Open Access Article
Open Access ArticleComparison of hypoglycemic effects of ripened pu-erh tea and raw pu-erh tea in streptozotocin-induced diabetic rats†
Qianzhi Dinga,
Wei Zhenga,
Bowei Zhangb,
Xiaojuan Chena,
Jie Zhanga,
Xu Panga,
Yong Zhangc,
Dexian Jiac,
Surui Peid,
Yuesheng Dongb and
Baiping Ma *a
*a
aBeijing Institute of Radiation Medicine, No. 27 Taiping Road, Haidian District, Beijing 100850, China. E-mail: mabaiping@sina.com; Tel: +86-010-66930282
bSchool of Life Science and Biotechnology, Dalian University of Technology, Dalian 116024, Liaoning, China
cBeijing University of Chinese Medicine, Beijing 100029, China
dAnnoroad Gene Technology Co., Ltd, Beijing 100176, China
First published on 23rd January 2019
Abstract
Pu-erh tea is produced from the leaves of large-leaf tea species (Camellia sinensis var. assamica) in the Yunnan province of China and divided into ripened pu-erh tea (RIPT, with pile-fermentation) and raw pu-erh tea (RAPT) according to processing methods. RIPT extract showed more potent anti-diabetic effects on two-hour postprandial blood glucose (2h-PBG) and fasting blood glucose (FBG) than RAPT extract. UHPLC-Q-TOF/MS and UHPLC-PDA analyses found that 17 newly formed components and the increased components after fermentation, such as quinic acid, gallic acid, caffeine, puerin I and so on, might be the main contributors to the enhanced activities of RIPT. In addition, the probiotic role of RIPT to some beneficial gut bacteria, such as lactobacillus, Prevotellaceae NK3B31 group, Alloprevotella and Prevotella, was observed in our study. These results might provide a clue to anti-diabetic mechanism and active components of pu-erh tea, and use as a functional beverage worth to be further studied.
1 Introduction
Diabetes mellitus is a complex metabolic disorder, which has become a growing global health problem and caused increasing premature mortality and healthcare costs.1 According to the International Diabetes Federation, there are an estimated 451 million individuals (age from 18 to 99) suffering from diabetes, and the number is predicted to be 693 million by 2045. The estimated healthcare expenditure relating to diabetes reached 850 billion USD for adults in 2017.2 Therefore, looking for anti-diabetic functional foods, which might have low side-effects and low cost, has a significance to global health.Tea is one of the most popular world-wide beverages and has various biological activities.3,4 In recent years, pu-erh tea becomes more and more popular in Southeast Asia due to its multiple health benefits, different tastes, and special flavor.3 Pu-erh tea is produced from the sun-dried leaves of Camellia sinensis var. assamica in Yunnan province, China. Due to the different processes, pu-erh tea can be categorized into raw pu-erh tea (RAPT, directly compressed and shaped into cake) and ripened pu-erh tea (RIPT, pile-fermented by microorganisms). The participation of the pile-fermentation process is the biggest difference between RIPT and RAPT, and brings great distinctions on infusion colors and tastes,5 even the chemical constituents and biological activities in our study. Currently, some studies have reported the anti-diabetic effect of pu-erh tea. Wang et al. found that flavanols from the water extract of pu-erh tea showed an inhibition effect on α-glucosidase, suggesting potential hypoglycemic effects.6 In the study of Deng et al., a single administration of polysaccharides from pu-erh tea decreased the postprandial blood sugar in mice, and the mechanism was due to the inhibition of α-glucosidase.7 Huang et al. reported that the 95% ethanol precipitate and ethyl acetate fractions of pu-erh tea showed remarkable inhibition against α-glycosidase in vitro.8 The above studies are only focused on the beneficial effects of RIPT, and the form of a single administration could not simulate the general drinking habit in daily life.
Therefore, a comparative study between RIPT and RAPT was carried out on pharmacological activity and chemical constituent in our research. The anti-diabetic activities of RIPT and RAPT were evaluated in streptozotocin-induced diabetic Wistar rats. Then, the constituents of the RIPT and RAPT were identified. Besides, as the low bioavailability of active constituents from tea9,10 and the growing evidences on the effect of gut microbiota on diabetes,11,12 the 16S rRNA analysis was carried out to investigate the effects of RIPT on the gut microbiota.
2 Materials and methods
2.1 Source of pu-erh tea samples
Six batches of samples, including three RIPTs and three RAPTs, were collected from the same production place, Yiwu district, Dai Autonomous Prefecture of Xishuangbanna, Yunnan province, China.2.2 Preparations of extracts from RIPT and RAPT
The tea extract was prepared with process similar to the general drinking habit in China. Each sample (1000 g) was extracted twice with 10 L and 7 L distilled water each for one hour at 100 °C, respectively. After filtration, two solutions were combined and concentrated at 55 °C, and then lyophilized.2.3 Animals and treatment
Male Wistar rats (180–220 g) were purchased from Beijing Vital River Laboratory Animal Technology Co., Ltd. (Beijing, China). The rats were provided with a standard rodent diet and free access to water, and were maintained at a temperature of 20–22 °C. The animals and protocols for this study were approved by international ethical guidelines and the Institutional Animal Care and Use Committee of Dalian Medical University, with the permission number of SCXK 2013-0003.The nongenetic diabetic rat model was established following the methods published previously.13,14 Seventy rats were fed with a high-fat, high-sugar diet (HFHSD) (D12451, 45% calories from fat) instead of standard chow, and had free access to water. On Day 14, the rats were injected intraperitoneally with 40 mg kg−1 streptozocin (STZ, Sigma-Aldrich Chemical Co., St. Louis, USA.). On Day 21 and Day 28, oral glucose tolerance tests (OGTTs) were performed. A total of 30 rats met the standard of diabetes, with 2h-PBG levels ranging from 16.7 mmol mL−1 to 24.0 mmol mL−1 in both OGTTs, and were selected as diabetic rats. Another 5 rats were fed a normal diet as the negative control.
In this set of experiments, normoglycemic rats were fed a standard chow diet and were given vehicle alone (0.5% CMC-Na) daily by oral gavage (NM). The diabetic rats fed with a HFHSD were divided into six groups (n = 5 for each group) and were treated with either vehicle (DM), 500 mg kg−1 of metformin in vehicle (Met), 600 mg kg−1 of extract from RIPT (IH), 120 mg kg−1 of extract from RIPT (IL), 800 mg kg−1 of extract from RAPT (AH), and 160 mg kg−1 of extract from RAPT (AL) in vehicle for 6 weeks.
