 Open Access Article
Open Access ArticleBiochemical characterization and biocatalytic application of a novel D-tagatose 3-epimerase from Sinorhizobium sp.†
Zhangliang Zhua,
Chao Lia,
Xin Liua,
Dengke Gaoa,
Xueyu Wanga,
Masaru Tanokura*ab,
Hui-Min Qin *a and
Fuping Lu*a
*a and
Fuping Lu*a
aKey Laboratory of Industrial Fermentation Microbiology of the Ministry of Education, Tianjin Key Laboratory of Industrial Microbiology, College of Biotechnology, Tianjin University of Science and Technology, National Engineering Laboratory for Industrial Enzymes, Tianjin 300457, P. R. China. E-mail: huiminqin@tust.edu.cn; lfp@tust.edu.cn; Fax: +86-22-60602298; Tel: +86-22-60601958
bLaboratory of Basic Science on Healthy Longevity, Department of Applied Biological Chemistry, Graduate School of Agricultural and Life Sciences, The University of Tokyo, 1-1-1 Yayoi, Bunkyo, Tokyo 113-8657, Japan. E-mail: amtanok@mail.ecc.u-tokyo.ac.jp
First published on 22nd January 2019
Abstract
Sinorhizobium sp. D-tagatose 3-epimerase (sDTE) catalyzes the conversion of D-tagatose to D-sorbose. It also recognizes D-fructose as a substrate for D-allulose production. The optimal temperature and pH of the purified sDTE was 50 °C and 8.0, respectively. Based on the sDTE homologous model, Glu154, Asp187, Gln213, and Glu248, form a hydrogen bond network with the active-site Mn2+ and constitute the catalytic tetrad. The amino acid residues around O-1, -2, and -3 atoms of the substrates (D-tagatose/D-fructose) are strictly conserved and thus likely regulate the catalytic reaction. However, the residues at O-4, -5, and -6, being responsible for the substrate-binding, are different. In particular, Arg65 and Met9 were found to form a unique interaction with O-4 of D-fructose and D-tagatose. The whole cells with recombinant sDTE showed a higher bioconversion rate of 42.5% in a fed-batch bioconversion using D-fructose as a substrate, corresponding to a production of 476 g L−1 D-allulose. These results suggest that sDTE is a potential industrial biocatalyst for the production of D-allulose in fed-batch mode.
Background
Rare sugars (i.e. xylitol, erythritol, L-ribose, D-tagatose, and D-allulose) are defined as monosaccharides and their derivatives are infrequently found in nature. They have great potential use in the food and pharmaceutical industries. For instance, D-allulose (D-psicose or D-ribo-2-hexulose) is considered an ideal substitute for sucrose as an ultra-low calorie sweetener.1 It is absorbed in the small intestine and excreted from the body via the urine.2–4 D-Allulose reduces the oxidation occurring via Maillard reaction during food processing, and thus increases the pleasantness of the food flavor.5 D-Tagatose can be used as a low-calorie sweetener in non-chronically administered drugs, mouth wash, and tooth paste, as well as a wide variety of health foods and dietary supplements.6 Furthermore, it is also a potential new antidiabetic and obesity control drug.7 Compared with the chemical synthesis of rare sugars, which generates chemical waste and by-products, as well as requires complex multi-step reactions and the protection–deprotection of functional groups,8 the biosynthesis of rare sugars by enzymatic pathways is more environmentally friendly, since it can be done under moderate reaction conditions, offering high specificity, and sustainability.In 2002, the creative izumoring strategy was established, through which all rare sugars and their derivatives could be cyclically converted via enzymatic pathways by epimerization, isomerization, and oxidization–reduction reactions.9–12 Notably, ketose 3-epimerase plays an irreplaceable role in the bioconversion of rare sugars, in which both D-tagatose 3-epimerase (DTE, EC 5.1.3.-) and D-allulose 3-epimerase (DAE, also named as DPE, EC 5.3.1) family enzymes can catalyze the reversible interconversion of D-allulose and D-sorbose with D-fructose and D-tagatose, respectively. Furthermore, the L-ribulose 3-epimerases (LREs) from M. loti and T. maritima, which showed the substrate specificity for L-ribulose, were also reported to convert D-fructose to D-allulose.13,14 DTE/DAE family enzymes require ions (Co2+ or Mn2+) for their catalytic activity.15,16 Genes encoding DTE/DAE family enzymes have been identified in P. cichorii,17 R. sphaeroides,18 A. tumefaciens,15 A. globiformis,19 Agrobacterium sp.,20 C. bolteae,21 C. cellulolyticum,16 C. scindens,22 Clostridium sp.,23 Desmospora sp.,24 F. plautii,25 Ruminococcus sp.,26 and T. primitia.27 However, only several crystal structures have been solved, including C. cellulolyticum DAE,28 A. tumefaciens DAE,29 P. cichorii DTE,30 M. loti LRE,31 and T. maritima LRE.14 The catalytic mechanism was also proposed based on the structural information. A C3–O3 proton-exchange mechanism was proposed, in which one Glu removes a proton from C-3 to generate a cis-enediolate intermediate via O-3, and then another Glu protonates C-3 from the opposite side.29
Compared with the enzymatic synthesis of rare sugars, bioconversion using whole cells eliminates the purification steps, and it is considered to be more commercially competitive. Furthermore, whole recombinant cells of Escherichia coli expressing DTE/DAE from C. bolteae,21 C. cellulolyticum,16 and A. tumefaciens32 were investigated, showing that whole cells have showed greater stability and resistance to environmental perturbations than the isolated enzymes. A novel putative D-tagatose 3-epimerase gene from Sinorhizobium sp. (sDTE, 882 bp, 293 amino acids) has been identified and is 32.1 kDa without a signal peptide. To understand the relationship between the 3-D structure and biochemical characterization of sDTE, we analyzed its structural model and substrate specificity. The biocatalytic production of D-allulose using resting cells was also investigated.
