Open Access Article
Open Access ArticleUpconversion fluorescent nanoparticles based-sensor array for discrimination of the same variety red grape wines†
Kewei
Wang
a,
Yanli
Li
a,
Haijie
Li
a,
Mingyuan
Yin
a,
Huilin
Liu
b,
Qiliang
Deng
 *a and
Shuo
Wang
*ac
*a and
Shuo
Wang
*ac
aKey Laboratory of Food Nutrition and Safety, Ministry of Education, Tianjin Key Laboratory of Food Nutrition and Safety, College of Chemical Engineering and Materials Science, Tianjin University of Science and Technology, Tianjin, 300457, P. R. China. E-mail: yhdql@tust.edu.cn; s.wang@tust.edu.cn
bBeijing Advanced Innovation Center for Food Nutrition and Human Health, Beijing Technology and Business University, 11 Fucheng Road, Beijing, 100048, China
cTianjin Key Laboratory of Food Science and Health, School of Medicine, Nankai University, 94 Weijin Road, Tianjin, 300071, China
First published on 5th March 2019
Abstract
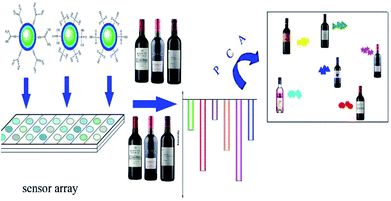
In this research, a novel fluorescent sensor array based on upconversion nanomaterials (UCNPs) for the discrimination of the same variety red grape wines from different manufacturers was developed. The sensor array was composed of six elements: one positively charged UCNPs modified with guanidine groups (UCNPs@GDN), two negatively charged UCNPs modified with sulfonic acid groups (UCNPs@SO3H) and phosphonic acid groups (UCNPs@PO(OH)2), respectively, and their mixture 1 (UCNPs@GDN + UCNPs@SO3H), mixture 2 (UCNPs@GDN + UCNPs@PO(OH)2) and mixture 3 {UCNPs@GDN + UCNPs@SO3H + UCNPs@PO(OH)2}. The discrimination mechanism is mainly attributed to the emission of those upconversion fluorescent nanoparticles being quenched by organic ingredients that usually exist in red grape wines. The discrimination of red grape wines was carried out by employing UCNPs@GDN, UCNPs@SO3H and UCNPs@PO(OH)2 in pH = 7.0 HEPES buffer, the mixture 1 and mixture 2 in pH = 9.0 PBS buffer, and mixture 3 in pH = 6.0 Tris–HCl buffer. Principal component analysis (PCA) of the data obtained from our established array showed obvious distinction among the nine red grape wines from different manufacturers. The present work is expected to inspire more marvellous research in the fields of UCNPs and red grape wines identification.
Introduction
Red grape wine is gaining popularity among people. Recently, red wine fraud is becoming widespread with increasing globalization. Some manufacturers made and sold counterfeit and shoddy products under the drive of interest. One of the most common types of fraud is that wines are adulterated, usually with the addition of cheaper raw materials (e.g. sugars,1 ethanol2), harmful chemicals (e.g. lead-infested syrup,3 industrial grade glycerol4) and other additives.5 Another is counterfeiting and relabelling of inferior and cheaper wines to more expensive brands.3 The struggle against wine fraud is becoming more and more important. Therefore, many different methods for counterfeit and shoddy wine identification have been developed within the past decades, among them the use of mass spectrometry,6 UV-visible (UV-vis),7 mid-infrared (IR) spectroscopy8 and gas chromatography.9 Even though these techniques have been demonstrated with success to a certain extent, some disadvantages including expensive equipment and preliminary procedures of sample purification have hampered the wide application of these techniques in rapid identification.A sensor array, a mimics biology differential sensing approach, differentiates similar analyte based on the non-specific interaction collections between cross-reactive receptors and analyte.10 Compared with traditional sensing methods,11,12 sensor array is composed of the same property many of or different kind sensor elements. And a unique result is got when the same object is inspected. This approach is suitable to distinguish falsification, varieties and sample classification. According to the kinds of output signals, it can be classified into three categories: optical, gravimetric, or electrical. In terms of red wine analysis, electronic sensor arrays have been applied to analysis the composition.13–15 In addition, several reports have focused on the identification of different types,16,17 different varieties18–20 and different aging times21,22 of red wines. Although electronic sensor array seems to be promising, tremendous efforts are still needed to circumvent unstability caused by the aging of the active surface of electrical sensors and sensitivity to humidity.10 Optical sensor arrays, the newly developed detection method, is attracting more and more attentions, and has been widely used to identify gas, organic small molecules, biomacromolecules and cells.23–25 Suslick et al.26 demonstrated a colorimetric sensor array based on nanoporous pigments produced by immobilizing pH indicators in siloxanes for the differentiation among 19 different toxic industrial chemicals. Hamilton et al.27 constructed an 8-porphryin fluorometric array for the recognition of metal and non-metal containing proteins, and demonstrated that the array resolution should be improved by increasing the number of porphyrin in order to differentiate more target proteins. Bunz et al. developed a series of sensor array based on water soluble fluorescent conjugated polymers for identification of cells28 white wines,29 nonsteroidal anti-inflammatory drugs,30 organic acids,31 aromatic carboxylic acids,32 fruit juices,33 whiskies,34 and saccharides.35 Anslyn et al.18 identified six varieties of red grape wines based on a colorimetric sensor array of peptides, metals and indicators, however, such sensor array is failed to discriminate the same variety of red grape wines. In most of these sensor arrays, however, small molecule organic compounds and conjugated polymers are usually utilized as signal element, for which it is difficult to avoid background fluorescence interference. To circumvent the drawback, various types of fluorophores with negligible background interference are urgently demand to construct more kinds of sensor arrays for various complex samples.