2.4 OGTT assay
After 14 hour fasting, the rats were orally administered with 2 g kg−1 glucose. Blood samples were collected from the tail vein and the levels of FBG and 2h-PBG were measured using the Roche glucometer at week 6.2.5 Biochemical analyses
The OGTT was performed every week. At the end of the study, the rats were sacrificed after 16 h of fasting. The blood samples were collected and centrifuged at 4000 g for 5 min at 4 °C to obtain the plasma. Fasting plasma insulin (FINS) levels were determined using ELISA kits (Abcam, USA). The values of hemoglobin A1c (HbA1c), total cholesterol (TC), and triglyceride (TG) in plasma were measured according to commercially available kits purchased from Jiancheng Institute of Biotechnology, Nanjing, China.2.6 Analysis of chemical constituents
In this study, 10 mg of powders were accurately weighed and suspended in 1 mL water, respectively. The samples were then vortexed 1 min, followed by centrifugation for 10 min at 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm at 4 °C. 2 μL of the ultra-performance liquid chromatography coupled with a hybrid quadrupole time-of-flight mass spectrometry (UHPLC-Q-TOF/MS) analysis.
000 rpm at 4 °C. 2 μL of the ultra-performance liquid chromatography coupled with a hybrid quadrupole time-of-flight mass spectrometry (UHPLC-Q-TOF/MS) analysis.
An Acquity UPLC system (Waters Corp., Milford, MA, USA) coupled with a Synapt MS system (Waters Corp., Milford, MA, USA) was used. The samples were separated on a Waters Acquity UPLC HSS T3 column (100 × 2.1 mm, 1.8 μm). The mobile phase consisted with (A) 0.1% formic acid solution (v/v) and (B) acetonitrile, while the gradient program was as follows: 98% (A) in 0–1 min; 98–96% (A) in 1–2 min; 96–95% (A) in 2–4 min; 95–93% (A) in 4–6 min; 93–92% (A) in 6–9 min; 92–90% (A) in 9–11 min; 90–86% (A) in 11–15 min; 86% (A) in 15–16 min; 86–83% (A) in 16–19 min; 83–76% (A) in 19–22 min; 76–60% (A) in 22–25 min; 60–43% (A) in 25–26 min; 43–42% (A) in 26–27 min; 42–5% (A) in 27–28 min; 5% (A) in 28–28.5 min. The mobile phase flow rate was 0.6 mL min−1 with the column temperature at 45 °C. The wavelength of PDA detector was set from 190 to 400 nm, and 280 nm was set at the monitoring wavelength.
The data acquisition mode was MSE. Each extract was directed to a trap mass spectrometer with an electrospray interface (ESI) operating in full scan MS mode from m/z 50 to 1500 Da. Mass spectra were acquired in both negative and positive modes with the source temperature was 100 °C, the desolvation temperature was 450 °C, and desolvation gas flow of 850 L h−1. The capillary voltage was 3 kV. At low CE scan, the cone voltage was 30 V, and the collision energy was 6 eV (trap) and 4 eV (transfer). At high CE scan, the cone voltage was 30 V, and the collision energy was 50–65 eV (trap) and 15 eV (transfer). Leucine-enkephalin was used as lock mass.
Standards, including gallic acid (>98%), catechin (>98%), catechingallate (>98%), epicatechin (>98%), epicatechingallate (>98%), epigallocatechin (>98%), epigallocatechin gallate (>98%), gallocatechingallate (>98%), (+)-gallocatechin (>98%), theophylline (>98%), procyanidin B1 (>97%), were purchased from Chengdu Biopurify Phytochemicals Ltd. Caffeine (>98%) was isolated previously. Its structure and purity were confirmed by NMR spectra and high-performance liquid chromatography coupled with evaporative light scattering detection (HPLC-ELSD).
Acetonitrile (HPLC grade) was purchased from Fisher Scientific Co. (Loughborough, UK). Distilled water was purchased from Watsons. Formic acid (HPLC grade) was purchased from Acros Co. Ltd. (St. Louis, MO, USA). Other reagents were obtained commercially in analytical purity (Beijing, China).
2.7 Feces collection, bacterial DNA extraction, PCR amplification and sequencing
At the end of the experiment, feces of rats from NM, DM, Met and IH were collected into sterilized plastic tubes and stored at −80 °C until tested.Bacterial DNA was extracted from the fecal contents of the rat. The purity and concentration of extracted bacterial DNA were tested by NanoPhotometer spectrophotometer and Qubit 2.0 Flurometer, respectively.
The V3–V4 hypervariable regions of the bacteria 16S rRNA gene were amplified with primers 341F (5′-CCTACGGGNGGCWGCAG-3′) and 805R (5′-GACTACHVGGGTATCTAATCC-3′) by thermocycler PCR system (GeneAmp 9700, ABI, USA). All PCR reactions were carried out in 30 μL reactions with 15 μL of Phusion High-Fidelity PCR Master Mix (New England Biolabs); 0.2 μmol mL−1 of forward and reverse primers, and about 10 ng template DNA. Thermal cycling consisted of initial denaturation at 95 °C for 3 min, followed by 25 cycles of denaturation at 95 °C for 30 s, annealing at 55 °C for 30 s, and elongation at 72 °C for 30 s. Then final extension at 72 °C for 10 min. Mix same volume of 1× loading buffer (contained SYB green) with PCR products and operate electrophoresis on 2% agarose gel for detection. Samples with bright main strip around 460 bp (V3 + V4) were chosen for further experiments. PCR products were mixed in equidensity ratios. Then, mixture PCR products were purified with GeneJET Gel Extraction Kit (Thermo Scientific). Sequencing libraries were generated using NEB Next Ultra DNA Library Prep Kit for Illumina (NEB, USA) following manufacturer's recommendations and index codes were added. The library quality was assessed on the Qubit@ 2.0 Fluorometer (Life Technologies, CA, USA) and Agilent Bioanalyzer 2100 system. At last, the library was sequenced on an Illumina MiSeq platform and 250 bp paired-end reads were generated.