Materials and methods
Cloning and expression
The sDTE gene (GenBank ID: NZ_CP016452.1; Protein ID: WP_069063284.1) was synthesized by Genewiz (Suzhou, China), and was inserted into the vector pET28a(+) (Novagen, Madison, WI, USA) with NdeI and EcoRI sites. The E. coli BL21 (DE3) cells harboring the sDTE gene were grown on Luria–Bertani (LB) medium supplemented 50 mg L−1 kanamycin at 37 °C until the OD600 value reached 0.6 to 0.8 and then the recombinant cells were induced at 16 °C for 20 h by an addition isopropyl β-D-1-thiogalactopyranoside (IPTG) to a final concentration of 0.5 mM. The cells were then collected from culture broth by centrifugation at 4 °C for 10 min at 5000 × g and washed twice using lysis buffer (20 mM Tris–HCl pH 8.0) containing 10 mM imidazole, 500 mM NaCl, and 1 mM dithiothreitol (DTT).Purification of the sDTE enzyme
The recombinant E. coli cells were re-suspended in lysis buffer, and then disrupted by sonication for 10 min (pulse on 2.5 s and pulse off 3.0 s). Cell debris was cleared by centrifugation (40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g, 30 min, 4 °C) and passage through a 0.45 μm filters. The supernatants were loaded onto a column containing Ni-NTA Superflow resin (Qiagen, Hilden, Germany) equilibrated with lysis buffer. After the unbound proteins were washed with 10 mL lysis buffer, the recombinant enzyme was eluted from the column with 15 mL elution buffer (20 mM Tris–HCl pH 8.0) containing 300 mM imidazole, 100 mM NaCl, and 1 mM DTT. The eluted solution was dialyzed overnight against 20 mM Tris–HCl pH 8.0, 1 mM DTT to remove imidazole and was further purified by ion exchange using Resource Q (volume: 6 mL, flow rate: 5 mL min−1, GE Healthcare). The molecular mass was estimated by gel filtration chromatography on a Superdex 200 Increase column 10/300 GL (volume: 24 mL, GE Healthcare) and equilibrated with buffer (20 mM Tris–HCl pH 8.0, 100 mM NaCl, and 1 mM DTT) at a flow rate of 0.5 mL min−1.33,34 The total protein concentration was determined using a BCA assay kit (Solarbio, China) and bovine serum albumin was used as the standard. The amount and purity of the target protein were determined by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and Coomassie Brilliant Blue staining. The purified enzyme was used for activity assays.
000 × g, 30 min, 4 °C) and passage through a 0.45 μm filters. The supernatants were loaded onto a column containing Ni-NTA Superflow resin (Qiagen, Hilden, Germany) equilibrated with lysis buffer. After the unbound proteins were washed with 10 mL lysis buffer, the recombinant enzyme was eluted from the column with 15 mL elution buffer (20 mM Tris–HCl pH 8.0) containing 300 mM imidazole, 100 mM NaCl, and 1 mM DTT. The eluted solution was dialyzed overnight against 20 mM Tris–HCl pH 8.0, 1 mM DTT to remove imidazole and was further purified by ion exchange using Resource Q (volume: 6 mL, flow rate: 5 mL min−1, GE Healthcare). The molecular mass was estimated by gel filtration chromatography on a Superdex 200 Increase column 10/300 GL (volume: 24 mL, GE Healthcare) and equilibrated with buffer (20 mM Tris–HCl pH 8.0, 100 mM NaCl, and 1 mM DTT) at a flow rate of 0.5 mL min−1.33,34 The total protein concentration was determined using a BCA assay kit (Solarbio, China) and bovine serum albumin was used as the standard. The amount and purity of the target protein were determined by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and Coomassie Brilliant Blue staining. The purified enzyme was used for activity assays.
MALDI-TOF mass spectrometry
A total of 1 μL of enzyme sample solution was spotted onto a MALDI target containing 1 μL saturated alpha-cyano-4-hydroxycinnamic acid (HCCA) in a solvent mixture comprising acetonitrile and water (70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30, v/v) with 0.01% trifluoroacetic acid. The sample was analyzed using an ultrafleXtreme TOF/TOF instrument (Bruker Daltonics, Germany) operating in linear mode. MALDI-TOF spectra were analyzed using FlexAnalysis software (Bruker Daltonics).
30, v/v) with 0.01% trifluoroacetic acid. The sample was analyzed using an ultrafleXtreme TOF/TOF instrument (Bruker Daltonics, Germany) operating in linear mode. MALDI-TOF spectra were analyzed using FlexAnalysis software (Bruker Daltonics).
