 Open Access Article
Open Access ArticleQuantification of azide groups on a material surface and a biomolecule using a clickable and cleavable fluorescent compound†
Rika Sakai,
Hiroki Iguchi and
Tatsuo Maruyama *
*
Department of Chemical Science and Engineering, Graduate School of Engineering, Kobe University, 1-1 Rokkodai, Nada-ku, Kobe 657-8501, Japan. E-mail: tmarutcm@crystal.kobe-u.ac.jp
First published on 18th February 2019
Abstract
Here, we propose a novel method for quantifying azide groups on a solid surface and a protein using a clickable and cleavable fluorescent compound. The clickable and cleavable fluorescent compound conjugates with the azide groups on the material surface and the fluorophore is then liberated into the solvent via a cleavage reaction, which can be simply quantified with a conventional fluorometer.
Introduction
A bioorthogonal reaction is a chemical reaction that can be selectively and reliably performed in a complex biological environment without non-favorable interactions and interference with the biological system.1–3 Huisgen 1,3-dipolar cycloaddition between an azide and an alkyne (the azide/alkyne cycloaddition click reaction), which is one of the most famous bioorthogonal reactions, proceeds in a high yield under mild aqueous conditions without byproducts.4–6 This chemistry has been applied to a number of biomolecules and synthetic compounds to create multifunction-integrated molecular systems. This click reaction has also been used to immobilize biomolecules, ligands, and polymers on substrate surfaces7–11 for microarray preparation,12 fabrication of biomolecule-conjugated nanoparticles13–16 and fluorescent labeling of living cells.17–19When click chemistry is applied to sensitive analytical and biological devices, quantification of the reactive sites plays an important role in controlling the amount of azide groups (or alkyne groups) present on a substrate surface, because the surface density of the functional molecules affects the performance of the substrate.20,21 However, to our knowledge, there have been no reports of quantification methods for clickable moieties (azide and alkyne groups) on substrate surfaces. Conventional surface analysis methods, such as X-ray photon spectroscopy (XPS), Fourier transform infrared spectroscopy and secondary-ion mass spectrometry are qualitative and provide information about the elements or functional groups not only on the outermost surface but also those at a certain depth (e.g., 10 nm for XPS) from the surface.22,23 These methods require a high vacuum environment or dry atmosphere, and the surfaces of polymeric materials vary depending on the environment. Therefore, it is of great importance to quantify the clickable moieties on the outermost surface in a practical environment (e.g., aqueous solution) for surface modification based on click chemistry.
In this study, we aimed to develop a method for quantification of the azide groups present on the outermost surface in an aqueous environment. We synthesized a clickable and cleavable fluorescent compound that conjugates with the azide groups on the substrate surface via Huisgen 1,3-dipolar cycloaddition (Fig. 1). Hydrolysis of the ester bond in the fluorescent compound liberates the fluorophore into the bulk solution, which allows the azide groups on a substrate surface to be quantified with a conventional fluorometer. The present methodology can be also applied to the quantification of azide groups conjugated with a protein.
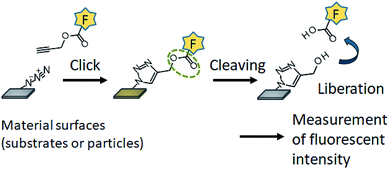 | ||
| Fig. 1 Schematic of quantification of the azide groups on substrate surfaces using the clickable and cleavable fluorescent compound. | ||
Experimental
Materials
2,4-Dihydroxybenzaldehyde, 2,2-dimethyl-1,3-dioxane-4,6-dione, piperidinium acetate, 2-hydroxy-4-methoxybenzaldehyde, propargyl-PEG4-alcohol, and azidobenzoic acid were purchased from Tokyo Chemical Industry (Tokyo, Japan). K2CO3, propargyl-bromide, CuSO4, β-casein from bovine milk, rhodamine B isothiocyanate (RITC), and Tris–EDTA buffer solution (pH 7) were purchased from Sigma-Aldrich (St. Louis, MO). DBCO-s-s-amine was purchased from Conju-Probe LLC (San Diego, CA). The other chemicals were purchased from Wako Pure Chemical Industries, Ltd. (Osaka, Japan). NH2-PEG-resin particles (10 μm) were purchased from Watanabe Chemical Industries (Hiroshima, Japan). Slide-A-Lyzer G2 dialysis cassettes (20k MWCO, 15 mL) and micro BCA protein assay were purchased from Thermo Fisher Scientific (Waltham, USA). High-quality deionized (DI) water (>15 MΩ cm) produced by an Elix-5 system (Millipore, Molsheim, France) was used in the experiments.1H-NMR (500 MHz, CDCl3): δ (ppm) 3.97 (s, 3H), 6.93 (d, 1H, J = 2.2 Hz), 7.03 (dd, 1H, J = 2.6, 8.9 Hz), 7.65 (d, 1H, J = 8.8 Hz), 8.86 (s, 1H), 12.18 (s, 1H). DART-TOF/MS (m/z): [M + H]+ calcd for C11H9O5, 221.0; found, 221.0. Yield: 83%.