2.8 Bioinformatics analysis
Raw reads were filtered to remove the adapter-polluted reads, low quality reads (average quality lower than 19 with PHRED algorithm) and the reads with N bases exceeding 5%. Then, the clean paired reads were spliced with the PEAR software15 into merged sequences based on sequence overlap. After merging, the sequence Chimeras were removed, and the sequences were clustered into operational taxonomic units (OTUs) by UCLUST16 with a threshold of 97% pairwise identity. Then, the Silva database (release 128, http://www.arb-silva.de) was used to annotate the taxonomic information. After OTUs annotation, the abundance information was normalized with a standard sequence number according to the sample which had the least sequences. Then, the normalized output data were applied to the subsequent analyses, such as alpha and beta diversity. The community richness and diversity estimations, such as Chao1, ACE and the Shannon index, were calculated by QIIME version 1.8.0 (ref. 17) and displayed utilizing R software. In the beta diversity analysis, the cluster analysis was utilized with principal coordinate analysis (PCoA) based on matrix of Taxonomic OTU, by using R software with ggplot2 and ade4 package. In the PCoA analysis, the distance matrices of weighted or unweighted UniFrac18 among samples were also needed.3 Statistical analysis
The data in the negative mode of principal component analysis (PCA) and orthogonal partial least squares discriminant analysis (OPLS-DA) were processed by the MarkerLynx V4.1 software (Waters Co., Milford, USA). The method parameters were set as follows: mass range 100–1500 Da, retention time range 0.2–25.0 min, mass tolerance 5.0 ppm, peak width at 5% height was 1.00 s, peak-to-peak baseline noise 0.00, noise elimination level was set at 6.00 and retention time tolerance was set at 0.01 min. The results were visualized in a score plot to show group clusters, and a variable importance in projection (VIP)-plot to show variables contributing to the classification.The statistical analysis was performed using SPSS 17.0. Comparisons between groups were analyzed using one-way ANOVA followed by Fisher LSD multiple comparison. p < 0.05 was considered statistically significant.
4 Results
4.1 Effects of RAPT and RIPT extracts on the diabetes indices and lipid profiles
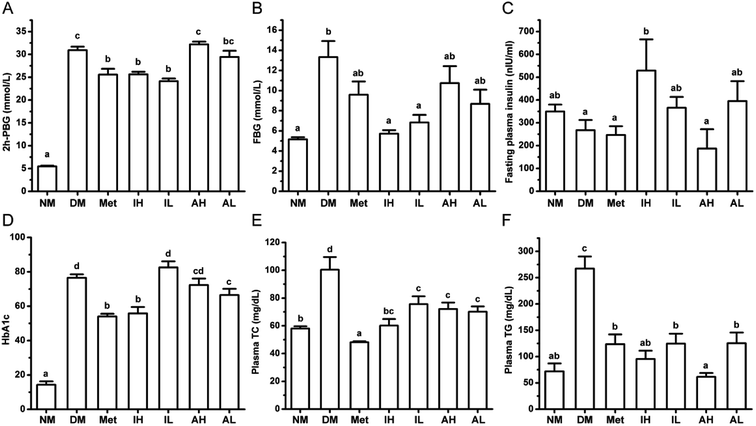
After a single injection of STZ and HFHSD induction, the levels of 2h-PBG and FBG were significantly higher in the DM group, compared with those of the NM group (Fig. 1A and B). Metformin, a positive control, significantly lowered the levels of 2h-PBG in diabetic rats. The 2h-PBG lowering effects in the Met, IH and IL groups were significantly observed compared with the DM group, while no significant 2h-PBG-lowering effect was observed in the AH and AL group. Compared with the DM group, the FBG level of the Met group was decreased but not significant. The RIPT administration significantly decreased the FBG level with a dose-dependent pattern in diabetic rat. Meanwhile no significantly differences were observed on the FBG levels of the AH and AL groups compared with the DM group.The level of FINS was significantly increased in the IL group compared with that of the DM group, but showed no significance when compared with that of the NM group (Fig. 1C). Moreover, the high level of HbA1c in diabetic rats was improved by RIPT administration which had similar effect to metformin (Fig. 1D).
The levels of plasma TC (Fig. 1E) and TG (Fig. 1F) were significantly higher in the DM group, compared to those of the NM group. After 6 week treatment, the metformin, RIPT and RAPT significantly decreased the plasma TC and TG in diabetic rats, respectively. The effect on plasma TG showed a dose-dependent pattern.
4.2 Chemical profiles and different markers of the extracts from RAPT and RIPT
The extract of RAPT and RIPT were analyzed in both negative and positive ion modes with the same LC conditions. Based on exact mass, fragment ions, retention times, published literatures, and comparison with the standard references, 66 major peaks in both negative and positive ion modes of the pu-erh tea were identified, as summarized in Table 1, including 45 components in the RIPT, and 49 chemical components in the RAPT. As shown in Table 1, 17 peaks were detected in RIPT only, including a series of puerins, such as peak 13, 29, 33, 34, 43, 46, 47, and 50. Meanwhile, 21 peaks were detected in RAPT only. The rest of 28 peaks were detected in both RIPT and RAPT, the base peak ion (BPI) chromatograms of the RAPT and RIPT by UHPLC-Q-TOF/MS are shown in Fig. 2.| No. | Rt (min) | Formula | [M − H]− experimental | [M − H]− theoretical | Error (mD) | Identification | Sourceb | VIP value | |