Upconversion nanoparticles (UCNPs) can absorb lower-energy photos and emit high-energy photons. Such materials have many advantages, such as high imaging sensitivity, low toxicity, deep tissue penetration, long lifetime fluorescence, less tissue photodamage, and negligible autofluorescence background. These advantages make the UCNPs particularly attractive for sensor, biological imaging36 and drug delivery.37 In the past few years, several groups have developed UCNPs-based fluorescence sensor to detect mycotoxins,38 water,39 glucose,40 diclofenac,41 explosives42 and protein.43,44 However, to the best of our knowledge, the feasibility of UCNPs as fluorophore in the field of sensor array has not been investigated.
At the present research, we attempted to construct a novel fluorescent sensor array based on UCNPs to identify the same variety red grape wines from different manufactures. To accomplish this goal, we modified UCNPs with phosphonic acid groups, sulfonic acid groups and guanidine groups, respectively, and the resulted fluorescent materials were chosen as fluorophore to construct a new fluorescent sensor array. The new optical sensor array is composed of six elements to identify the same variety of red grape wines. As showed in Scheme 1, when the sensor array meets with red grape wines from different manufactures, their fluorescence intensity could be partly quenched.
Materials and methods
Materials and chemicals
S-Ethylisothiourea hydrobromide and 1-octadecene (ODE, 90%) were purchased from TCI chemical (Tokyo, Japan). Tetraethyl orthosilicate (98%), Y(CH3COO)3·4H2O (99.9%), Yb(CH3COO)3·4H2O (99.9%), Er(CH3COO)3·XH2O (99.9%) and vinylphosphonic acid were obtained from Sigma Aldrich (St Louis, USA). 3-Mercaptopropyltriethoxysilane was purchased from J&K chemical (Beijing, China). Aminopropyltriethoxysilane (AMEO), allyltriethoxysilane and oleic acid (OA, 90%) were got from Alfa aesar Co. Ltd (Massachusetts, USA). 2,2-Azobis(isobutyronitrile) (AIBN) and Triton X-100 were purchased from GFCO chemical (Hongkong, China). Hydrogen peroxide (30%), ethanol absolute, cyclohexane (95%), methanol (99.5%), tannic acid and ammonia solution (wt 30%) were obtained from Sinopharm chemical reagent Co. Ltd (Shanghai, China). All red grape wines were purchased from their origin (seeing Table S2† for details). All other chemical reagents were of analytical grade and used as received.Synthesis of UCNPs
NaYF4:Yb (20%), Er (2%) nanoparticles were synthesized according to the literature previously reported with a slight modification.45 Briefly, 1 mM Ln (CH3COO)3 (Ln = Y, Yb, Er) was added to 100 mL three-neck flask containing OA (6.00 mL) and ODE (17.00 mL). The solution was heated to 100 °C under vigorous stirring, evacuated, heated to 160 °C and kept for 30 min, then cooled to room temperature. Methanol solution (10.00 mL) containing NaOH (100.00 mg) and NH4F (148.00 mg) was slowly added to the reaction mixture. After kept for 30 min at room temperature, the system was heated to 100 °C to remove methanol. Then, the solution was heated to 300 °C, and kept for 1 h. Finally, the solution was cooled down to room temperature, and the resulted UCNPs were collected by centrifugation, washed with ethanol three times, and dried in air. All reactions were carried out in argon except evacuation.Synthesis of UCNPs@GDN
UCNPs@GDN was prepared following our previously report.42 UCNPs@NH2 (20.00 mg) and S-ethylisothiourea hydrobromide (10.00 mg) were added to 100 mL flask containing 10 mM of phosphate buffered saline (PBS, 30.00 mL), then the solution was heated to 70 °C under argon atmosphere. After 3 h, the solution was cooled to room temperature. UCNPs@GDN was obtained by centrifugation, washed with ethanol, followed by water several times.Synthesis of UCNPs@SO3H
The UCNPs@SO3H was fabricated as the following procedure. UCNPs (0.12 mM) was dispersed in the mixture of 18.00 mL cyclohexane and 0.30 mL Triton X-100 in 50.00 mL flask under stirring. After kept the system for 10 min, 0.24 mL ammonia solution (wt 30%) and 1.20 mL Triton X-100 were added. Subsequently, the system was sealed and sonicated for 20 min to form a transparent solution. Tetraethyl orthosilicate (120 μL) was slowly dropped into the reaction mixture. After 2 h, 75.0 μL 3-mercaptopropyltriethoxysilane was added, and the solution was kept for 48 h under stirring. The resulted product was collected by centrifugation, washed and dried in air. The product was dispersed in distilled water. Then, 10.00 mL hydrogen peroxide (30%) was slowly dropped. After stirred for 24 h at room temperature, the UCNPs@SO3H was obtained by centrifugation, washed three times with water and dried in air.Synthesis of UCNPs@PO(OH)2