![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/b_char_e002.gif) C–CMRN. Propargyl bromide (0.4 mmol) was added to a dehydrated DMF solution (7 mL) containing 7-methoxycoumarin-3-carboxylic acid (0.2 mmol) and K2CO3 (0.6 mmol). The solution was stirred at room temperature for 24 h. Crude C
C–CMRN. Propargyl bromide (0.4 mmol) was added to a dehydrated DMF solution (7 mL) containing 7-methoxycoumarin-3-carboxylic acid (0.2 mmol) and K2CO3 (0.6 mmol). The solution was stirred at room temperature for 24 h. Crude C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) C–CMRN was obtained as a yellow powder after filtration and evaporation. A HPLC system (LC-20AT, Shimadzu, Kyoto, Japan) equipped with an Inertsil ODS-3 column (10 mm × 250 mm, GL Science, Tokyo, Japan) was used to purify C
C–CMRN was obtained as a yellow powder after filtration and evaporation. A HPLC system (LC-20AT, Shimadzu, Kyoto, Japan) equipped with an Inertsil ODS-3 column (10 mm × 250 mm, GL Science, Tokyo, Japan) was used to purify C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) C–CMRN. The mobile phase was water containing 0.1 wt% trifluoroacetic acid (TFA) and acetonitrile containing 0.1 wt% TFA and the flow rate was 5 mL min−1. The eluted compounds were observed by measuring the absorbance at 350 nm. The gradient started at 30% acetonitrile and 70% water, and the proportion of acetonitrile increased to 100% over 30 min. Isocratic 100% acetonitrile was maintained for 10 min.
C–CMRN. The mobile phase was water containing 0.1 wt% trifluoroacetic acid (TFA) and acetonitrile containing 0.1 wt% TFA and the flow rate was 5 mL min−1. The eluted compounds were observed by measuring the absorbance at 350 nm. The gradient started at 30% acetonitrile and 70% water, and the proportion of acetonitrile increased to 100% over 30 min. Isocratic 100% acetonitrile was maintained for 10 min.1H-NMR (500 MHz, CDCl3): δ (ppm) 3.91 (s, 3H), 4.93 (s, 2H), 6.83 (s, 1H), 6.91 (dd, 1H, J = 2.5, 8.8 Hz), 7.52 (d, 1H, J = 8.8 Hz), 8.58 (s, 1H), DART-TOF/MS (m/z): [M + H]+ calcd for C14H11O5, 259.1; found, 259.1. Yield: 12%.
![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/b_char_e002.gif) C–CMRN. A DMSO solution (0.4 mL) containing 4 mmol C
C–CMRN. A DMSO solution (0.4 mL) containing 4 mmol C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) C–CMRN and 4 mmol azidobenzoic acid was mixed and stirred with an aqueous solution (1 mL) of 0.4 mmol CuSO4 and 2 mmol sodium ascorbate at 25 °C in a water bath for 3 h. This solution was evaporated and analyzed by HPLC (Fig. S1†). The HPLC fractions of the reaction products were collected and lyophilized.
C–CMRN and 4 mmol azidobenzoic acid was mixed and stirred with an aqueous solution (1 mL) of 0.4 mmol CuSO4 and 2 mmol sodium ascorbate at 25 °C in a water bath for 3 h. This solution was evaporated and analyzed by HPLC (Fig. S1†). The HPLC fractions of the reaction products were collected and lyophilized.1H-NMR (500 MHz, DMSO-d6): δ (ppm) 3.96 (s, 3H), 5.57 (s, 2H), 6.83 (s, 1H), 6.91 (dd, 1H, J = 2.5, 8.8 Hz), 7.52 (d, 1H, J = 8.5 Hz),7.90 (d, 2H, J = 8.8 Hz), 8.25 (d, 2H, J = 9.2 Hz), 8.09 (d, 2H, J = 8.9 Hz), 8.69 (s, 1H), 8.86 (s, 1H).
