strains belonging to the phyla Firmicutes and Proteobacteria. Members of the phyla Firmicutes and Proteobacteria may also rapidly deplete nutrients from the media that are essential for the growth of Actinobacteria, leading to their eventual failure to grow in the subsequent cultivation. Another possible reason for the absence of Actinobacteria is the sensitivity of members of this phylum to certain media constituents, such as ammonia, amino acids, phosphate, vitamins and salts, or antibiotics produced by other bacteria growing in the cultivation.33,36
In the present study, the relative abundance of tag gene sequences in the samples was closely affiliated with members of the genera Citrobacter and Pseudomonas (phylum Proteobacteria) and Lysinibacillus (phylum Firmicutes), indicating that these newly identified bacterial species may play a role in the biodegradation of PLA/PHB-blended nonwovens. Several species belonging to the genus Citrobacter, such as C. amalonaticus, C. koseri, and C. freundii, have been reported to be capable of using polyhydroxy compounds as a sole carbon source. Citrobacter species can also be differentiated by their abilities to convert tryptophan to indole, ferment lactose, and utilize malonate.14 Pseudomonas is capable of growing in diesel and jet fuel and can degrade natural rubber,37 while Lysinibacillus can degrade xylan.38 Although these phylogenetic lineages present in the domesticated cultivation have been reported to play important roles in the degradation of organic matter or polymers, they have not been previously suggested to be associated with the degradation of the PLA/PHB-blended nonwovens.
The results obtained in this study are useful for designing cultivation strategies to accelerate the biodegradation of renewable artificial polymers and may further expand our understanding of the biodegradation mechanisms in domesticated systems. These dominant bacterial strains could be isolated and developed for use in the efficient degradation and recycling of renewable PLA/PHB-blended nonwovens. The isolation of novel strains and the characterization of enzymes produced by these microorganisms will allow for a better understanding of the degradation and cycling of renewable polymer blends in natural environments, such as soil and compost.
Conclusions
In this study, a method for the rapid degradation of PLA/PHB-blended nonwovens using domesticated bacterial strains was demonstrated. The principal components of the bacterial communities in the domesticated cultivation suspensions were identified to reveal the taxa that promoted the biodegradation process. Proteobacteria and Firmicutes were the 2 dominant bacterial participants, accounting for 80.43 and 19.44% of the total classified microbial sequences involved in the biodegradation process, respectively, and may contribute to the high biodegradation of the PLA/PHB-blended nonwovens.Conflicts of interest
The authors have no conflicts of interest.Acknowledgements
This research was financially supported by the Joint Funds of the National Natural Science Foundation of China (Research project No: U1301231) and the Natural Science Foundation of Guangdong (Research project No: 2013B090600028).References
- T. An, S. Schaeffer, S. Li, S. Fu, J. Pei, H. Li, J. Zhuang, M. Radosevich and J. Wang, Carbon fluxes from plants to soil and dynamics of microbial immobilization under plastic film mulching and fertilizer application using 13C pulse-labeling, Soil Biol. Biochem., 2015, 80, 53–61 CrossRef CAS.
- W. Sikorska, P. Dacko, M. Sobota, J. Rydz, M. Musio and M. Kowalczuk, Degradation Study of Polymers from Renewable Resources and their Compositions in Industrial Composting Pile, Macromol. Symp., 2010, 272, 132–135 CrossRef.
- P. Kumar, Role of Plastics on Human Health, Indian J. Pediatr., 2018, 85, 384–389 CrossRef PubMed.
- M. Haward, Plastic pollution of the world's seas and oceans as a contemporary challenge in ocean governance, Nat. Commun., 2018, 9, 667 CrossRef PubMed.
- X. Ping, K. Wang, M. Jia and M. Yang, Biodegradation and mechanical property of polylactic acid/thermoplastic starch blends with poly(ethylene glycol), J. Wuhan Univ. Technol., Mater. Sci. Ed., 2013, 28, 157–162 CrossRef.
- D. V. Cong, T. Hoang, N. V. Giang, N. T. Ha, T. D. Lam and M. Sumita, A novel enzymatic biodegradable route for PLA/EVA blends under agricultural soil of Vietnam, Mater. Sci. Eng., C, 2012, 32, 558–563 CrossRef CAS.