Site-directed mutagenesis
The mutant sDTE were constructed by one-step PCR reactions using a pair of complementary primers (Table S1†). The amplified PCR products were treated with DpnI to digest the template plasmid, and the DpnI-treated mixture is then used to transform into E. coli JM109 cells to yield plasmids encoding genes. The mutation constructs were verified by DNA sequencing (Genewiz, China). The plasmids containing mutant genes were introduced into E. coli BL21 (DE3) for protein expression and the mutant proteins were purified by the method for wild-type enzyme described above.CD spectroscopy
The circular dichroism (CD) spectra (MOS-450, Biologic, Claix, Charente, France) were recorded in the far-UV (190–260 nm) with a 1 mm path-length cell at 25 °C. Four spectra of the far-UV CD of were recorded at a scan rate of 1 nm s−1 with a response of 1 s and bandwidth of 0.1 nm in 20 mM Tris–HCl (pH 8.0), 100 mM NaCl, and 1 mM DTT containing sDTE (0.10 mg mL−1). The reported spectra were the averages of four scans that were corrected for buffer blanks. Analysis of the protein secondary structure was performed using the program SELCON3 (http://www.dichroweb.cryst.bbk.ac).35Confirmation of D-tagatose and D-sorbose using HPLC
The product D-sorbose was determined using high-performance liquid chromatography (HPLC; Agilent 1260, USA) equipped with an evaporative light-scattering detector (ELSD 6000, Alltech Technology Ltd., Canada) and a Prevail Carbohydrate ES column-W (5 μm, 4.6 × 250 mm, Agela Technologies, China). The column was eluted at 40 °C with acetonitrile (85%) at 1 mL min−1.Enzyme activity assay
The activity was assayed by determining D-sorbose formation using D-tagatose as the substrate.16,17 The activities of the wild-type and mutant enzymes were conducted in 20 mM Tris–HCl buffer (pH 8.0) containing 10 g L−1 D-tagatose, 1 μM Mn2+, and an appropriate enzyme (0.5 μM), at 50 °C for 10 min. The reaction mixtures were incubated in a boiling water bath for 5 min and then D-sorbose was measured using HPLC.To investigate the optimal temperature, the activity was determined by incubating sDTE at 20 to 70 °C. The optimal pH of the purified enzyme was determined at 50 °C for 10 min across a pH range of 5.5 to 11.0, using four buffers systems (MES: pH 5.5–6.5, PBS: 7.0–8.0, Tris–HCl: 8.5–9.0, CAPS: pH 9.5–11.0). To determine the effect of various metal ions on the activity of sDTE, the enzyme solution purified was treated with EDTA for 8 h and dialyzed against 20 mM Tris–HCl buffer (pH 8.0). The reaction system contains the following ions at 1 mM: Ca2+, Mg2+, Mn2+, Co2+, Zn2+, Cu2+, Fe2+, and Fe3+, respectively. The conversion of the wild-type enzyme in 20 mM Tris–HCl buffer at 50 °C without adding metals was defined as 100%. The activity was then measured as described above. All assays were repeated three separate times, and the data are shown as the means ± S.D.
Structural homologous model of sDTE
The homology homologous model of sDTE was generated using Modeller 9.9.36 The crystal structure of Pseudomonas cichorii DTE (PcDTE, PDB ID: 4XSL, 1.6 Å) was chosen as template, which has 29% sequence identity to the target protein sDTE. A sequence alignment was automatically generated between PcDTE and sDTE using align2d command. Subsequently, homology modeling was performed using the automodel command. Thereafter, each model was optimized and simulated annealing MD simulations were used to refine the structure. Finally, the best model was chosen from using the values of the Modeller objective function and the DOPE assessment scores. The generated model structures were visualized by utilizing the PyMoL software (http://www.pymol.org).37Bioconversion of D-fructose into D-allulose using isolated sDTE enzyme and resting cells
Enzymatic synthesis of D-allulose was carried out in a 10 mL reaction system containing 5 μM purified sDTE, and 750 g L−1 D-fructose at optimal conditions. The progress of the enzymatic conversion was determined every 10 min for 60 min. For the high production of D-allulose from D-fructose, recombinant E. coli BL21 (DE3) whole cells harboring sDTE were cultivated as described above, harvested, and washed twice with 20 mM Tris–HCl (pH 8.0). The biotransformation was conducted in 20 mM Tris–HCl (pH 8.0) containing 750 g L−1 D-fructose and 30 mg of cells (wet weight per mL). Samples comprising 1.0 mL of the resting cell suspension were drawn every 10 min for 60 min. A fed-batch strategy was used to investigate the product yield with an initial D-fructose concentration of 750 g L−1. Sampling was drawn every 20 min for 180 min. Cells were disrupted by sonication, and were centrifuged at 40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g and 4 °C for 30 min to remove the cell debris. The samples were diluted for HPLC analysis. All experiments were carried out in triplicates using E. coli BL21 (DE3) cells without the sDTE gene as controls.
000 × g and 4 °C for 30 min to remove the cell debris. The samples were diluted for HPLC analysis. All experiments were carried out in triplicates using E. coli BL21 (DE3) cells without the sDTE gene as controls.