![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/b_char_e002.gif) C–EG3–CMRN. Propargyl-PEG4-alcohol (0.2 mmol) was added to a solution of 7-methoxycoumarin-3-carboxylic acid (0.4 mmol), 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide, hydrochloride (WSC, 0.24 mmol) and N,N-dimethyl-4-aminopyridine (DMAP, 0.2 mmol) in DMF (1 mL). The solution was stirred at room temperature for 3 h. The product was purified by HPLC. After evaporation and freeze-drying, the product was obtained as a yellow powder.
C–EG3–CMRN. Propargyl-PEG4-alcohol (0.2 mmol) was added to a solution of 7-methoxycoumarin-3-carboxylic acid (0.4 mmol), 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide, hydrochloride (WSC, 0.24 mmol) and N,N-dimethyl-4-aminopyridine (DMAP, 0.2 mmol) in DMF (1 mL). The solution was stirred at room temperature for 3 h. The product was purified by HPLC. After evaporation and freeze-drying, the product was obtained as a yellow powder.1H-NMR (500 MHz, CDCl3): δ (ppm) 2.41 (t, 1H), 3.70–3.73 (m, 8H), 3.84 (t, 2H), 3.91 (s, 3H), 4.19 (d, 2H, J = 2.2 Hz), 7.88 (t, 2H, J = 9 Hz), 6.82 (s, 1H), 6.90 (dd, 2H, J = 2.5, 8.5 Hz), 7.51 (d, 1H, J = 8.9 Hz), 8.53 (s, 1H). DART-TOF/MS (m/z): [M + H]+ calcd for C20H23O8, 391.1; found, 391.2. Yield: 21%.
![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/b_char_e002.gif) C–EG3–CMRN. A solution of C
C–EG3–CMRN. A solution of C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) C–EG3–CMRN (2.5 μmol) and azidobenzoic acid (2.5 μmol) in DMSO (0.4 mL) was mixed and stirred with an aqueous solution (1.6 mL) of CuSO4 (6 μmol) and sodium ascorbate (30 μmol) at 37 °C for 3 h. The solution was analyzed as described above (click reaction of C
C–EG3–CMRN (2.5 μmol) and azidobenzoic acid (2.5 μmol) in DMSO (0.4 mL) was mixed and stirred with an aqueous solution (1.6 mL) of CuSO4 (6 μmol) and sodium ascorbate (30 μmol) at 37 °C for 3 h. The solution was analyzed as described above (click reaction of C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) C–EG3–CMRN).
C–EG3–CMRN).1H-NMR (500 MHz, DMSO-d6): δ (ppm) 3.54–3.61 (m, 8H), 3.71 (t, 2H), 3.88 (s, 3H), 4.34 (t, 2H, J = 2.2 Hz), 4.61 (s, 2H), 6.98 (dd, 1H, J = 2.5, 8.8 Hz), 7.01 (s, 1H),7.82 (d, 1H, J = 8.6 Hz), 7.98 (d, 2H, J = 8.5 Hz), 8.09 (d, 2H, J = 8.9 Hz), 8.69 (s, 1H), 8.86 (s, 1H). Yield: 96%.
![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) C–EG3–CMRN with azidobenzoic acid. The mixture was stirred at 37 °C for 3 h and analyzed by HPLC.
C–EG3–CMRN with azidobenzoic acid. The mixture was stirred at 37 °C for 3 h and analyzed by HPLC.![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/b_char_e002.gif) C–CMRN. Azidated resin particles (0, 0.1875, 0.375, or 0.75 mg) were dispersed in water (1.8 mL) containing CuSO4 (1 μmol) and sodium ascorbate (4 μmol). The dispersion was added to 1 mM C
C–CMRN. Azidated resin particles (0, 0.1875, 0.375, or 0.75 mg) were dispersed in water (1.8 mL) containing CuSO4 (1 μmol) and sodium ascorbate (4 μmol). The dispersion was added to 1 mM C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) C–EG3–CMRN in DMSO (0.1 mL) and shaken at 40 °C for 24 h. After centrifugation, the supernatant was removed and the resin particles were washed three times with methanol and water. The resin particles were dispersed in NaOH aqueous solution (1 M, 0.2 mL) and then shaken at 37 °C for 24 h to liberate CMRN from the resin particles. After centrifugation, HCl solution (1 M, 0.2 mL) and phosphate buffer (0.1 M, pH 7.4, 2 mL) were added to neutralize the supernatant. The fluorescence intensity of the neutralized solution was measured with a fluorescence spectrophotometer. The excitation and emission wavelengths were 335 and 406.5 nm, respectively. The excitation and emission band widths were 5 nm. The standard curve of a liberated fluorophore in phosphate buffer is shown in Fig. S2a.† Quantification was performed in triplicate, unless specified otherwise.