|---|---|---|---|---|---|---|---|---|---|
| RAPT | RIPT | ||||||||
| a Detected in the positive mode.b “+” means the compound was detected in that source, while “−” means not. | |||||||||
| 1 | 0.48 | C7H12O6 | 191.0576 | 191.0556 | 2.0 | Quinic acid | + | + | 2.52395 |
| 2 | 0.91 | C14H16O10 | 343.0668 | 343.0665 | 0.3 | Theogallin or its isomers | + | + | NA |
| 3 | 0.95 | C13H16O10 | 331.0689 | 331.0665 | 2.4 | Gallic acid-4-O-glucoside or its isomers | + | − | 2.703 |
| 4 | 1.21 | C13H16O10 | 331.0667 | 331.0665 | 0.2 | Gallic acid-4-O-glucoside or its isomers | + | + | 4.92352 |
| 5 | 1.36 | C7H6O5 | 169.0154 | 169.0137 | 1.7 | Gallic acid | + | + | 2.69024 |
| 6 | 1.47 | C13H16O10 | 331.0637 | 331.0665 | −2.8 | Gallic acid-4-O-glucoside or its isomers | + | − | NA |
| 7 | 1.77 | C14H16O10 | 343.0679 | 343.0665 | 1.4 | Theogallin | + | + | 4.2577 |
| 8 | 2.33 | C14H16O10 | 343.0670 | 343.0665 | 0.5 | Theogallin or its isomers | + | + | 3.26224 |
| 9 | 2.95 | C15H14O7 | 305.0668 | 305.0661 | 0.7 | Gallocatechin | + | + | 3.911 |
| 10 | 3.18 | C7H8N4O2 | 181.0706 | 181.0726 | −2.0 | Theophylline or its isomersa | + | + | NA |
| 11 | 4.02 | C16H18O9 | 353.0873 | 353.0873 | 0.0 | Chlorogenic acid | + | + | 2.61641 |
| 12 | 4.28 | C7H8N4O2 | 179.0559 | 179.0569 | −1.0 | Theophylline or its isomers | + | + | NA |
| 13 | 4.54 | C22H24O7 | 399.1442 | 399.1444 | −0.3 | Puerin A or its isomers | − | + | NA |
| 14 | 5.12 | C16H14O9 | 349.0559 | 349.0560 | −0.1 | 6-Carboxyl-(−)-gallocatechin or its isomers | − | + | NA |
| 15 | 5.8 | C15H14O7 | 305.0660 | 305.0661 | −0.1 | Epigallocatechin | + | + | 4.30649 |
| 16 | 6.1 | C15H14O6 | 289.0713 | 289.0712 | 0.1 | Catechin | + | + | 4.13423 |
| 17 | 6.24 | C37H30O18 | 761.1342 | 761.1354 | −1.2 | Theasinensin B | + | − | NA |
| 18 | 6.48 | C16H18O9 | 353.0862 | 353.0873 | −1.1 | 3-O-Caffeoylquinic acid or its isomers | + | + | NA |
| 19 | 6.76 | C20H20O14 | 483.0780 | 483.0775 | 0.5 | 1,6-Di-O-galloyl-β-D-glucopyranose or its isomers | + | − | NA |
| 20 | 6.88 | C8H10N4O2 | 195.0895 | 195.0882 | 1.3 | Caffeinea | + | + | NA |
| 21 | 7.01 | C20H20O14 | 483.0781 | 483.0775 | 0.6 | 1,6-Di-O-galloyl-β-D-glucopyranose or its isomers | + | − | 2.84282 |
| 22 | 7.22 | C16H18O9 | 353.0859 | 353.0873 | −1.4 | 3-O-Caffeoylquinic acid or its isomers | + | + | NA |
| 23 | 7.56 | C30H26O12 | 577.1343 | 577.1346 | −0.3 | Procyanidin B1 | + | − | 4.41889 |
| 24 | 8.04 | C30H26O12 | 577.1352 | 577.1346 | 0.6 | The isomer of procyanidin B1 | + | − | NA |
| 25 | 8.21 | C37H30O17 | 745.1422 | 745.1405 | 1.7 | Epicatechin-(4β-8)-epigallocatechin-3-O-gallate or its isomers | + | − | NA |
| 26 | 8.42 | C30H26O12 | 577.1387 | 577.1346 | 4.1 | Procyanidin B2 | + | − | NA |
| 27 | 8.51 | C16H14O9 | 349.0556 | 349.0560 | −0.4 | 6-Carboxyl-(−)-gallocatechin or its isomers | − | + | NA |
| 28 | 9.04 | C37H30O17 | 745.1418 | 745.1405 | 1.3 | Epicatechin-(4β-8)-epigallocatechin-3-O-gallate or its isomers | + | + | NA |
| 29 | 9.3 | C21H23NO8 | 416.1342 | 416.1345 | −0.3 | Puerin VII | − | + | NA |
| 30 | 9.3 | C16H14O8 | 333.0603 | 333.0610 | −0.7 | 8-Carboxyl-(+)-catechin or its isomers | — | + | 2.89648 |
| 31 | 9.49 | C15H14O6 | 289.0690 | 289.0712 | −2.2 | Epicatechin | + | + | 4.72175 |
| 32 | 9.86 | C22H18O11 | 457.0789 | 457.0771 | 1.8 | Epigallocatechin gallate | + | + | 3.74396 |
| 33 | 10.76 | C21H23NO8 | 416.1320 | 416.1345 | −2.5 | Puerin V | − | + | NA |
| 34 | 11.62 | C21H23NO8 | 416.1364 | 416.1345 | 1.9 | Puerin VIII | − | + | NA |
| 35 | 11.64 | C37H30O16 | 729.1465 | 729.1456 | 0.9 | Epicatechin-(4β-8)-epicatechin 3-gallate | + | − | NA |
| 36 | 11.71 | C22H18O11 | 457.0784 | 457.0771 | 1.3 | Gallocatechin gallate | + | − | 4.03217 |
| 37 | 12.14 | C27H24O18 | 635.0878 | 635.0884 | −0.6 | 1,4,6-Tri-O-galloyl-β-D-glucopyranose or its isomers | + | − | NA |
| 38 | 12.22 | C29H48O | 411.3631 | 411.3627 | 0.4 | α-Spinasterol or its isomers | − | + | NA |
| 39 | 12.31 | C27H30O15 | 593.1515 | 593.1506 | 0.9 | Kaempferol-3-O-rutinoside or its isomers | − | + | NA |
| 40 | 12.42 | C37H30O16 | 729.1478 | 729.1456 | 2.2 | Epicatechin-(4α-8)-epicatechin-3′-gallate | + | − | NA |
| 41 | 12.5 | C14H11NO6 | 288.0509 | 288.0508 | 0.1 | N-(3,4-Dihydroxybenzoyl)-3,4-dyhydrobenzamide | − | + | NA |