Allyltriethoxysilane (0.12 mM), vinylphosphonic acid (0.12 mM) and AIBN (0.03 mM) were added to 50 mL flask containing cyclohexane (18.00 mL). The mixture was stirred for 12 h at 60 °C, and the poly(allyltriethoxysilane-co-vinylphosphonic acid) was obtained. UCNPs (24.72 mg) were dispersed in cyclohexane containing 0.30 mL Triton X-100. After vigorously stirred for 10 min, 0.24 mL ammonia solution (wt 30%) and 1.20 mL Triton X-100 were added into the solution, the system was sealed and sonicated for 20 min to form a transparent solution. Then, 120 μL of tetraethylorthosilicate was slowly dropped into the reaction mixture. After 2 h, 6.00 mL poly(allyltriethoxysilane-co-vinylphosphonic acid) was dropped into the previous mixture, and stirred for 48 h. Finally, UCNPs@PO(OH)2 was collected by centrifugation, washed three times with water and dried in air.Characterizations
The size and shape of the as-prepared products were characterized by transmission electron microscopy (TEM) (2010 FEF, JEOL, Japan) with an attached energy-dispersive X-ray spectroscope (EDS). Powder X-ray diffraction (XRD) patterns of the dried powders were measured on a Siemens D5005 X-ray powder diffractometer (Bruker, Germany) at a scanning rate of 1° min−1 in the 2θ range from 20° to 70°. Fourier transform infrared (FT-IR) spectra (4000–400 cm−1) were carried out by using a Vector 22 FT-IR spectrophotometer (Bruker, Germany). Fluorescence spectra were performed on an F-2500 Fluorescence spectrometer (Hitachi, Japan) equipped with an external 980 nm laser (Beijing Viasho Technology Co.) instead of internal excitation source. The datas of zeta potential were got on a ZS 90 zeta potentiometer (Malvern, UK). The picture was drawn by Origin 8.Fluorescence response of sensor array to ingredients of red grape wine
PBS buffer (10 mM) was made with the pH range from 4.0 to 11.0 by dissolving sodium phosphate dibasic dodecahydrate into water and adjusting with disodium hydrogen phosphate dodecahydrate solution to desired pH. Other buffer solutions such as Tris–HCl (10 mM) with the pH range from 4.0 to 9.0 and HEPES buffer (10 mM) with the pH range from 7.0 to 11.0 were prepared in the similar way.The solution containing 0.20 mg mL−1 of materials were prepared by adding fluorescent materials to proper buffer. Here, UCNPs@GDN, UCNPs@SO3H and UCNPs@PO(OH)2 were added to HEPES buffer (pH 7.0). (UCNPs@GDN + UCNPs@SO3H) and (UCNPs@GDN + UCNPs@PO(OH)2) were dispersed in PBS buffer (pH 9.0). {UCNPs@GDN + UCNPs@SO3H + UCNPs@PO(OH)2} was put into Tris–HCl buffer (pH 6.0). Then 1.00 mL of solution containing 0.20 mg mL−1 of fluorescent materials was mixed with 1.00 mL of solution containing different wine ingredients (shown in Table S1†). The fluorescence intensity of the system was then checked at the emission wavelength of 551 nm with the excitation wavelength of 980 nm.
Fluorescence response of sensor array to tannic acid
The solution containing 0.20 mg mL−1 of materials were prepared by adding fluorescent materials (UCNPs@GDN, UCNPs@SO3H, UCNPs@PO(OH)2, mixture 1, mixture 2 or mixture 3) to proper buffer. Then 1.00 mL of solution containing 0.20 mg mL−1 of fluorescent materials was mixed with 1.00 mL of tannic acid (5.0 μM) solution. The fluorescence intensity was then checked at the emission wavelength of 551 nm with the excitation wavelength of 980 nm.Identifying of red grape wines based on fluorescence sensing array
UCNPs@GDN, UCNPs@SO3H and UCNPs@PO(OH)2 were dispersed in HEPES buffer solution (pH = 7.0), respectively. Mixture 1 and mixture 2 were dispersed in PBS buffer solution (pH = 9.0), respectively. Proper amount fluorescent materials (mixture 3) was dispersed in Tris–HCl buffer solution (pH = 6.0). Then, 0.20 mL red grape wines were mixed with 1.80 mL buffer solution containing 0.20 mg mL−1 of fluorescent materials. The fluorescence intensity was then checked at the emission wavelength of 551 nm with the excitation wavelength of 980 nm.Principal component analysis (PCA)
Principal component analysis (PCA) is a method of mathematical transformation. It transforms a set of related variables into another group of irrelevant variables through linear transformation. The central idea of principal component analysis is to reduce the dimensionality of a data set. This reduction is achieved by transforming to a set of principal components, which are ordered so that the first few retain most of the variation present in all original variables.Results and discussion
Characterization of upconversion fluorescent materials
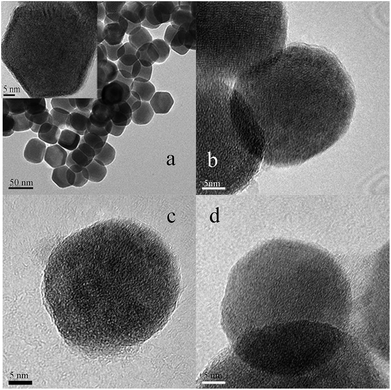