Results and discussion
Characterization of sDTE
The recombinant sDTE was successfully overexpressed in E. coli BL21 (DE3), and was purified using Ni-NTA Superflow resin, followed by anion-exchange chromatography on a Resource Q column (Fig. 1A). The molecular mass of sDTE was analyzed by a size exclusion column, and a single peak corresponding to a dimer was observed (Fig. 1B). The molecular mass was further estimated by MALDI-TOF mass spectrometry, revealing a mass of approximately 68.83 kDa (Fig. 1C). This value was about two times the molecular mass of recombinant sDTE (34.44 kDa), including a tag of 21 residues (MGSSHHHHHHHSSGLVPRGSH) at the N-terminus. Therefore, the recombinant enzyme is a dimer, which is the same as the DTEase from P. cichorii.17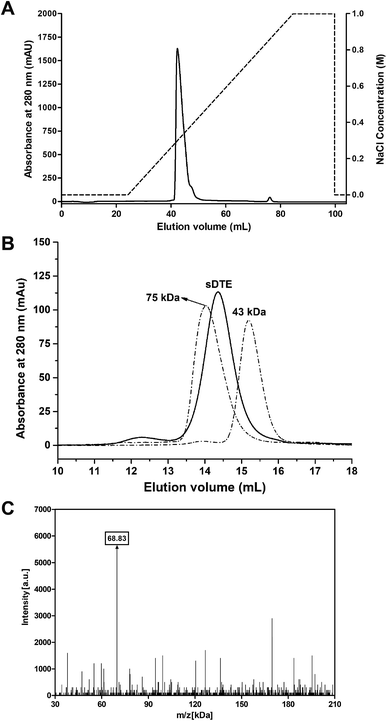 | ||
| Fig. 1 Purification of sDTE by anion-exchange (A) and size-exclusion chromatography (B). Protein markers of ovalbumin (43.0 kDa) and conalbumin (75.0 kDa) were used. (C) MALDI-TOF spectra of sDTE. | ||
The secondary structural features of purified sDTE were shown in Fig. 2A and B using CD spectroscopy, which showed a positive peak around 198 nm. The secondary structural content of the sDTE was 35.1% α-helix, 11% β-sheet, and 31.3% unstructured regions, respectively. The reaction product was analyzed by HPLC, which revealed a single peak of D-sorbose, with a retention time of 19.26 min (Fig. 2C).
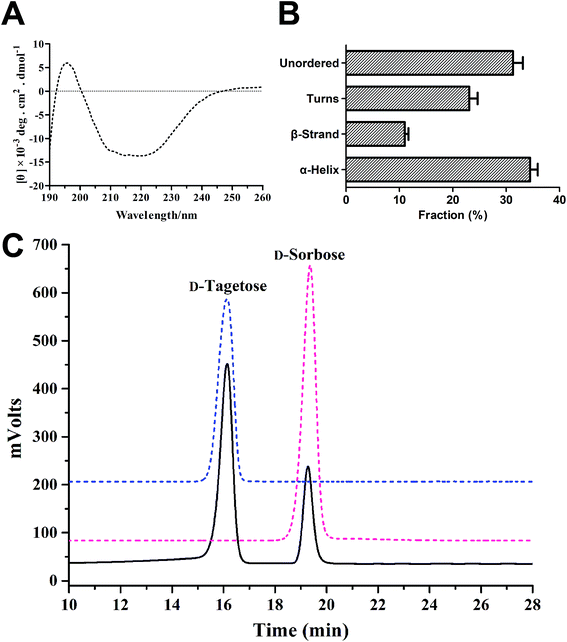 | ||
| Fig. 2 CD spectra (A) and secondary structure assignments (B) of sDTE; (C) confirmation of the enzymatic conversion of D-tagatose using HPLC. | ||
The optimum pH and temperature for sDTE are shown in Fig. 3. Recombinant enzyme was optimally active in Tris–HCl buffer pH 8.0 and showed activity at pH values from 5.5 to 11.0, indicating that the sDTE had an extensive pH range (Fig. 3A). The sDTE showed a maximum activity at 50 °C, and the activity significantly decreased at temperatures over 60 °C (Fig. 3B). The effects of various metal ions on the activity of sDTE are displayed in Fig. 3C. The activity of sDTE increased to 156% and 143% in the presence of 1 mM MnCl2 and CoCl2, respectively, suggesting that Mn2+ and Co2+ might participate in the catalytic reaction. The activity of sDTE was unchanged in the presence of MgCl2, but it decreased when CaCl2, FeCl3, CuCl2, ZnCl2, and FeCl2 were added to the reaction system. The results thus support the previously published view that DTE family members are metalloenzymes.15,16
The kinetic parameters of sDTE with four substrates were determined by non-linear regression fitting of Michaelis–Menten equation with GraphPad Prism 5.0 software (GraphPad Software, San Diego, CA). As shown in Fig. 4 and Table 1, sDTE was characterized by Km values of between 26 and 83 mM, and catalytic efficiencies kcat/Km of between 0.41 and 3.78 s−1 mM−1. sDTE showed a high activity toward D-tagatose, with a Michaelis constant of 26 mM and catalytic efficiency of 3.78 s−1 mM−1, which was four times of that of D-fructose, with a Michaelis constant of 54.7 mM and catalytic efficiency of 0.91 s−1 mM−1. sDTE showed low catalytic activity toward D-sorbose, the value of kcat/Km was 0.41 s−1 mM−1. For D-allulose, the kcat, Km, and kcat/Km are 78.4 s−1, 39.8 mM, and 1.97 s−1 mM−1, respectively, which is familiar to that of other DTEases (Table 2).15,16,19,23 Enzymes can be affected by temperature and pH in a variety of ways, for example, complex effects on the chemistry of catalysis and also on the structure, folding and stability of the protein.38 However, we only determined kcat and Km at optimum temperature and pH. We compared the enzymatic properties of sDTE with family enzymes in Table 2. sDTE shows optimal pH and temperature at 8.0 and 50 °C, and requires metal ions (Co2+ or Mn2+) for enzymatic activities. sDTE is a dimer (68 kDa) in solution, which is different from other enzymes (tetramer). sDTE showed the substrate specificity toward D-tagatose with a higher productivity (476 g L−1 D-allulose) in a fed-batch bioconversion than other enzymes.