C–EG3–CMRN in DMSO (0.1 mL) and shaken at 40 °C for 24 h. After centrifugation, the supernatant was removed and the resin particles were washed three times with methanol and water. The resin particles were dispersed in NaOH aqueous solution (1 M, 0.2 mL) and then shaken at 37 °C for 24 h to liberate CMRN from the resin particles. After centrifugation, HCl solution (1 M, 0.2 mL) and phosphate buffer (0.1 M, pH 7.4, 2 mL) were added to neutralize the supernatant. The fluorescence intensity of the neutralized solution was measured with a fluorescence spectrophotometer. The excitation and emission wavelengths were 335 and 406.5 nm, respectively. The excitation and emission band widths were 5 nm. The standard curve of a liberated fluorophore in phosphate buffer is shown in Fig. S2a.† Quantification was performed in triplicate, unless specified otherwise.![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) C–EG3–CMRN (5 μmol) in DMSO (0.4 mL) was added to the solution and the solution was stirred at 37 °C for 3 h. After the reaction, excess C
C–EG3–CMRN (5 μmol) in DMSO (0.4 mL) was added to the solution and the solution was stirred at 37 °C for 3 h. After the reaction, excess C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) C–EG3–CMRN and Cu(I) catalyst were removed by dialysis for 2 days. NaOH solution (1 M, 0.5 mL) was added to the solution (0.5 mL) and the solution was then shaken at 40 °C for 2 h to liberate CMRN fluorophore from β-casein. HCl solution (1 M, 0.5 mL) and NaHCO3 buffer (1 M, pH 8, 2 mL) were added to neutralize the solution. The fluorescent intensity of the solution was measured with a fluorescent spectrometer. The excitation and emission bands were 335 and 406.5 nm, respectively. The standard curve of a liberated fluorophore in carbonate buffer is shown in Fig. S2b.† Quantification was performed in triplicate, unless specified otherwise.
C–EG3–CMRN and Cu(I) catalyst were removed by dialysis for 2 days. NaOH solution (1 M, 0.5 mL) was added to the solution (0.5 mL) and the solution was then shaken at 40 °C for 2 h to liberate CMRN fluorophore from β-casein. HCl solution (1 M, 0.5 mL) and NaHCO3 buffer (1 M, pH 8, 2 mL) were added to neutralize the solution. The fluorescent intensity of the solution was measured with a fluorescent spectrometer. The excitation and emission bands were 335 and 406.5 nm, respectively. The standard curve of a liberated fluorophore in carbonate buffer is shown in Fig. S2b.† Quantification was performed in triplicate, unless specified otherwise.Results and discussion
First, we designed and synthesized two different types of clickable and cleavable fluorescent compounds: C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) C–CMRN and C
C–CMRN and C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) C–EG3–CMRN (Fig. 2a). The alkyne group acts as the clickable part and the ester bond acts as the cleavable part in hydrolysis. C
C–EG3–CMRN (Fig. 2a). The alkyne group acts as the clickable part and the ester bond acts as the cleavable part in hydrolysis. C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) C–CMRN has no spacer between the ester bond and alkyne, but C
C–CMRN has no spacer between the ester bond and alkyne, but C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) C–EG3–CMRN has an oligo (ethylene glycol) spacer. The click reactivities of C
C–EG3–CMRN has an oligo (ethylene glycol) spacer. The click reactivities of C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) C–CMRN and C
C–CMRN and C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) C–EG3–CMRN were investigated in a homogeneous solution. Azidobenzoic acid was used as a model azide compound and the click reaction was performed with a copper catalyst in a mixture of water/dimethylformamide (DMF) (5/1, v/v). High-performance liquid chromatography (HPLC) measurements of C