In this research, the novel type materials with strong upconversion fluorescence have been produced for distinguishing the same variety red grape wines. In order to achieve this goal, UCNPs with a small particle size (from 40 to 50 nm) were prepared by solvo-thermal method. The resultant UCNPs were further modified by reversed micro-emulsion method to obtain one positively charged material (UCNPs@GDN) and two negatively charged materials (UCNPs@SO3H and UCNPs@PO(OH)2). The fluorescence spectra of those nanomaterials were obtained by laser excitation under the same test conditions. They all have fluorescence peaks at 365 nm, 413 nm, 478 nm, 551 nm and 665 nm, and the fluorescence peak at 551 nm has the largest peak value (Fig. S1†). Thus, the change of fluorescence value at 551 nm was used as the reference of this experiment. And the size and shape of the as-prepared UCNPs were characterized by TEM. We can observe that these nanoparticles possess obvious crystal stripes, and their morphology were hexagonal, monodisperse, uniform, and narrow in size distribution (Fig. 1(a)). After modified with sulfonic acid, a thin layer of materials was formed on the surface of UCNPs (Fig. 1(b)). These results indicated that sulfonic acid groups have been grafted to the surface of UCNPs. Similar results were also obtained for UNCPs functionalized with phosphonic acid groups (Fig. 1(c)) and guanidine groups (Fig. 1(d)), respectively. These present results may demonstrate that all target fluorescent materials have been successfully obtained.In order to further determine the crystal structure of the resulted fluorescent materials, XRD analysis was carried out. We could see that UCNPs display the diffraction peaks positioned at 2θ of 29.74°, 30.64°, 34.82°, 39.58°, 43.36°, 46.26°, 51.86°, 52.92°, 53.54°, 55.12°, 61.12° and 62.14° (Fig. S2(a)†). These values are agreed well with the standard alignment card (JCPDS card, the card number is 16-0334), the result indicated that the crystal of UCNPs was hexagonal structure. The characteristic peak position and peak shape of UCNPs@SO3H, UCNPs@PO(OH)2 and UCNPs@GDN had no obvious change (Fig. S(2b–d)†), these results indicated that the modification process did not affect the structure of the crystal. However, we observed the characteristics peak intensity decreased due to the existence of a thin layer of functional materials introduced by the modification process.
The resulted fluorescent materials were further characterized by FT-IR to determinate the functional groups. As showed in Fig. S3(a),† the peaks at 1638 cm−1 and 1457 cm−1 belong to the stretching vibration peaks of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O and C–O, respectively. The peaks at 2975 cm−1 and 2927 cm−1 are the stretching vibration of methyl and methylene, respectively. The peak at 3446 cm−1 is the stretching vibration of –OH, showing that OA molecule exists on the surface of the resulted UCNPs. In Fig. S3(b),† the peaks at 1399 cm−1 and 1747 cm−1 belong to the stretching vibration of S–O and S
O and C–O, respectively. The peaks at 2975 cm−1 and 2927 cm−1 are the stretching vibration of methyl and methylene, respectively. The peak at 3446 cm−1 is the stretching vibration of –OH, showing that OA molecule exists on the surface of the resulted UCNPs. In Fig. S3(b),† the peaks at 1399 cm−1 and 1747 cm−1 belong to the stretching vibration of S–O and S![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, respectively. The peaks at 2895 cm−1 and 2827 cm−1 are the stretching vibration of methyl and methylene, respectively. The peak at 3626 cm−1 is the stretching vibration of –OH. The peak at 1089 cm−1 is the stretching vibration of Si–O–Si. In Fig. S3(c),† the peaks at 1632 cm−1 and 1203 cm−1 are the stretching vibration peaks of P–O and P
O, respectively. The peaks at 2895 cm−1 and 2827 cm−1 are the stretching vibration of methyl and methylene, respectively. The peak at 3626 cm−1 is the stretching vibration of –OH. The peak at 1089 cm−1 is the stretching vibration of Si–O–Si. In Fig. S3(c),† the peaks at 1632 cm−1 and 1203 cm−1 are the stretching vibration peaks of P–O and P![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, respectively. The peaks at 2924 cm−1 and 2854 cm−1 are the stretching vibration of methyl and methylene, respectively. The peak at 3439 cm−1 is the stretching vibration of –OH. The peak at 1088 cm−1 is the stretching vibration of Si–O–Si. In Fig. S3(d),† the peaks at 1395 cm−1 and 1645 cm−1 are the stretching vibration of C–N and C
O, respectively. The peaks at 2924 cm−1 and 2854 cm−1 are the stretching vibration of methyl and methylene, respectively. The peak at 3439 cm−1 is the stretching vibration of –OH. The peak at 1088 cm−1 is the stretching vibration of Si–O–Si. In Fig. S3(d),† the peaks at 1395 cm−1 and 1645 cm−1 are the stretching vibration of C–N and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N. The peaks at 2926 cm−1 and 2887 cm−1 are the stretching vibration of methyl and methylene, respectively. The peak at 3625 cm−1 is the stretching vibration of N–H. The peak at 1086 cm−1 is the stretching vibration of Si–O–Si. All these results indicated that the target fluorescent materials have been successfully prepared.