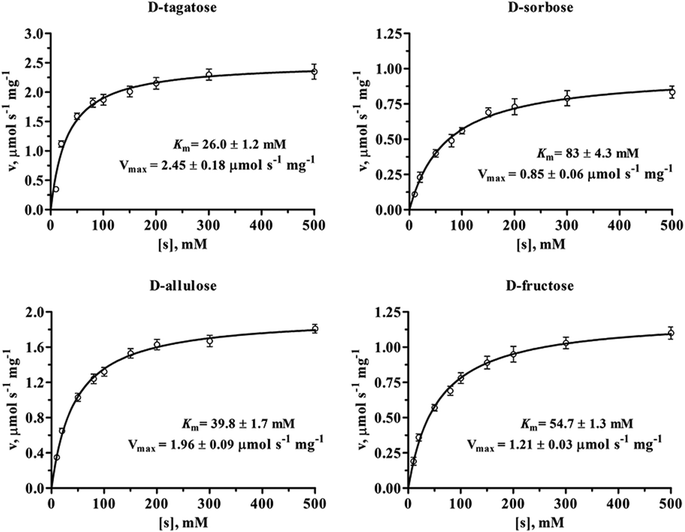 | ||
| Fig. 4 Michaelis–Menten plots of sDTE toward D-tagatose, D-sorbose, D-allulose and D-fructose. All assays were repeated three times, and the data are shown as mean ± S.D. | ||
| Substrates | Km (mM) | kcat (s−1) | kcat/Km (s−1 mM−1) | Relative activity (%) |
|---|---|---|---|---|
| a All assays were repeated three times, and the data are shown as mean ± S.D. | ||||
| D-Fructose | 54.7 ± 1.3 | 50 ± 0.82 | 0.91 | 46 ± 0.96 |
| D-Allulose | 39.8 ± 1.7 | 78.4 ± 2.3 | 1.97 | 75 ± 2.2 |
| D-Tagatose | 26.0 ± 1.2 | 98.3 ± 0.92 | 3.78 | 100 ± 3.1 |
| D-Sorbose | 83 ± 4.3 | 34.0 ± 0.92 | 0.41 | 19 ± 0.6 |
| Strains for enzyme source | A. tumefaciens15 | R. sphaeroides18 | Ruminococcus sp.26 | Agrobacterium sp.20 | Sinorhizobium sp.a | |
|---|---|---|---|---|---|---|
| a This study.b Produced from D-fructose with initial concentration of 700 g L−1.c Produced from D-fructose with initial concentration of 500 g L−1.d Produced from D-fructose with initial concentration of 750 g L−1.e Produced from D-fructose using a fed-batch bioconversion.f NR, not reported. | ||||||
| Optimum pH | 8.0 | 8.5 | 7.5–8.0 | 7.5–8.0 | 8.0 | |
| Optimum temperature (°C) | 50 | 35 | 60 | 55–60 | 50 | |
| Metal ions required | Mn2+ | Mn2+ | Mn2+ | Co2+ | Mn2+ | |
| Most suitable substrate | D-Allulose | D-Fructose | D-Allulose | D-Allulose | D-Tagatose | |
| MW of native protein | 132 (tetramer) | 128 (tetramer) | 128 (tetramer) | 132 (tetramer) | 68 (dimer) | |
| kcat (s−1) | D-Allulose | 39.7 | 20.5 | 40.5 | NR | 78.4 |
| D-Fructose | 34.5 | 42.6 | 59.4 | 106.7 | 50 | |
| Km (mM) | D-Allulose | 12 | 215 | 48 | NR | 39.8 |
| D-Fructose | 24 | 78 | 216 | 110 | 54.7 | |
| kca/Km (mM−1 s−1) | D-Allulose | 3.3 | 0.1 | 0.84 | NR | 1.97 |
| D-Fructose | 1.44 | 0.55 | 0.28 | 0.98 | 0.91 | |
| Productivity of D-allulose (g L−1) | 230b | 118b | 125c | 150c | 260d | |
| 476e | ||||||
Homology model of sDTE
Based on the crystal structure of P. cichorii D-tagatose 3-epimerase, which has 29% sequence identity to sDTE,30 the structural homology model of sDTE was generated. We evaluated the 3D model of sDTE that the secondary structural content of the homology model was 42.8% α-helix and 9.4% β-sheet, respectively (https://servicesn.mbi.ucla.edu/SAVES/). These data are a little different between CD spectroscopy and homology model because pH, temperature, and the buffer may affect CD spectroscopy of sDTE. The structure of sDTE (Fig. 5A) constituted of eight β-strands, ten α-helices, and one 310 helix, in which eight characteristic units of β-strand/α-helix at the core of structural motif formed canonical TIM-barrel (β/α)8 fold with a cluster of β-strands surrounded by α-helices. The additional two α-helices of α6 and α9 and one 310 helix were packed together along with the TIM barrel. The active site is located at the top of the TIM barrel.The structural model structure of sDTE showed that four residues, Glu154, Asp187, Gln213, and Glu248, coordinated the Mn2+ metal ion (Fig. 5B). Glu154 and Gln213 were located in the β5 and β7 stands of the TIM-barrel, respectively. Asp187 and Glu248 were located in the loop region of active site. They form a hydrogen bond network of catalytic tetrad of sDTE for the hydride transfer reaction. These four residues are partly conserved in the DTE/DAE/LRE superfamily (Fig. S1†), and three of the residues Glu154, Asp187, and Glu248 were absolutely conserved. However, Gln215 was not conserved, and replaced by a histidine, in P. cichorii DTE, A. tumefaciens DAE, C. cellulolyticum DAE, and T. maritima LRE.14–17 The carbonyl oxygen O-2 and hydroxyl group O-3 in the substrate D-fructose/D-tagatose coordinated Mn2+ in a bidentate manner (Fig. 5B). This structure model is consistent with the previously proposed mechanism of deprotonation/protonation by two Glu residues of Glu154 and Glu248.28–30 In briefly, The Glu248 residue coordinating the Mn2+ removes a proton from C3 to generate a cis-enediolate intermediate, after which Glu154 protonates C3 from the opposite side.
Substrate specificity of sDTE