C–EG3–CMRN were investigated in a homogeneous solution. Azidobenzoic acid was used as a model azide compound and the click reaction was performed with a copper catalyst in a mixture of water/dimethylformamide (DMF) (5/1, v/v). High-performance liquid chromatography (HPLC) measurements of C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) C–CMRN indicate simultaneous synthesis of compound 1′ (at a retention time (RT) of 15.3 min) and production of 7-methylcoumarin-3-carboxylic acid (Fig. S1†), indicating that hydrolysis of the ester bond is probably due to chelation of a Cu ion at the neighboring site. HPLC measurements of C
C–CMRN indicate simultaneous synthesis of compound 1′ (at a retention time (RT) of 15.3 min) and production of 7-methylcoumarin-3-carboxylic acid (Fig. S1†), indicating that hydrolysis of the ester bond is probably due to chelation of a Cu ion at the neighboring site. HPLC measurements of C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) C–EG3–CMRN reveal that the peak at RT = 14.6 min disappears and a peak at RT = 7.7 min appears (Fig. 2b). 1H-NMR analysis of these fractions reveals that the peaks at RT = 14.6 and 7.7 min correspond to C
C–EG3–CMRN reveal that the peak at RT = 14.6 min disappears and a peak at RT = 7.7 min appears (Fig. 2b). 1H-NMR analysis of these fractions reveals that the peaks at RT = 14.6 and 7.7 min correspond to C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) C–EG3–CMRN and compound 1 (the product of the click reaction), respectively. These results indicate that C
C–EG3–CMRN and compound 1 (the product of the click reaction), respectively. These results indicate that C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) C–EG3–CMRN can be used as a substrate in the azide/alkyne click reaction and that the reaction proceeds in high yield (>96%).
C–EG3–CMRN can be used as a substrate in the azide/alkyne click reaction and that the reaction proceeds in high yield (>96%).
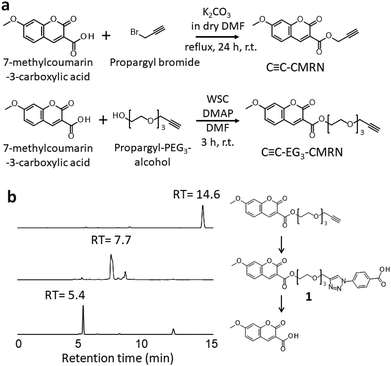 | ||
Fig. 2 (a) Synthesis of the clickable and cleavable fluorescent compounds C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) C–CMRN and C C–CMRN and C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) C–EG3–CMRN. (b) HPLC profiles of C C–EG3–CMRN. (b) HPLC profiles of C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) C–EG3–CMRN after the click reaction and after subsequent hydrolysis. C–EG3–CMRN after the click reaction and after subsequent hydrolysis. | ||
Next, we investigated hydrolysis of the ester bond of C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) C–EG3–CMRN. NaOH aqueous solution was added to the solution containing compound 1 to give a final NaOH concentration of 0.1 M, and the mixture was then stirred at 37 °C for 3 h. HPLC measurements show that the peak of compound 1 (at RT = 7.7 min) completely disappears and a new peak of 7-methylcoumarin-3-carboxylic acid appears at RT = 5.4 min (Fig. 2b). These results demonstrate that compound 1, which is the product of the C
C–EG3–CMRN. NaOH aqueous solution was added to the solution containing compound 1 to give a final NaOH concentration of 0.1 M, and the mixture was then stirred at 37 °C for 3 h. HPLC measurements show that the peak of compound 1 (at RT = 7.7 min) completely disappears and a new peak of 7-methylcoumarin-3-carboxylic acid appears at RT = 5.4 min (Fig. 2b). These results demonstrate that compound 1, which is the product of the C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) C–EG3–CMRN/azidobenzoic acid click reaction, can be hydrolyzed by aqueous NaOH to liberate its fluorophore.
C–EG3–CMRN/azidobenzoic acid click reaction, can be hydrolyzed by aqueous NaOH to liberate its fluorophore.