N. The peaks at 2926 cm−1 and 2887 cm−1 are the stretching vibration of methyl and methylene, respectively. The peak at 3625 cm−1 is the stretching vibration of N–H. The peak at 1086 cm−1 is the stretching vibration of Si–O–Si. All these results indicated that the target fluorescent materials have been successfully prepared.
In order to further determinate the functional groups modified on UCNPs, we performed X-ray energy spectrum analysis for all materials. As showed in Fig. S4(a),† the UCNPs samples contain C, O, F, Na, Y, Yb, Er, Cu and Ca elements. The Cu and Ca elements come from the copper mesh. Y, Yb and Er elements are the rare earth elements doped via the reaction. The presence of C and O elements indicated that oleic acid molecules might be linked on the surface of UCNPs. Compared with Fig. S4(a),† the appearing of S element in Fig. S4(b),† P element in Fig. S4(c)† and N element in Fig. S4(d)† indicated that sulfonic acid groups, phosphonic acid group and guanidine have been successfully modified on UCNPs, respectively.
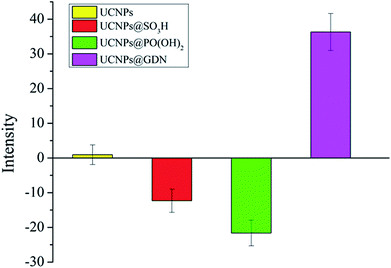
In this research, the surface potentials of resulted fluorescent materials have also been examined. As shown in Fig. 2, UCNPs and UCNPs@GDN displayed the positive surface potentials with +0.94 mV and +36.3 mV, respectively. UCNPs@SO3H and UCNPs@PO(OH)2 displayed the negative surface potentials, which are −12.3 mV and −21.6 mV, respectively. The results further confirmed that the guanidine groups, sulfonic acid groups and phosphonic acid groups have been successfully modified onto the surface of the fluorescent materials.
Fluorescence response patterns
For the discrimination of red grape wines, we set out for a suitable sensor array composed of six elements: one positively charged material (UCNPs@GDN), two negatively charged materials (UCNPs@SO3H and UCNPs@PO(OH)2) and their mixture 1 (UCNPs@GDN + UCNPs@SO3H), mixture 2 (UCNPs@GDN + UCNPs@PO(OH)2) and mixture 3 {UCNPs@GDN + UCNPs@SO3H + UCNPs@PO(OH)2}. The detailed information of the same variety of red grape wines from different manufactures was listed in Table S2.†The response of the sensory array to wine ingredients was first investigated. The PBS buffer solution at pH = 7.0 containing fluorescent materials (0.2 mg mL−1) was mixed with each of wine ingredients (added the concentration of each ingredient shown in Table S1†). After three minutes, the fluorescence intensity was recorded. As showed in Fig. S5,† the sensory array exhibited different quenched degree to the different ingredients. Among these ingredients, tannic acid caused the significant fluorescence quenching. Thus, the fluorescence response of the sensory array caused by the mixture of tannic acid plus different ingredient was further examined. As showed in Fig. 3, the fluorescence sensory array exhibited significant difference in terms of quenched fluorescence intensity to different mixtures. Thus, tannic acid plays an important role in fluorescence recognition. The quenching reason may be mainly attributed to the electron migration between the fluorescence materials and tannic acid.
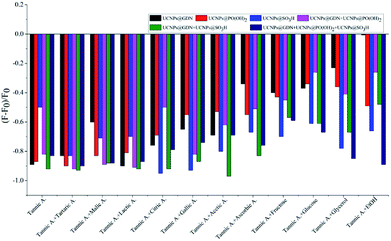 | ||
| Fig. 3 Fluorescence response pattern (F − F0/F0) obtained from the fluorescent materials treated with different wine ingredients plus tannic acid. | ||
The effects of the concentration of tannic acid to the detection system subsequently were investigated. The different buffer solutions (PBS buffer, HEPES buffer and Tris–HCl buffer) at pH = 7.0 containing fluorescent materials (0.2 mg mL−1) were mixed with different concentrations of tannic acid (0.1 μM, 0.5 μM, 1.0 μM, 5.0 μM, 10.0 μM and 50.0 μM). After three minutes, the fluorescence intensity was recorded. As showed in Fig. S6,† the fluorescence intensity of all materials decreased with the concentration increasing. When the concentration of tannic acid reached 5.0 μM, the fluorescence quenching (F − F0/F0) of all sensing elements was in the range from −0.2 to −0.69. In order to avoid other objects caused excessively high or too low fluorescent quenching, 5.0 μM of tannic acid was chosen for the following experiment.