The hydroxyl group O-1 of the substrate D-tagatose/D-fructose formed hydrogen bonds with Arg219 and Glu160 (Fig. 6). The Mn2+ ion is coordinated by O-2 and -3 of the substrate, possibly neutralizing the oxyanion O-2 oxyanion a putative cis-enediolate intermediate. O-3 is stabilized further by hydrogen bonds with the hydroxyl groups of Glu154 and Glu248. When modeling with D-fructose or D-tagatose, we found that D-fructose has the same configurations of O-1, -2, and -3 as D-tagatose (Fig. 6A). Furthermore, the interactions between D-fructose/D-tagatose at the O-1, -2, and -3 positions and the enzyme were very similar to those observed in other DTE/DAE family enzymes.28–30 Therefore, sDTE strictly regulates the catalytic reaction at the positions 1, 2, and 3. However, the residues interacting with D-tagatose/D-fructose at the hydroxyl groups of O-4, -5, and -6 positions were not conserved among the enzymes of this family. The substrate–enzyme interactions at O-4, -5, and -6 are essential for the substrate specificity. They control the spatial orientation and conformation of the various substrates. In particular, Arg65 formed a unique interaction with O-4 of D-fructose and D-tagatose. Notably, O-4 of D-fructose formed two hydrogen bonds with NE and NH of Arg65. However, O-4 of D-tagatose was stabilized by Met9. Arg65 and Met9 were not conserved in the DTE/DAE family, and the mutation of R65G and M9A abolished the catalytic activity toward D-tagatose. The relative activities of R65G and M9A toward D-tagatose were 7.3% and 5.1%, respectively compared with wild-type sDTE (Fig. S2†). On the other hand, O-5 of D-tagatose interacted with Glu35 while O-5 of D-fructose was anchored by the main chain of Gly104. The structural information supports the biochemical data, since sDTE showed high activity toward D-tagatose and D-fructose. When comparing the superimposed of structural models of sDTE and C. cellulolyticum DAE, we found that D-allulose formed different hydrogen bond interactions with sDTE than D-tagatose (Fig. 6B), in which O-4 could interact with Gly104 and O-5 was recognized by Arg65. There was no interaction between O-6 of D-allulose and sDTE. Some residues, such as Met9, Phe15, Leu37, Leu111, Phe189, and Phe257 formed a hydrophobic environment for the substrate recognition (Fig. 6C).Bioconversion of D-fructose into D-allulose using isolated sDTE enzyme and resting cells
The biocatalytic synthesis of D-allulose using D-fructose as substrate was performed using isolated enzyme, as well as whole-cell batch, and fed-batch strategies (Fig. 7). Under enzymatic catalysis and batch bioconversion, the productivity of D-allulose gradually increased and reached a plateau at 40 min. The final productivity of D-allulose reached 190 and 226 g L−1, respectively (Fig. 7A and B). The conversion rates were 25.3 and 30.1%, respectively. For high levers of product yield, the D-allulose titers were improved using a fed-batch strategy (Fig. 7C). The reaction system was added with D-fructose every 60 min to continue the bioconversion. The yields of D-allulose showed a steady increase during the first two fed-batch processes, with productivity values of 223 and 372 g L−1 (bioconversion before 120 min). The maximum D-allulose concentrations (476 g L−1) was obtained at 180 min during the third fed-batch fermentation process (bioconversion from 120 min to 180 min). The conversion rate reached 42.5%. Therefore, fed-batch bioconversion with E. coli BL21 (DE3) expressing sDTE is an effective strategy to improve the D-allulose yield.The bioconversion rate of D-allulose using A. tumefaciens DAE, at 32.9%, compares favorably with other enzymes from this family, and was markedly higher than the 20% achieved with P. cichorii DTE.15,17 Yang et al. reported that turnover rate of D-allulose using A. tumefaciens DAE in K. marxianus was about 25.3%.39 R. sphaeroides DTE shows the highest activity on D-fructose, with a D-allulose bioconversion rate of 23% (40 °C, pH 9.0), which is also lower than that of A. tumefaciens DAE.40 In our research, sDTE showed a D-allulose bioconversion of 25.3 and 30.1% in pure-enzyme and whole-cell batch strategies, respectively. Furthermore, the bioconversion rate of sDTE-expressing cells reached 42.5% in fed-batch fermentation, which was higher than that of A. tumefaciens DAE.