| 42 | 12.76 | C16H14O8 | 333.0601 | 333.0610 | −0.9 | 8-Carboxyl-(+)-catechin or its isomers | − | + | 2.56147 |
| 43 | 12.98 | C21H23NO8 | 416.1363 | 400.1396 | 1.8 | Puerin VI | − | + | NA |
| 44 | 13.19 | C21H20O13 | 479.0820 | 479.0826 | −0.6 | Myricetin-3-O-β-D-galactopyranoside | + | − | NA |
| 45 | 13.49 | C27H24O18 | 635.0880 | 635.0884 | −0.4 | 1,4,6-Tri-O-galloyl-β-D-glucopyranose or its isomers | + | — | 3.39172 |
| 46 | 13.59 | C21H22NO7 | 400.1398 | 400.1396 | 0.2 | Puerin I | − | + | NA |
| 47 | 14.07 | C21H22NO7 | 400.1403 | 400.1396 | 0.7 | Puerin II | − | + | 2.57155 |
| 48 | 14.28 | C26H28O14 | 563.1409 | 563.1401 | 0.8 | Apigenin-6-C-α-L-arabinopyranosyl-8-C-β-D-glucopyranoside | + | + | 2.80337 |
| 49 | 14.56 | C22H18O10 | 441.0818 | 441.0822 | −0.4 | Epicatechin gallate | + | + | 4.5466 |
| 50 | 14.84 | C21H22NO7 | 400.1391 | 400.1396 | −0.5 | Puerin III | − | + | 3.78329 |
| 51 | 15.13 | C14H6O8 | 300.9951 | 300.9984 | −3.3 | Ellagic acid | + | + | 4.0874 |
| 52 | 15.36 | C27H30O16 | 609.1477 | 609.1456 | 2.1 | The isomer of rutin | + | + | NA |
| 53 | 15.47 | C22H18O10 | 441.0835 | 441.0822 | 1.3 | Catechin gallate | + | − | 4.36602 |
| 54 | 15.87 | C27H30O16 | 609.1472 | 609.1456 | 1.6 | Rutin | + | − | NA |
| 55 | 15.92 | C21H22NO7 | 400.1387 | 400.1396 | −0.9 | Puerin IV | − | + | 4.23005 |
| 56 | 16.04 | C21H20O12 | 463.0873 | 463.0877 | −0.4 | Quercetin-3-O-glucoside | + | + | NA |
| 57 | 16.5 | C33H40O20 | 755.2036 | 755.2035 | 0.1 | Quercetin-4′-O-α-L-rhamnopyranosyl-3-O-α-L-rhamnopyranosyl-(1→6)-β-D-glucopyranoside or its isomers | + | − | NA |
| 58 | 17.05 | C27H30O15 | 593.1529 | 593.1506 | 2.3 | Kaempferol-3-O-rutinoside or its isomers | + | + | NA |
| 59 | 17.42 | C33H40O20 | 755.2067 | 755.2035 | 3.2 | Quercetin-4′-O-α-L-rhamnopyranosyl-3-O-α-L-rhamnopyranosyl-(1→6)-β-D-glucopyranoside or its isomers | + | − | NA |
| 60 | 17.44 | C21H20O11 | 447.0927 | 447.0927 | 0.0 | Luteolin-7-O-glucoside | + | − | NA |
| 61 | 17.76 | C22H18O9 | 425.0832 | 425.0873 | −4.1 | Epiafzelechin-3-O-gallate | + | − | NA |
| 62 | 18.26 | C33H40O19 | 739.2134 | 739.2086 | 4.8 | Kaempferol-3-O-[α-L-rhamnopyranosyl-(1→3)-α-L-rhamnopyranosyl-(1→6)-β-D-glucopyranoside] | − | + | NA |
| 63 | 18.28 | C27H30O15 | 593.1534 | 593.1506 | 2.8 | Kaempferol-3-O-rutinoside or its isomers | + | + | NA |
| 64 | 18.49 | C21H20O11 | 447.0919 | 447.0927 | −0.8 | Kaempferol-3-O-glucoside | + | + | 2.78936 |
| 65 | 22.11 | C15H10O7 | 301.0346 | 301.0348 | −0.2 | Quercetin | + | + | NA |
| 66 | 23.84 | C15H10O6 | 285.0419 | 285.0399 | 2.0 | Kaempferol | + | + | NA |
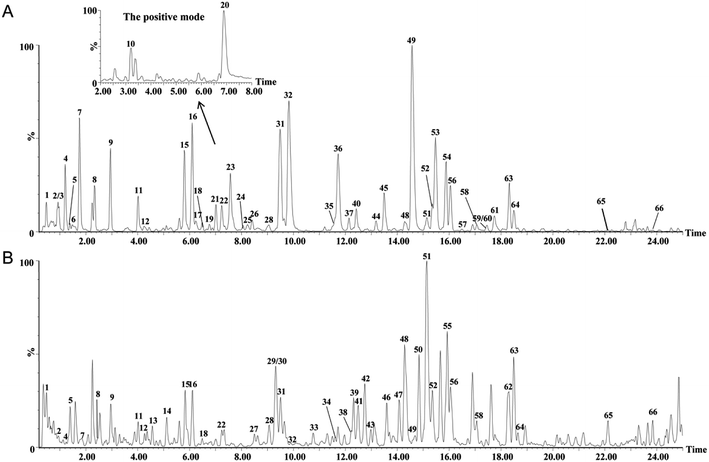 | ||
| Fig. 2 The BPI chromatograms of the extracts from the negative mode in RAPT (A) and RIPT (B) analyzed by UHPLC-Q-TOF/MS. | ||
As the great difference between the chromatograms of the RAPT and RIPT, the multivariate statistical methods, including PCA and OPLS-DA, were carried out to find the different components during the pile-fermentation process, additionally. As shown in Fig. 3A, samples were segregated into two groups from the score plot. And the components with the VIP value greater than 2.5 were considered as potential different markers (Fig. 3B), as a result, 26 major different components were identified during the pile-fermentation process (Table 1), including some puerins, such as puerin II (peak 47), III (peak 50), and IV (peak 55).
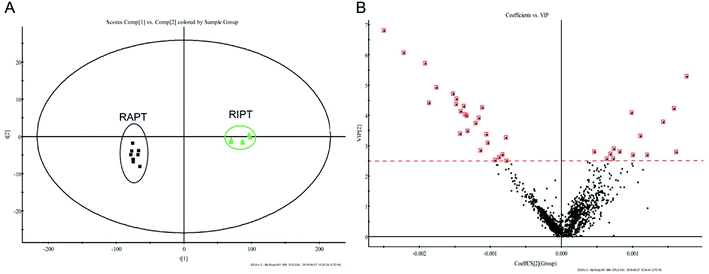 | ||
| Fig. 3 PCA (A), VIP (B) plot of the extracts from the negative mode in RIPT and RAPT analyzed by UHPLC-Q-TOF/MS. | ||
UHPLC-PDA was used to explain the changes of relative contents caused by pile-fermentation process. A database based on identified components and PDA chemometric data were built. Although quercetin and kaempferol were also mutual components in two kinds of pu-erh tea, the relative content of these two components was not compared as their low content in pu-erh tea and the low response value at 280 nm. Therefore, this database consisted of 26 identified components, and the result was shown in Table 2. As the result, the contents of shared peaks, such as epigallocatechin, catechin, epicatechin and epigallocatechin gallate, were decreased after fermentation except for quinic acid, gallic acid, theophylline or its isomers, caffeine, apigenin-6-C-α-L-arabinopyranosyl-8-C-β-D-glucopyranoside, and ellagic acid. At the same time, puerin I-VIII, a kind of characteristic components, were produced during the fermentation. The corresponding reactions were complex.