The sensor elements and pH were also optimized. In PBS buffer, the maximum fluorescence quenching (F − F0/F0) of UCNPs@GDN was −0.47 at pH 5.0, UCNPs@SO3H −0.71 at pH 11.0, UCNPs@PO(OH)2 −0.59 at pH 7.0, mixture 1 −0.76 at pH 9.0, mixture 2 −0.74 at pH 9.0, mixture 3 −0.73 at pH 6.0, respectively (Fig. S7(a)†). In Tris–HCl buffer, the maximum fluorescence quenching (F − F0/F0) were −0.43 at pH 7.0, −0.67 at pH 6.0, −0.65 at pH 5.0, −0.75 at pH 4.0, −0.72 at pH 6.0, −0.90 at pH 6.0 for UCNPs@GDN, UCNPs@SO3H, UCNPs@PO(OH)2, mixture 1, mixture 2 and mixture 3, respectively (Fig. S7(b)†). In HEPES buffer, the maximum fluorescence quenching (F − F0/F0) of sensor elements UCNPs@GDN, UCNPs@SO3H, UCNPs@PO(OH)2, mixture 1, mixture 2 and mixture 3 were −0.73 at pH 7.0, −0.75 at pH 7.0, −0.80 at pH 7.0, −0.65 at pH 9.0, −0.73 at pH 11.0 and −0.73 at pH 10.0, respectively (Fig. S7(c)†). In summary, the results obtained for all sensor elements in the three buffers system at different pH indicated that the optimum differentiation conditions for red grape wines were: UCNPs@GDN, UCNPs@SO3H and UCNPs@PO(OH)2 in HEPES buffer system (pH 7.0), mixture 1 and mixture 2 in PBS buffer system (9.0), and mixture 3 in Tris–HCl buffer system (pH 6.0).
Finally, the effect of incubation times (1 min, 2 min, 3 min, 4 min and 5 min) on the fluorescent quenching was also investigated. As showed in Fig. S8 and S9,† the fluorescent intensity of all the materials showed a slight decrease with the prolong of incubation time. When the reaction time was 3 min, the maximum degree fluorescence quenching was observed. When the incubation time was extended, the fluorescence quenching was basically unchanged.
Based on the above experimental results, different volume red grape wines (5 vol%, 10 vol%, 15 vol%, 20 vol%, 25 vol% and 30 vol%) were added into three buffer (PBS buffer, HEPES buffer and Tris–HCl buffers) containing 0.20 mg mL−1 fluorescent materials. After three minutes, the fluorescence intensity of the system was recorded. As showed in Fig. S10† (left), the quenching of the materials treated with red grape wine showed a decrease with the volume of red grape wine increased. The fluorescence quenching of fluorescent materials caused by red grape wine was similar with that of tannic acid on fluorescent materials when the volume of red grape wine was 10 vol%.
Then, the effect of incubation times (1 min, 2 min, 3 min, 4 min and 5 min) to the fluorescent quenching was also investigated. The fluorescent intensity of all the fluorescent materials showed a slight decrease with the prolong of incubation time, the fluorescence quenching value reached the highest when the reaction time was 3 min (Fig. S10†).
Under the optimized conditions, the sensor array composed of six elements was employed to identify the same variety red grape wines from nine countries. As shown in Fig. 4, every sensor element shows different fluorescent response to red grape wines from different manufactures. The sensor array which composed of six sensor elements show different fluorescent response to identical red grape wine, and different red grape wines from different manufactures, respectively. Then, PCA, a statistical technique that maximizes the ratio of between-class variance to within-class variance, was used to differentiate quantitatively the fluorescence-response patterns (6 sensor elements × 9 wines × 6 replicates) (Table S3†) of the sensor array for these red grape wines.
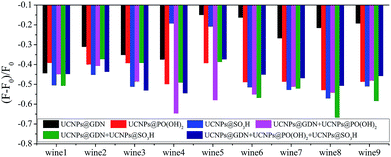 | ||
| Fig. 4 Fluorescence response pattern (F − F0/F0) obtained from sensor array treated with red grape wines. | ||
After the above analysis, six principal components (40.81%, 29.09%, 23.43%, 4.26%, 1.81% and 0.60%) were generated that represent linear combinations of the response matrices obtained from the fluorescence-response patterns. Nine red grape wines were separated complete in the 2D canonical score plot for the first two factors (factor 1: 40.81%, factor 2: 29.09%) and wine 7 is closely to wine 9 (Fig. S11†). However, as 3D canonical score plot (Fig. 5) composed of the three maximum factors (factor 1: 40.81%, factor 2: 29.09%, factor 3: 23.43%; the sum of those was 93.3%) showed, wine 7 is separated from wine 9. And others were separated completely. Thus, the 54 training cases (9 wines × 6 replicates) were separated into nine respective groups which the one group represents one wine, respectively, with 100% accuracy according to the jackknifed classification matrix derived from then analysis of subsets of the datasets. In order to validate the efficiency of our sensing system, we established tests with randomly chosen red grape wines of our training set. The new cases were classified into different groups generated from the training matrix, based on the shortest Mahalanobis distance to the respective group. Only 1 of 45 unknown red grape wines was misclassified, representing an accuracy of 98% (Table S5†).