Conclusions
The purified D-tagatose 3-epimerase from Sinorhizobium sp. catalyses mainly the epimerization of D-tagatose to D-sorbose at the C3 position. sDTE showed optimal activity in Tris–HCl buffer at pH 8.0 and 50 °C. The binding-site residues of sDTE interacting with the O-4, -5, and -6 positions of the substrate are different, and Arg65 formed an interaction with O-4 of D-fructose and D-tagatose in a unique recognition mode. Furthermore, both the isolated sDTE enzyme and resting cells E. coli expressing sDTE showed high bioconversion rates in the production of D-allulose. Furthermore, the fed-batch strategy was an effective way to improve the D-allulose yield. The research on sDTE provides a valuable tool for commercial-scale manufacture of rare sugars in the future.Author contributions
T. M., F. L., and H.-M. Q. designed the research; Z. Z., C. L., and X. L. performed the experiments; Z. Z., D. G., and X. W. analyzed data; H.-M. Q., F. L., and M. T. wrote the paper. All authors have approved the final version of the manuscript.Funding
This work was supported by National Key Research and Development Project of China (2017YF0400304), National Natural Science Foundation of China (31771911), the Innovative Research Team of Tianjin Municipal Education Commission (TD13-5015), Natural Science Foundation of Tianjin (16JCQNJC09200) and the Overseas High-level Talents Program of Tianjin University of Science and Technology to H.-M. Qin, China.Ethics approval and consent to participate
No animals or human subjects were used in the above research.Consent for publication
Our manuscript does not contain any individual data in any form.Conflicts of interest
The authors declare no competing financial interests.References
- W. Zhang, S. Yu, T. Zhang, B. Jiang and W. Mu, Trends Food Sci. Technol., 2016, 54, 127–137 CrossRef.
- A. Hossain, F. Yamaguchi, T. Matsuo, I. Tsukamoto, Y. Toyoda, M. Ogawa, Y. Nagata and M. Tokuda, Pharmacol. Ther., 2015, 155, 49–59 CrossRef CAS PubMed.
- T. Matsuo, T. Tanaka, M. Hashiguchi, K. Izumori and H. Suzuki, Asia Pac. J. Clin. Nutr., 2003, 12, 225–231 CAS.
- R. L. Whistler, P. P. Singh and W. C. Lake, Carbohydr. Res., 1974, 34, 200–202 CrossRef CAS PubMed.
- S. O'Charoen, S. Hayakawa and M. Ogawa, Int. J. Food Sci. Technol., 2015, 50, 194–202 CrossRef.
- D.-K. Oh, Appl. Microbiol. Biotechnol., 2007, 76, 1–8 CrossRef CAS PubMed.
- Y. Lu, G. V. Levin and T. W. Donner, Diabetes, Obes. Metab., 2008, 10, 109–134 CrossRef CAS PubMed.
- M. Emmadi and S. S. Kulkarni, Nat. Prod. Rep., 2014, 31, 870–879 RSC.
- T. B. Granström, G. Takata, M. Tokuda and K. Izumori, J. Biosci. Bioeng., 2004, 97, 89–94 CrossRef.
- K. Izumori, Naturwissenschaften, 2002, 89, 120–124 CrossRef CAS PubMed.
- K. Izumori, J. Biotechnol., 2006, 124, 717–722 CrossRef CAS PubMed.
- W. Mu, W. Zhang, Y. Feng, B. Jiang and L. Zhou, Appl. Microbiol. Biotechnol., 2012, 94, 1461–1467 CrossRef CAS PubMed.
- K. Uechi, G. Takata, Y. Fukai, A. Yoshihara and K. Morimoto, Biosci., Biotechnol., Biochem., 2013, 77, 511–515 CrossRef CAS PubMed.
- S.-M. Shin, T.-P. Cao, J.-M. Choi, S.-B. Kim, S.-J. Lee, S. H. Lee and D.-W. Lee, Appl. Environ. Microbiol., 2017, 83, e03291-16 CrossRef PubMed.
- H. J. Kim, E. K. Hyun, Y. S. Kim, Y. J. Lee and D. K. Oh, Appl. Environ. Microbiol., 2006, 72, 981–985 CrossRef CAS PubMed.