| Peak no. | Rt (min) | Peak area | Changes in RIPTb | p-value | |
|---|---|---|---|---|---|
| RIPT | RAPT | ||||
| a All of the results were expressed as the mean ± SD (n = 3). p value was calculated by two-tailed t test.b Five key increased components were emphasized with bold arrows. | |||||
| 1 | 0.48 | 5413.78 ± 644.21 | 819.11 ± 162.87 |  |
5.4 × 10−13 |
| 2 | 0.91 | 743.89 ± 402.95 | 1627.22 ± 680.07 | ↓ | 4.0 × 10−3 |
| 4 | 1.21 | 540.11 ± 223.95 | 5509.22 ± 1136.93 | ↓ | 7.5 × 10−10 |
| 5 | 1.36 | 27![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 794.89 ± 14 794.89 ± 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100.32 100.32 |
20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 960.56 ± 4531.10 960.56 ± 4531.10 |
 |
1.9 × 10−1 |
| 7 | 1.77 | 1665.11 ± 266.70 | 31![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 399.89 ± 1296.43 399.89 ± 1296.43 |
↓ | 4.5 × 10−21 |
| 8 | 2.33 | 129.56 ± 113.96 | 3462.00 ± 777.84 | ↓ | 8.8 × 10−10 |
| 9 | 2.95 | 98.11 ± 8.16 | 1196.22 ± 127.16 | ↓ | 1.8 × 10−14 |
| 10 | 3.18 | 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 456.22 ± 248.83 456.22 ± 248.83 |
5320.44 ± 1293.96 |  |
3.0 × 10−9 |
| 11 | 4.02 | 193.78 ± 79.17 | 2829.00 ± 259.68 | ↓ | 2.7 × 10−15 |
| 12 | 4.28 | 600.33 ± 155.54 | 171.89 ± 45.19 | ↑ | 6.2 × 10−7 |
| 15 | 5.8 | 88.44 ± 2.96 | 1756.33 ± 492.03 | ↓ | 2.2 × 10−8 |
| 16 | 6.1 | 299.33 ± 65.45 | 5488.33 ± 560.78 | ↓ | 6.5 × 10−15 |
| 18 | 6.48 | 128.67 ± 41.45 | 1274.00 ± 200.88 | ↓ | 1.4 × 10−11 |
| 20 | 6.88 | 149![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 441.78 ± 3758.53 441.78 ± 3758.53 |
98![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 945.78 ± 8242.66 945.78 ± 8242.66 |
 |
1.5 × 10−11 |
| 22 | 7.22 | 191.89 ± 18.11 | 2735.89 ± 203.26 | ↓ | 5.3 × 10−17 |
| 28 | 9.04 | 486.44 ± 103.20 | 1557.89 ± 96.01 | ↓ | 1.3 × 10−13 |
| 31 | 9.49 | 214.78 ± 111.37 | 9497.00 ± 1907.87 | ↓ | 1.2 × 10−10 |
| 32 | 9.86 | 68.11 ± 50.40 | 39![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 900.56 ± 9011.16 900.56 ± 9011.16 |
↓ | 4.8 × 10−10 |
| 48 | 14.28 | 277.67 ± 42.04 | 186.44 ± 53.65 |  |
1.0 × 10−3 |
| 49 | 14.56 | 188.89 ± 100.50 | 49![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 858.67 ± 7605.17 858.67 ± 7605.17 |
↓ | 1.3 × 10−12 |
| 51 | 15.13 | 3704.67 ± 817.51 | 2069.56 ± 156.84 |  |
2.3 × 10−5 |
| 52 | 15.36 | 281.78 ± 56.62 | 351.33 ± 58.48 | ↓ | 2.1 × 10−2 |
| 56 | 16.04 | 387.00 ± 175.11 | 1230.89 ± 127.26 | ↓ | 3.0 × 10−9 |
| 58 | 17.05 | 173.78 ± 77.95 | 248.78 ± 58.63 | ↓ | 3.5 × 10−2 |
| 63 | 18.28 | 395.44 ± 217.17 | 1378.11 ± 704.96 | ↓ | 1.0 × 10−3 |
| 64 | 18.49 | 375.56 ± 171.71 | 761.44 ± 353.08 | ↓ | 9.4 × 10−3 |
4.3 The overall structural changes of the gut microbiota regulated by RIPT intervention
After 6 week administration, as the outstanding pharmaceutical effect of IH, feces of rats in the IH group togethering with those from the NM, DM and Met groups were collected for the gut microbiota analysis. A total of 957![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 056 high-quality sequences from V3–V4 hypervariable region of the bacterial 16S rRNA gene were collected from 20 samples. The number of average sequence was 47
056 high-quality sequences from V3–V4 hypervariable region of the bacterial 16S rRNA gene were collected from 20 samples. The number of average sequence was 47![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 852, while the maximum number and minimum number were 53
852, while the maximum number and minimum number were 53![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 882 and 39
882 and 39![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 395, respectively. For the length, the average number was 450 bp. With a threshold of 97% pairwise identity, all the sequences were clustered into 667 OTUs. Then, 144 genera or the next higher taxonomic ranks were identified from Silva Database.
395, respectively. For the length, the average number was 450 bp. With a threshold of 97% pairwise identity, all the sequences were clustered into 667 OTUs. Then, 144 genera or the next higher taxonomic ranks were identified from Silva Database.
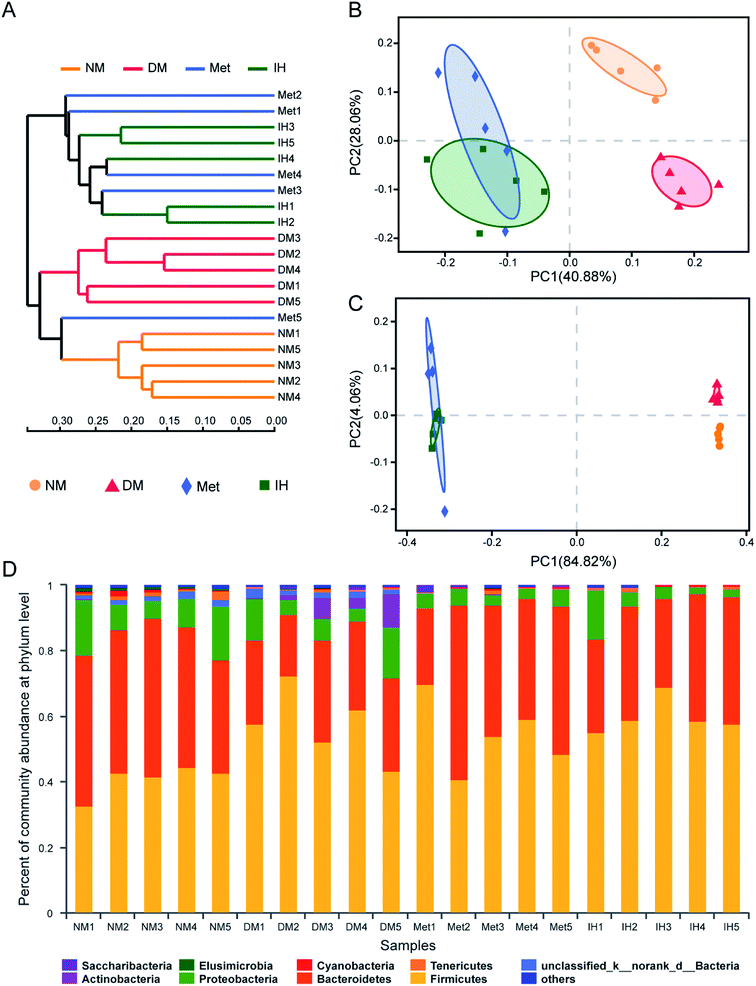
To identify the similarities of gut microbiota between the samples of the NM, DM, Met and IH groups, Bray–Curtis distance was performed based on OTU abundance and presented as a hierarchical clustering tree using the data (Fig. 4A). The result showed that samples from each group could be grouped into three clusters, including the NM, DM and treatment (groups treated by IH or metformin) clusters. Among the clusters, the grouped IH and Met samples indicated that they showed similar bacteria compositions. Meanwhile, weighted and unweighted Unifrac distances both indicated samples were significantly separated into three groups, including the NM, DM and treatment (groups treated by IH or metformin) groups (Fig. 4B and C). This result was consistent with the results of hierarchical clustering tree. Besides, PC1 values were respectively 40.88% and 84.82% in PCoA plots of weighted and unweighted Unifrac distances, indicating PC1 could reflect the treatment effect on shaping the structure of gut microbiota.