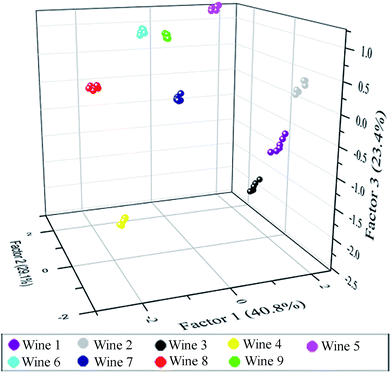 | ||
| Fig. 5 3D canonical score plot obtained with the sensor array of six elements treated with red grape wines. Each point represents the response pattern for a single red wine to the sensor array. | ||
Conclusions
In conclusion, a six-element sensor array consisting of UCNPs@GDN, UCNPs@SO3H, UCNPs@PO(OH)2, UCNPs@GDN + UCNPs@SO3H, UCNPs@GDN + UCNPs@PO(OH)2 and UCNPs@GDN + UCNPs@SO3H + UCNPs@PO(OH)2 has been constructed and employed to discriminate the same variety of red grape wines from different manufactures. Differentiation the nine red grape wines were performed at pH = 7.0 HEPES buffer for UCNPs@GDN, UCNPs@SO3H and UCNPs@PO(OH)2, at pH = 9.0 PBS buffer for mixture 1 {UCNPs@GDN + UCNPs@SO3H}, and mixture 2 {UCNPs@GDN + UCNPs@PO(OH)2}, and at pH = 6.0 Tris–HCl buffer for mixture 3 {UCNPs@GDN + UCNPs@SO3H + UCNPs@PO(OH)2}. All the nine red grape wines were clearly differentiated by the established sensor array. The present fluorescence sensor array shows great potential in distinguishing a wide range of targets through pattern recognition.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors are grateful for the financial support provided by the Ministry of Science and Technology of China (Project No. 2016YFD0401101) and the National Natural Science Foundation of China (Project No. 21375094).Notes and references
- N. Dordevic, F. Camin, R. M. Marianella, G. J. Postma, L. M. C. Buydens and R. Wehrens, Aust. J. Grape Wine Res., 2013, 19, 324–330 CrossRef CAS.
- E. I. Geana, R. Popescu, D. Costinel, O. R. Dinca, I. Stefanescu, R. E. Ionete and C. Bala, Food Control, 2016, 62, 1–9 CrossRef CAS.
- L. Holmberg, Wine fraud, 2010 Search PubMed.
- V. Dixit, J. C. Tewari, B.-K. Cho and J. M. K. Irudayaraj, Appl. Spectrosc., 2005, 59, 1553–1561 CrossRef CAS.
- V. Constanta, G. Rapeanu and G. Bahrim, Identification of Romanian Wine Adulteration from Vrancea County, 2009 Search PubMed.
- A. Ziółkowska, E. Wąsowicz and H. H. Jeleń, Food Chem., 2016, 213, 714–720 CrossRef.
- A. G. Mignani, L. Ciaccheri, B. Gordillo, A. A. Mencaglia, M. L. González-Miret, F. J. Heredia and B. Culshaw, Sens. Actuators, B, 2012, 171–172, 458–462 CrossRef CAS.
- D. Picque, P. Lieben, G. Corrieu, R. Cantagrel, O. Lablanquie and G. Snakkers, J. Agric. Food Chem., 2006, 54, 5220–5226 CrossRef CAS.
- D. González-Arjona, G. López-Pérez, V. González-Gallero and A. G. González, J. Agric. Food Chem., 2006, 54, 1982–1989 CrossRef.
- J. R. Askim, M. Mahmoudi and K. S. Suslick, Chem. Soc. Rev., 2013, 42, 8649–8682 RSC.
- Q. Zhang, Y. Qiao, F. Hao, L. Zhang, S. Wu, Y. Li, J. Li and X. Song, Fabrication of a Biocompatible and Conductive Platform Based on a Single Stranded DNA/Graphene Nanocomposite for Direct Electrochemistry and Electrocatalysis, Chem.–Eur. J., 2010, 16, 8133–8139 CrossRef CAS.
- L. Tang and J. Li, Plasmon–Based Colorimetric Nanosensors for Ultrasensitive Molecular Diagnostics, ACS Sens., 2017, 2, 857–875 CrossRef CAS.
- M. L. Rodriguez-Mendez, C. Apetrei, M. Gay, C. Medina-Plaza, J. A. de Saja, S. Vidal, O. Aagaard, M. Ugliano, J. Wirth and V. Cheynier, Food Chem., 2014, 155, 91–97 CrossRef CAS.
- A. M. Simoes da Costa, I. Delgadillo and A. Rudnitskaya, Talanta, 2014, 129, 63–71 CrossRef CAS.
- A. Rudnitskaya, L. M. Schmidtke, A. Reis, M. R. Domingues, I. Delgadillo, B. Debus, D. Kirsanov and A. Legin, Food Chem., 2017, 229, 20–27 CrossRef CAS.
- W. Novakowski, M. Bertotti and T. R. L. C. Paixão, Microchem. J., 2011, 99, 145–151 CrossRef CAS.
- E. Witkowska Nery and L. Kubota, Anal. Chim. Acta, 2016, 918, 60–68 CrossRef.
- P. Umali, S. E. LeBoeuf, R. Newberry, S. Kim, L. Tran, W. A. Rome, T. Tian, D. Taing, J. Hong, M. Kwan, H. Heymann and E. Anslyn, Chem. Sci., 2011, 2, 439–445 RSC.
- M. Gutiérrez, C. Domingo, J. Vila-Planas, A. Ipatov, F. Capdevila, S. Demming, S. Büttgenbach, A. Llobera and C. Jiménez-Jorquera, Sens. Actuators, B, 2011, 156, 695–702 CrossRef.
- L. T. Gallagher, J. Seok Heo, M. A. Lopez, B. M. Ray, J. Xiao, P. Umali, A. Zhang, S. Dharmarajan, H. Heymann and E. Anslyn, Supramol. Chem., 2012, 24, 1–7 CrossRef.
- V. Parra, T. Hernando, M. L. Rodríguez-Méndez and J. A. de Saja, Electrochim. Acta, 2004, 49, 5177–5185 CrossRef CAS.