- W. Mu, F. Chu, Q. Xing, S. Yu, L. Zhou and B. Jiang, J. Agric. Food Chem., 2011, 59, 7785–7792 CrossRef CAS PubMed.
- H. Itoh, H. Okaya, A. R. Khan, S. Tajima, S. Hayakawa and K. Izumori, Biosci., Biotechnol., Biochem., 1994, 58, 2168–2171 CrossRef CAS.
- Z. Qi, Z. Zhu, J.-W. Wang, S. Li, Q. Guo, P. Xu, F. Lu and H.-M. Qin, Microb. Cell Fact., 2017, 16, 193 CrossRef PubMed.
- A. Yoshihara, T. Kozakai, T. Shintani, R. Matsutani, K. Ohtani, T. Iida, K. Akimitsu, K. Izumori and P. K. Gullapai, J. Biosci. Bioeng., 2017, 123, 170–176 CrossRef CAS PubMed.
- W.-C. Tseng, C.-N. Chen, C.-T. Hsu, H.-C. Lee, H.-Y. Fang, M.-J. Wang, Y.-H. Wu and T.-Y. Fang, Int. J. Biol. Macromol., 2018, 112, 767–774 CrossRef CAS PubMed.
- M. Jia, W. M. Mu, F. F. Chu, X. M. Zhang, B. Jiang, L. L. Zhou and T. Zhang, Appl. Microbiol. Biotechnol., 2014, 98, 717–725 CrossRef CAS PubMed.
- W. L. Zhang, D. Fang, Q. C. Xing, L. Zhou, B. Jiang and W. M. Mu, PLoS One, 2013, 8, e62987 CrossRef CAS PubMed.
- W. M. Mu, W. L. Zhang, D. Fang, L. Zhou, B. Jiang and T. Zhang, Biotechnol. Lett., 2013, 35, 1481–1486 CrossRef CAS PubMed.
- W. Zhang, D. Fang, T. Zhang, L. Zhou, B. Jiang and W. Mu, J. Agric. Food Chem., 2013, 61, 11468–11476 CrossRef CAS PubMed.
- C.-S. Park, T. Kim, S.-H. Hong, K.-C. Shin, K.-R. Kim and D.-K. Oh, PLoS One, 2016, 11, e0160044 CrossRef PubMed.
- Y. Zhu, Y. Men, W. Bai, X. Li, L. Zhang, Y. Sun and Y. Ma, Biotechnol. Lett., 2012, 34, 1901–1906 CrossRef PubMed.
- W. Zhang, T. Zhang, B. Jiang and W. Mu, J. Sci. Food Agric., 2016, 96, 49–56 CrossRef CAS PubMed.
- H.-C. Chan, Y. Zhu, Y. Hu, T.-P. Ko, C.-H. Huang, F. Ren, C.-C. Chen, Y. Ma, R.-T. Guo and Y. Sun, Protein Cell, 2012, 3, 123–131 CrossRef CAS PubMed.
- K. Kim, H.-J. Kim, D.-K. Oh, S.-S. Cha and S. Rhee, J. Mol. Biol., 2006, 361, 920–931 CrossRef CAS PubMed.
- H. Yoshida, M. Yamada, T. Nishitani, G. Takada, K. Izumori and S. Kamitori, J. Mol. Biol., 2007, 374, 443–453 CrossRef CAS PubMed.
- K. Uechi, H. Sakuraba, A. Yoshihara, K. Morimoto and G. Takata, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2013, 69, 2330–2339 CrossRef CAS PubMed.
- C. S. Park, K. C. Shin and D. K. Oh, J. Biosci. Bioeng., 2016, 121, 186–190 CrossRef CAS PubMed.
- H.-M. Qin, T. Miyakawa, A. Inoue, R. Nishiyama, A. Nakamura, A. Asano, T. Ojima and M. Tanokura, Chem. Commun., 2018, 54, 555–558 RSC.
- H.-M. Qin, T. Miyakawa, A. Inoue, R. Nishiyama, A. Nakamura, A. Asano, Y. Sawano, T. Ojima and M. Tanokura, J. Biol. Chem., 2017, 292, 2182–2190 CrossRef CAS PubMed.
- L. Whitmore and B. A. Wallace, Biopolymers, 2008, 89, 392–400 CrossRef CAS PubMed.
- A. Šali and T. L. Blundell, J. Mol. Biol., 1993, 234, 779–815 CrossRef PubMed.
- W. Humphrey, A. Dalke and K. Schulten, J. Mol. Graphics, 1996, 14, 33–38 CrossRef CAS PubMed.
- A. Cornish-Bowden, Fundamentals of Enzyme Kinetics, Portland Press, London, 1995 Search PubMed.
- P. Yang, X. Zhu, Z. Zheng, D. Mu, S. Jiang, S. Luo, Y. Wu and M. Du, World J. Microbiol. Biotechnol., 2018, 34, 65 CrossRef PubMed.
- L. Zhang, W. Mu, B. Jiang and T. Zhang, Biotechnol. Lett., 2009, 31, 857–862 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: The data collected upon which this article is based upon are all included in this manuscript and the additional files associated with it. See DOI: 10.1039/c8ra10029b |
| This journal is © The Royal Society of Chemistry 2019 |