- Y. X. Weng, Y. J. Jin, Q. Y. Meng, L. Wang, M. Zhang and Y. Z. Wang, Biodegradation behavior of poly(butylene adipate-co-terephthalate) (PBAT), poly(lactic acid) (PLA), and their blend under soil conditions, Polym. Test., 2013, 32, 918–926 CrossRef CAS.
- H. Kang, B. Qiao, R. Wang, Z. Wang, L. Zhang, J. Ma and P. Coates, Employing a novel bioelastomer to toughen polylactide, Polymer, 2013, 54, 2450–2458 CrossRef CAS.
- S. Zhang, X. Feng, S. Zhu, H. Qian, K. Han, Y. Ma and M. Yu, Novel toughening mechanism for polylactic acid (PLA)/starch blends with layer-like microstructure via pressure-induced flow (PIF) processing, Mater. Lett., 2013, 98, 238–241 CrossRef CAS.
- V. Nagarajan, A. K. Mohanty and M. Misra, Perspective on Polylactic Acid (PLA) based Sustainable Materials for Durable Applications: Focus on Toughness and Heat Resistance, ACS Sustainable Chem. Eng., 2016, 4, 2899–2916 CrossRef CAS.
- M. P. Arrieta, J. López, A. Hernández and E. Rayón, Ternary PLA–PHB–Limonene blends intended for biodegradable food packaging applications, Eur. Polym. J., 2014, 50, 255–270 CrossRef CAS.
- F. Acevedo, P. Villegas, V. Urtuvia, J. Hermosilla, R. Navia and M. Seeger, Bacterial polyhydroxybutyrate for electrospun fiber production, Int. J. Biol. Macromol., 2017, 106, 692–697 CrossRef PubMed.
- Y. Tokiwa and T. Suzuki, Hydrolysis of polyesters by lipases, Nature, 1977, 270, 76–78 CrossRef CAS PubMed.
- U. Witt, R. J. Müller and W. D. Deckwer, New biodegradable polyester-copolymers from commodity chemicals with favorable use properties, J. Environ. Polym. Degrad., 1995, 3, 215–223 CrossRef CAS.
- Y. X. Weng, X. L. Wang and Y. Z. Wang, Biodegradation behavior of PHAs with different chemical structures under controlled composting conditions, Polym. Test., 2011, 30, 372–380 CrossRef CAS.
- E. Rudnik and D. Briassoulis, Comparative Biodegradation in Soil Behaviour of two Biodegradable Polymers Based on Renewable Resources, J. Polym. Environ., 2011, 19, 18–39 CrossRef CAS.
- Y. Sadef, T. G. Poulsen and K. Bester, Modeling organic micro pollutant degradation kinetics during sewage sludge composting, Waste Management, 2014, 34, 2007–2013 CrossRef CAS PubMed.
- H. Sun, E. Terhonen, K. Koskinen, L. Paulin, R. Kasanen and F. O. Asiegbu, Bacterial diversity and community structure along different peat soils in boreal forest, Appl. Soil. Ecol., 2014, 74, 37–45 CrossRef.
- M. K. Gouda, A. E. Swellam and S. H. Omar, Biodegradation of Synthetic Polyesters (BTA and PCL) with Natural Flora in Soil Burial and Pure Cultures under Ambient Temperature, Res. J. Environ. Earth Sci., 2012, 4, 325–333 CAS.
- F. Chen, K. Zhang, H. Zheng, X. Lin, Z. A. Ouyang and T. U. Naimei, Analyzing the Effect of Mixed Decomposition of Conifer and Broadleaf Litters on Soil Microbial Communities by Using PCR-DGGE, Chin. J. Appl. Environ. Biol., 2011, 17, 145–150 CrossRef CAS.
- H. Zhao, Z. Cui, X. Wang, L. S. Turng and X. Peng, Processing and characterization of solid and microcellular poly(lactic acid)/polyhydroxybutyrate-valerate (PLA/PHBV) blends and PLA/PHBV/Clay nanocomposites, Composites, Part B, 2013, 51, 79–91 CrossRef CAS.
- C. Luan and C. You, A novel experimental investigation into sintered neck tensile strength of ash at high temperatures, Powder Technol., 2015, 269, 379–384 CrossRef CAS.