We then attempted to quantify the azide groups on resin particles using C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) C–EG3–CMRN. In a previous study, we reported that the resin particles have 65 nmol mg−1 amino groups.24 Before quantification, we produced azide groups on the resin particles by the condensation reaction between azidobenzoic acid and the amino groups on the resin particles (azidation, Fig. 3a). The azide groups on the resin particle surfaces were quantified using C
C–EG3–CMRN. In a previous study, we reported that the resin particles have 65 nmol mg−1 amino groups.24 Before quantification, we produced azide groups on the resin particles by the condensation reaction between azidobenzoic acid and the amino groups on the resin particles (azidation, Fig. 3a). The azide groups on the resin particle surfaces were quantified using C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) C–EG3–CMRN. Azidated resin particles (0, 0.19, 0.38, or 0.75 mg) were dispersed in water (1.8 mL) containing CuSO4 (1 μmol) and sodium ascorbate (4 μmol). The dispersion was mixed with dimethyl sulfoxide (DMSO, 0.1 mL) containing 1 mM C
C–EG3–CMRN. Azidated resin particles (0, 0.19, 0.38, or 0.75 mg) were dispersed in water (1.8 mL) containing CuSO4 (1 μmol) and sodium ascorbate (4 μmol). The dispersion was mixed with dimethyl sulfoxide (DMSO, 0.1 mL) containing 1 mM C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) C–EG3–CMRN and shaken at room temperature for 24 h. After washing the resin particles, the resin particles were dispersed in NaOH aqueous solution (final concentration of 0.1 M) to liberate the CMRN fluorophore from the resin particles. After centrifugation and neutralization, the fluorescence intensity of the solution was measured with a conventional fluorescence spectrophotometer. There is high linearity between the amount of particles and the fluorescence intensity of the liberated fluorophore (Fig. 2b). The quantified value of the azide groups on the resin particles is 55 nmol mg−1, which is comparable with the amount of amino groups on the resin particles. The slightly smaller value for the azide groups is because of the yield of azidation of the amino groups on the resin particles. These results suggest successful quantification of the azide groups on the resin particles using the proposed method. The standard curve (Fig. S4†) shows that the limit of detection of the present method was 5 nmol ml−1, which means that azide groups of 5 pmol on a surface was detected when using 1 mL aqueous solution of liberated fluorophore.
C–EG3–CMRN and shaken at room temperature for 24 h. After washing the resin particles, the resin particles were dispersed in NaOH aqueous solution (final concentration of 0.1 M) to liberate the CMRN fluorophore from the resin particles. After centrifugation and neutralization, the fluorescence intensity of the solution was measured with a conventional fluorescence spectrophotometer. There is high linearity between the amount of particles and the fluorescence intensity of the liberated fluorophore (Fig. 2b). The quantified value of the azide groups on the resin particles is 55 nmol mg−1, which is comparable with the amount of amino groups on the resin particles. The slightly smaller value for the azide groups is because of the yield of azidation of the amino groups on the resin particles. These results suggest successful quantification of the azide groups on the resin particles using the proposed method. The standard curve (Fig. S4†) shows that the limit of detection of the present method was 5 nmol ml−1, which means that azide groups of 5 pmol on a surface was detected when using 1 mL aqueous solution of liberated fluorophore.
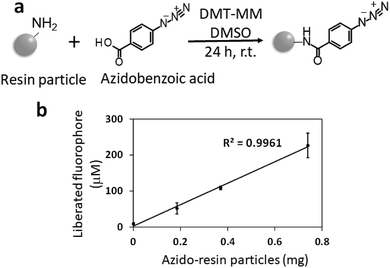 | ||
Fig. 3 (a) Schematic of azidation of the amino groups on the resin particles. (b) Quantification of the azide groups on the resin particles using C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) C–EG3–CMRN. C–EG3–CMRN. | ||
While the above investigations employed Cu(I) as a catalyst for 1,3-dipolar cycloaddition of azide and alkyne, a Cu-free click reaction (strain-promoted azide–alkyne cycloaddition) using a cycloalkyne has attracted a broad range of attention because of no use of a catalyst.25 We then attempted to quantify the azide groups on resin surfaces using a Cu-free click reaction (Fig. S3†). Although a dibenzocyclooctyne (DBCO) derivative is a useful functional molecule for a Cu-free click reaction, its water solubility is very low. We conjugated DBCO-s-s-amine with azide groups on resin surfaces and then the resultant amino groups were conjugated with rhodamine B isothiocyanate (RITC). The disulfide bond was cleaved by a reducing agent (TCEP) to liberate the fluorophore, which was measured using a fluorometer. Although the liberated fluorophore increased linearly with the amount of azidated resin particles (Fig. S5†), we detected only 4.2 nmol azide groups per 1 mg resin. The remarkably low amount of azide groups quantified might be explained by the following possible reasons. (i) The sterically bulky structure of DBCO-s-s-amine affected its conjugation with an azide group on a surface, leading to a low conjugation ratio. (ii) A mixture of THF/water, which was used to dissolve DBCO-s-s-amine for a Cu-free click reaction, altered the surface composition of resin, resulting in the inhibition of the azide group quantification. These results suggest that a relatively small fluorescent compound, which has water solubility and avoids steric hindrance, is appropriate for the quantification of azide groups on a surface. The following experiments were carried out using C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) C–EG3–CMRN.