- M. García, M. J. Fernández, J. L. Fontecha, J. Lozano, J. P. Santos, M. Aleixandre, I. Sayago, J. Gutiérrez and M. C. Horrillo, Talanta, 2006, 68, 1162–1165 CrossRef.
- M. De, S. Rana, H. Akpinar, O. R. Miranda, R. R. Arvizo, U. H. F. Bunz and V. M. Rotello, Nat. Chem., 2009, 1, 461–465 CrossRef CAS PubMed.
- L. You, D. Zha and E. V. Anslyn, Chem. Rev., 2015, 115, 7840–7892 CrossRef CAS.
- J. Han, H. Cheng, B. Wang, M. S. Braun, X. Fan, M. Bender, W. Huang, C. Domhan, W. Mier, T. Lindner, K. Seehafer, M. Wink and U. H. F. Bunz, Angew. Chem., Int. Ed., 2017, 56, 15246–15251 CrossRef CAS.
- S. H. Lim, L. Feng, J. W. Kemling, C. J. Musto and K. S. Suslick, Nat. Chem., 2009, 1, 562–567 CrossRef CAS.
- H. Zhou, L. Baldini, J. Hong, A. J. Wilson and A. D. Hamilton, J. Am. Chem. Soc., 2006, 128, 2421–2425 CrossRef CAS.
- A. Bajaj, O. R. Miranda, R. Phillips, I.-B. Kim, D. J. Jerry, U. H. F. Bunz and V. M. Rotello, J. Am. Chem. Soc., 2010, 132, 1018–1022 CrossRef CAS.
- J. Han, M. Bender, K. Seehafer and U. Bunz, Angew. Chem., Int. Ed., 2016, 55, 7689–7692 CrossRef CAS.
- J. Han, B. Wang, M. Bender, S. Kushida, K. Seehafer and U. Bunz, ACS Appl. Mater. Interfaces, 2016, 9, 790–797 CrossRef.
- J. Han, M. Bender, S. Hahn, K. Seehafer and U. Bunz, Chem.–Eur. J., 2016, 22, 3230–3233 CrossRef CAS.
- J. Han, B. Wang, M. Bender, K. Seehafer and U. Bunz, ACS Appl. Mater. Interfaces, 2016, 8, 20415–20421 CrossRef CAS.
- J. Han, B. Wang, M. Bender, K. Seehafer and U. H. F. Bunz, Analyst, 2017, 142, 537–543 RSC.
- J. Han, C. Ma, B. Wang, M. Bender, M. Bojanowski, M. Hergert and K. Seehafer, Chem, 2017, 2, 817–824 CAS.
- U. Bunz, M. Bojanowski, M. Bender and K. Seehafer, Chem.–Eur. J., 2017, 23, 12253–12258 CrossRef.
- H. Dong, S.-R. Du, X.-Y. Zheng, G.-M. Lyu, L.-D. Sun, L.-D. Li, P.-Z. Zhang, C. Zhang and C.-H. Yan, Chem. Rev., 2015, 115, 10725–10815 CrossRef CAS.
- J. yu, W. Yin, T. Peng, Y. Chang, Y. Zu, J. Li, X. He, X. Ma, Z. Gu and Y. Zhao, Nanoscale, 2017, 9, 4497–4507 RSC.
- S. Wu, N. Duan, X. Ma, Y. Xia, H. Wang, Z. Wang and Q. Zhang, Anal. Chem., 2012, 84, 6263–6270 CrossRef CAS.
- S. Guo, X. Xie, L. Huang and W. Huang, ACS Appl. Mater. Interfaces, 2016, 8(1), 847–853 CrossRef CAS.
- J. Yuan, Y. Cen, X.-J. Kong, S. Wu, C.-L. Liu, R.-Q. Yu and X. Chu, ACS Appl. Mater. Interfaces, 2015, 7, 10548–10555 CrossRef CAS.
- A. Hlavacek, Z. Farka, M. Huebner, V. Horáčková, D. Nemecek, R. Niessner, P. Skládal, D. Knopp and H. Gorris, Anal. Chem., 2016, 88, 6011–6027 CrossRef CAS.
- W. Wang, H. Li, M. Yin, K. Wang, Q. Deng, S. Wang and Y. Zhang, Sens. Actuators, B, 2017, 255, 1422–1429 CrossRef.
- T. Guo, Q. Deng, G. Fang, D. Gu, Y. Yang and S. Wang, Biosens. Bioelectron., 2016, 79, 341–346 CrossRef CAS.
- T. Guo, Q. Deng, G. Fang, Y. Yun, Y. Hu and S. Wang, Biosens. Bioelectron., 2016, 85, 596–602 CrossRef CAS.
- J. Zhao, Z. Lu, Y. Yin, C. McRae, J. A. Piper, J. M. Dawes, D. Jin and E. M. Goldys, Nanoscale, 2013, 5, 944–952 RSC.
Footnote |
| † Electronic supplementary information (ESI) available: Experimental procedures; characterizations (XRD, FT-IR and EDS) for all UCNPs materials; fluorescence response of UCNPs materials treated with tannic acid and red grape wines vs. concentrations and times; additional PCA data and plots. See DOI: 10.1039/c8ra09959f |
| This journal is © The Royal Society of Chemistry 2019 |