- J. M. Vergès and J. I. Morales, The gigapixel image concept for graphic SEM documentation. Applications in archeological use-wear studies, Micron, 2014, 65, 15–19 CrossRef PubMed.
- W. Guo, J. Tao, C. Yang, Q. Zhao, C. Song and S. Wang, The rapid evaluation of material biodegradability using an improved ISO 14852 method with a microbial community, Polym. Test., 2010, 29, 832–839 CrossRef CAS.
- G. Muyzer, E. C. de Waal and A. G. Uitterlinden, Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA, Appl. Environ. Microbiol., 1993, 59, 695–700 CAS.
- B. Dong, J. Yi, L. Dai and X. Dai, Evaluation of Several DNA Extraction Methods for Obtaining Total Community DNA from Anaerobic Digestion Sludge, Procedia Environ. Sci., 2013, 18, 856–863 CrossRef CAS.
- F. Guo and T. Zhang, Profiling bulking and foaming bacteria in activated sludge by high throughput sequencing, Water Res., 2012, 46, 2772–2782 CrossRef CAS PubMed.
- L. E. S. D. Miera, P. Arroyo, E. D. L. Calabuig, J. Falagán and G. Ansola, High-throughput sequencing of 16S RNA genes of soil bacterial communities from a naturally occurring CO2 gas vent, Int. J. Greenhouse Gas Control, 2014, 29, 176–184 CrossRef.
- A. M. Bolger, M. Lohse and B. Usadel, Trimmomatic: a flexible trimmer for Illumina sequence data, Bioinformatics, 2014, 30, 2114–2120 CrossRef CAS PubMed.
- R. C. Edgar, UPARSE: highly accurate OTU sequences from microbial amplicon reads, Nat. Methods, 2013, 10, 996 CrossRef CAS PubMed.
- I. Armentano, E. Fortunati, N. Burgos, F. Dominici, F. Luzi, S. Fiori, A. Jiménez, K. Yoon, J. Ahn, S. Kang and J. M. Kenny, Processing and characterization of plasticized PLA/PHB blends for biodegradable multiphase systems, eXPRESS Polym. Lett., 2015, 9, 583–596 CrossRef CAS.
- M. N. Kim and T. P. Sang, Degradation of poly(L-lactide) by a mesophilic bacterium, J. Appl. Polym. Sci., 2010, 117, 67–74 CAS.
- P. Sangwan and D. Y. Wu, New insights into polylactide biodegradation from molecular ecological techniques, Macromol. Biosci., 2008, 8, 304–315 CrossRef CAS PubMed.
- P. Sangwan, C. Way and D. Y. Wu, New insight into biodegradation of polylactide (PLA)/clay nanocomposites using molecular ecological techniques, Macromol. Biosci., 2009, 9, 677–686 CrossRef CAS PubMed.
- Z. Saadi, A. Rasmont, G. Cesar, H. Bewa and L. Benguigui, Fungal Degradation of Poly(l-lactide) in Soil and in Compost, J. Polym. Environ., 2012, 20, 273–282 CrossRef CAS.
- W. Amner, C. Edwards and A. J. Mccarthy, Improved medium for recovery and enumeration of the farmer's lung organism, Saccharomonospora viridis, Appl. Environ. Microbiol., 1989, 55, 2669 CAS.
- A. Linos, U. S. A. Keller and R. Reichelt, A Gram-negative bacterium, identified as Pseudomonas aeruginosa AL98, is a potent degrader of natural rubber and synthetic cis-1,4-polyisoprene, FEMS Microbiol. Lett., 2000, 182, 155–161 CrossRef CAS PubMed.
- C. S. Lee, Y. T. Jung, S. Park, T. K. Oh and J. H. Yoon, Lysinibacillus xylanilyticus sp. nov., a xylan-degrading bacterium isolated from forest humus, Int. J. Syst. Evol. Microbiol., 2010, 60, 281–286 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Fig. S1: DGGE electrophoresis pattern (A) of different subculture bacteria samples and the analysis diagram (B) with Quantity One software. Fig. S2: the average temperature and relativity humidity during the test period (data sources: Guangzhou Panyu Meteorological Observatory). Table S1: DSC values in the similarity matrix calculated from Quantity One software (%). See DOI: 10.1039/c8ra10591j |
| This journal is © The Royal Society of Chemistry 2019 |