In order to understand the detail microbial changes, the gut microbiota structure was analyzed at different taxonomic levels including phylum and genus level (Fig. 4D). At the phylum level, most of the microbiota was from Firmicutes (51.96% in all identified reads) and Bacteroidetes (35.30% in all identified reads). After being induced by STZ and HFHSD, the increased relative abundance of Firmicutes and the decreased relative abundance of Bacteroidetes were observed in diabetic rats. Therefore, the Bacteroidetes/Firmicutes (B/F) ratio was decreased in the DM group (0.48 ± 0.16) compared to that of the NM group (1.08 ± 0.23), and was slightly reverted in the Met and IH groups (Met, 0.79 ± 0.36; IH, 0.57 ± 0.12). The treatment with metformin or RIPT increased Bacteroidetes relative abundance and B/F ratio. Due to the differences between samples, no significant changes were observed. Besides, the Actinobacteria relative abundance was significantly increased in the DM group (4.286% ± 3.97%) and reverted by metformin or RIPT treatment (Met, 0.08003% ± 0.0747%; IH, 0.04494% ± 0.02207%).
When compared to the relative abundances of the NM group at the genus level, 26 genera in the DM group were significantly increased, and 29 genera in the DM group were significantly decreased (Table S1†). Among 26 increased genera, 11 genera were significantly decreased by RIPT, and 7 genera were significantly decreased by metformin. Among 29 decreased genera, 4 genera were significant increased by RIPT, and the same 4 genera were also significantly increased by metformin. Some known beneficial genera, such as lactobacillus, Prevotellaceae NK3B31 group, Alloprevotella and Prevotella_1, were enriched by RIPT (Table 3).
| Genus | Relative abundance | |||
|---|---|---|---|---|
| NM | DM | Met | IH | |
| a All of the results were expressed as the mean ± SD (n = 5). Values sharing a common letter (a and b) in each comparison did not show any statistically significant differences accessed by one-way ANOVA followed by Tukey's post hoc test (p < 0.05). | ||||
| Lactobacillus | 0.23 ± 0.14a | 0.75 ± 0.26a | 5.00 ± 4.37a | 19.22 ± 13.17b |
| Prevotellaceae_NK3B31_group | 1.78 ± 0.49 ab | 0.70 ± 0.48a | 5.10 ± 2.51b | 4.96 ± 3.90b |
| Alloprevotella | 5.21 ± 3.31a | 0.56 ± 0.53a | 3.36 ± 5.38a | 2.03 ± 1.97a |
| Prevotella_1 | 0.85 ± 0.66a | 0.58 ± 0.83a | 4.36 ± 6.14a | 4.51 ± 3.05a |
5 Discussions
In recent years, the anti-diabetic effect of pu-erh tea were widely reported and recognized. Du et al. reported that the pu-erh tea extract had beneficial effects on glucose homeostasis and insulin resistance improvement.4 In this study, the extract of RIPT and RAPT showed alleviation effect of hyperglycemia and hyperlipidemia with more potent activity in RIPT extract. Therefore, the differences on components of two pu-erh teas and modulation effect of RIPT on gut microbiota, which might cause the more effective hypoglycemic result after RIPT treatment, were further investigated.Compared RIPT with RAPT, the key difference is the manufacture process, specially the microbial pile-fermentation. Many microbes, including Aspergillus spp., Penicllium, Rhizopus, Saccharomyces, and Bacterium, play a key role in the post-fermentation process of RIPT manufacture, while RAPT is processed with natural aging only. As various effects of different microbes, some components could be dramatically changed during the fermentation process. In RAPT, the main components are catechins, tea polyphenols, soluble sugar.5 On the contrary, the contents of caffeine, gallic acid was increased after fermentation.19 Researches have shown that the gallated catechins would have a gradually decomposing process to produce catechins and gallic acid during pile-fermentation, therefore, the content of gallic acid was increased. However, catechins would further reacted during fermentation.20 In our study, these changes are consistent with published researches. 19 Components were detected in RIPT only, including puerin I-VIII, 8-carboxyl-(+)-catechin or its isomers, etc. in this work. As reported by Wang et al., puerin I-VIII came from catechins and theanine through fungal fermentation, and could be quality control markers and authentication for RIPT.21 The puerin II, III, and IV were also recognized as markers with VIP value greater than 2.5 for pile-fermentation process in this study. In addition, puerin I was reported to exhibit hypoglycemic and hypolipidemic effect through inhibiting α-glycosidase and activating low-density lipoprotein receptor.22 Therefore, puerins, especially the puerin I, might contribute to the more potent anti-diabetic effect in RIPT. When compared HPLC-PDA result of mutual components detected in RIPT and RAPT, 6 components, including quinic acid, gallic acid, theophylline or its isomers, caffeine, apigenin-6-C-α-L-arabinopyranosyl-8-C-β-D-glucopyranoside, and ellagic acid, were increased after pile-fermentation. As published research, caffeine could promote the expression of glucose transporter 4 to dispose glucose and result in alleviating diabetes.23 In our study, caffeine was significantly increased after pile-fermentation, which would be related to the better hypoglycemic effect of RIPT. Arya et al. reported that quinic acid down-regulated hyperglycemia, hyperlipidemia, and insulin resistance on diabetic rats induced by STZ.24 Meanwhile, gallic acid was also report to attenuate insulin resistance and enhances glucose uptake through activating peroxisome proliferator-activated receptor γ and glucose transporter 4, respectively. The increase of gallic acid content in the RIPT in our study would also be relevant to the better hypoglycemic effect. Therefore, these components increased after fermentation might also contribute to the enhanced activities of RIPT. Besides, the increase of insoluble polyphenols and polysaccharides in RIPT was also reported in previous study.5 They might affect the gut microbiota, and then alleviate diabetes.