C–EG3–CMRN.
In the last decade, clickable moieties have often been introduced to biomolecules (e.g., proteins and nucleic acids) to create functionally engineered biomolecules and immobilize them on various surfaces.26–30 Therefore, we then investigated quantification of azide groups introduced to a protein. β-Casein was chosen as a model protein and azide groups were introduced to the amino groups of β-casein by N-hydroxysuccinimide ester chemistry (Fig. 4a). β-Casein has 12 amino groups.31 The TNBS method32,33 shows that the azidation procedure conjugates 86% of the amino groups with azidobenzoic acid. Azidated β-casein (50 nmol) in an aqueous buffer (pH 7.0, 1.2 mL) containing CuSO4 (2.4 μmol) and sodium ascorbate (12 μmol) was mixed with DMSO (0.4 mL) containing C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) C–EG3–CMRN (5 μmol) and then shaken at 37 °C for 3 h. After dialysis to remove excess C
C–EG3–CMRN (5 μmol) and then shaken at 37 °C for 3 h. After dialysis to remove excess C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) C–EG3–CMRN and the Cu catalyst, NaOH aqueous solution was added to the solution to liberate the CMRN moiety from azidated β-casein.
C–EG3–CMRN and the Cu catalyst, NaOH aqueous solution was added to the solution to liberate the CMRN moiety from azidated β-casein.
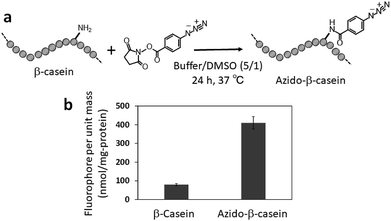 | ||
| Fig. 4 (a) Schematic of azidation of the amino groups on β-casein. (b) Quantification of the azide groups on β-casein. | ||
Fluorescence measurement of the solution shows 410 nmol CMRN moieties per milligram of casein (Fig. 4b), meaning that 81% of the amino groups of β-casein are azidated. The obtained azidation ratio is comparable with the result of the TNBS method.
Conclusions
In conclusion, we have proposed a novel approach to quantify the azide groups on a material surface and on a protein using a clickable and cleavable fluorescent compound (C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) C–EG3–CMRN). This is the first time that quantification of the azide groups on a solid material and a biomolecule has been achieved. Quantitative analysis of the clickable moieties on a surface will play an important role in surface chemistry and extend the applicability of the azide/alkyne cycloaddition click reaction on surfaces and at interfaces.
C–EG3–CMRN). This is the first time that quantification of the azide groups on a solid material and a biomolecule has been achieved. Quantitative analysis of the clickable moieties on a surface will play an important role in surface chemistry and extend the applicability of the azide/alkyne cycloaddition click reaction on surfaces and at interfaces.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
We thank Prof. A. Mori for his technical help. This work was supported by Special Coordination Funds for Promoting Science and Technology, Creation of Innovation Centers for Advanced Interdisciplinary Research Areas (Innovative Bioproduction Kobe), MEXT, Japan and also by JSPS KAKENHI (16H04577, 18K18976 and 18H04556).Notes and references
- E. M. Sletten and C. R. Bertozzi, Acc. Chem. Res., 2011, 44, 666–676 CrossRef CAS PubMed.
- C. P. Ramil and Q. Lin, Chem. Commun., 2013, 49, 11007–11022 RSC.
- N. K. Devaraj, ACS Cent. Sci., 2018, 4, 952–959 CrossRef CAS PubMed.
- R. Huissgen, Angew. Chem., Int. Ed., 1963, 2, 565–632 CrossRef.
- V. V. Rostovtsev, L. G. Green, V. V. Fokin and K. B. Sharpless, Angew. Chem., Int. Ed., 2002, 41, 2596–2599 CrossRef CAS PubMed.
- C. W. Tornoe, C. Christensen and M. Meldal, J. Org. Chem., 2002, 67, 3057–3064 CrossRef CAS PubMed.
- B. P. Duckworth, J. H. Xu, T. A. Taton, A. Guo and M. D. Distefano, Bioconjugate Chem., 2006, 17, 967–974 CrossRef CAS PubMed.
- W. H. Binder and R. Sachsenhofer, Macromol. Rapid Commun., 2008, 29, 952–981 CrossRef CAS.
- L. A. Canalle, S. S. van Berkel, L. T. de Haan and J. C. M. van Hest, Adv. Funct. Mater., 2009, 19, 3464–3470 CrossRef CAS.
- G. A. Hudalla and W. L. Murphy, Langmuir, 2009, 25, 5737–5746 CrossRef CAS PubMed.
- X. Deng, C. Friedmann and J. Lahann, Angew. Chem., Int. Ed., 2011, 50, 6522–6526 CrossRef CAS PubMed.
- X. L. Sun, C. L. Stabler, C. S. Cazalis and E. L. Chaikof, Bioconjugate Chem., 2006, 17, 52–57 CrossRef CAS PubMed.
- J. F. Lutz and Z. Zarafshani, Adv. Drug Delivery Rev., 2008, 60, 958–970 CrossRef CAS PubMed.
- K. Patel, S. Angelos, W. R. Dichtel, A. Coskun, Y. W. Yang, J. I. Zink and J. F. Stoddart, J. Am. Chem. Soc., 2008, 130, 2382–2383 CrossRef CAS PubMed.
- Y. Zhou, S. X. Wang, K. Zhang and X. Y. Jiang, Angew. Chem., Int. Ed., 2008, 47, 7454–7456 CrossRef CAS PubMed.
- W. R. Algar, D. E. Prasuhn, M. H. Stewart, T. L. Jennings, J. B. Blanco-Canosa, P. E. Dawson and I. L. Medintz, Bioconjugate Chem., 2011, 22, 825–858 CrossRef CAS PubMed.
- N. J. Agard, J. M. Baskin, J. A. Prescher, A. Lo and C. R. Bertozzi, ACS Chem. Biol., 2006, 1, 644–648 CrossRef CAS PubMed.
- S. T. Laughlin, J. M. Baskin, S. L. Amacher and C. R. Bertozzi, Science, 2008, 320, 664–667 CrossRef CAS PubMed.
- X. H. Ning, J. Guo, M. A. Wolfert and G. J. Boons, Angew. Chem., Int. Ed., 2008, 47, 2253–2255 CrossRef CAS PubMed.
- A. Shimomura, T. Nishino and T. Maruyama, Langmuir, 2013, 29, 932–938 CrossRef CAS PubMed.
- K. Nishimori, S. Kitahata, T. Nishino and T. Maruyama, Langmuir, 2018, 34, 6396–6404 CrossRef CAS PubMed.
- R. F. Roberts, D. L. Allara, C. A. Pryde, D. N. E. Buchanan and N. D. Hobbins, Surf. Interface Anal., 1980, 2, 5–10 CrossRef CAS.
- N. J. Harrick, Internal reflection spectroscopy, Interscience Publishers, New York, 1967 Search PubMed.
- S. Shiota, S. Yamamoto, A. Shimomura, A. Ojida, T. Nishino and T. Maruyama, Langmuir, 2015, 31, 8824–8829 CrossRef CAS PubMed.
- N. J. Agard, J. A. Prescher and C. R. Bertozzi, J. Am. Chem. Soc., 2004, 126, 15046–15047 CrossRef CAS PubMed.
- F. de Moliner, N. Kielland, R. Lavilla and M. Vendrell, Angew. Chem., Int. Ed., 2017, 56, 3758–3769 CrossRef CAS PubMed.
- C. D. Spicer, E. T. Pashuck and M. M. Stevens, Chem. Rev., 2018, 118, 7702–7743 CrossRef CAS PubMed.
- Y. Zou, L. Zhang, L. Yang, F. Zhu, M. M. Ding, F. Lin, Z. Wang and Y. W. Li, J. Controlled Release, 2018, 273, 160–179 CrossRef CAS PubMed.
- A. C. Braun, M. Gutmann, T. Luhmann and L. Meinel, J. Controlled Release, 2018, 273, 68–85 CrossRef CAS PubMed.
- O. Boutureira and G. J. L. Bernardes, Chem. Rev., 2015, 115, 2174–2195 CrossRef CAS PubMed.
- J. M. Girardet, J. L. Courthaudon, S. Campagna, V. Puyjalon, D. Lorient and G. Linden, Int. Dairy J., 1999, 9, 409–410 CrossRef CAS.
- M. L. Kakade and I. E. Liener, Anal. Biochem., 1969, 27, 273–280 CrossRef CAS PubMed.
- P. Cayot and G. Tainturier, Anal. Biochem., 1997, 249, 184–200 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: The experimental procedures, and additional results. See DOI: 10.1039/c8ra09421g |
| This journal is © The Royal Society of Chemistry 2019 |