To date, the increasing evidences showed that polyphenols and polysaccharides played a probiotic role to modulate gut microbiota, and then ameliorate metabolic syndrome.25,26 The relationships among tea components, diabetes and gut microbiota attracted our attention. Because of the best hypoglycemic activity of RIPT, the feces of rats from the IH group were collected together with those from the NM, DM, and Met groups, and their modulating effects on gut microbiota were investigated. According to the Bray–Curtis distance and PCoA plots, the overall structure of gut microbiota from the IH group was much more similar to that from the Met group, indicating that the gut microbiota in these two groups might have the similar contribution to anti-diabetic effect. As Wu et al. reported, the metformin-altered gut microbiota had benefits to the anti-diabetic effect.27 When looking into the changes at the phylum level, the increased B/F ratio, which was observed in our study, was reported to exhibit positive effects on attenuating insulin-resistance, diabetes, and obesity.28,29 Besides, the phylum Actinobacteria, which was enriched in the DM group and reversed in the IH group in our study, was reported as a pathogen bacteria which had a higher abundance in the women with gestational diabetes mellitus.30 When investigating the changes at the genus level, the probiotic role of RIPT to some health-promoting genera, including lactobacillus, Prevotellaceae NK3B31 group, Alloprevotella and Prevotella_1, were observed in our study. These increased genera were also reported as probiotics which could ameliorate diabetes through increasing insulin release in glucose tolerant individuals,31 producing beneficial short chain fatty acids32 or promoting glucose metabolism.33 These beneficial effects from gut microbiota potentially related to the increased probiotic components from RIPT, and then enhenced the anti-diabetic effect compared with that of RAPT.
6 Conclusion
This comparative study between RIPT and RAPT exhibited that RIPT extract had more potent anti-diabetic effect than RAPT extract. The differences in chemical profiles were also found by UHPLC-Q-TOF/MS analysis, and might contribute to the activity difference. Among them, 17 newly formed components and the increased components after fermentation, such as quinic acid, gallic acid, caffeine, puerin I and so on, might be the main contributors to the enhanced activities. In addition, the gut microbiota potentially contributed to the enhancement of hypoglycemic activity in RIPT through its probiotic role to some health-promoting genera, including Lactobacillus, Prevotellaceae NK3B31 group, Alloprevotella, etc. These results indicated that RIPT could be a functional beverage for alleviating diabetes. The mechanisms involved key components and gut microbiota modulation are needed to be further studied.Conflicts of interest
The authors have declared no conflicts of interest.Abbreviations
| BPI | Base peak ion |
| FBG | Fasting blood glucose |
| FINS | Fasting plasma insulin |
| GC-MS | Gas chromatography-mass spectrometer |
| HFHSD | High-fat, high-sugar diet |
| HPLC-ELSD | High-performance liquid chromatography coupled with evaporative light scattering detection |
| OGTT | Oral glucose tolerance test |
| OPLS-DA | Orthogonal partial least squares discriminant analysis |
| OTUs | Operational taxonomic units |
| PCA | Principal component analysis |
| PCoA | Principal coordinate analysis |
| PDA | Photo-diode array |
| SCFA | Short chain fatty acid |
| STZ | Streptozocin |
| TC | Total cholesterol |
| TG | Triglyceride |
| UHPLC-Q-TOF/MS | Ultra-performance liquid chromatography coupled with a hybrid quadrupole time-of-flight mass spectrometry |
| VIP | Variable importance in projection |
| 2h-PBG | Two-hour postprandial blood glucose |
Acknowledgements
The authors thank Annoroad Gene Tech. (Beijing) Co., Ltd for their excellent technical assistance. This study was supported by the National Natural Science Foundation of China (No. 81773867).References
- W. H. Organization, Global report on diabetes, World Health Organization, 2016 Search PubMed.
- N. H. Cho, J. E. Shaw, S. Karuranga, Y. Huang, J. D. da Rocha Fernandes, A. W. Ohlrogge and B. Malanda, Diabetes Res. Clin. Pract., 2018, 138, 271–281 CrossRef CAS PubMed.
- L. K. Lee and K. Y. Foo, Food Res. Int., 2013, 53, 619–628 CrossRef CAS.
- W. H. Du, S. M. Peng, Z. H. Liu, L. Shi, L. F. Tan and X. Q. Zou, J. Agric. Food Chem., 2012, 60, 10126–10132 CrossRef CAS PubMed.
- H. P. Lv, Y. J. Zhang, Z. Lin and Y. R. Liang, Food Res. Int., 2013, 53, 608–618 CrossRef CAS.
- X. Wang, Q. Liu, H. Zhu, H. Wang, J. Kang, Z. Shen and R. Chen, Acta Pharm. Sin. B, 2017, 7, 342–346 CrossRef PubMed.
- Y. T. Deng, S. Y. Linshiau, L. F. Shyur and J. K. Lin, Food Funct., 2015, 6, 1539–1546 RSC.
- Q. Huang, S. Chen, H. Chen, Y. Wang, Y. Wang, D. Hochstetter and P. Xu, Food Chem. Toxicol., 2013, 53, 75–83 CrossRef CAS PubMed.
- P. C. H. Hollman, J. Sci. Food Agric., 2000, 80, 1081–1093 CrossRef CAS.
- S. Sang, J. D. Lambert and C. S. Yang, J. Sci. Food Agric., 2010, 86, 2256–2265 CrossRef.
- G. Blandino, R. Inturri, F. Lazzara, R. M. Di and L. Malaguarnera, Diabetes Metab., 2016, 42, 303–315 CrossRef CAS PubMed.
- L. Zhao, Nat. Rev. Microbiol., 2013, 11, 639 CrossRef CAS PubMed.
- R. S. Danda, N. M. Habiba, H. Rincon-Choles, B. K. Bhandari, J. L. Barnes, H. E. Abboud and P. E. Pergola, Kidney Int., 2005, 68, 2562–2571 CrossRef CAS PubMed.
- B. Zhang, W. Sun, N. Yu, J. Sun, X. Yu, X. Li, Y. Xing, D. Yan, Q. Ding, Z. Xiu, B. Ma, L. Yu and Y. Dong, J. Funct. Foods, 2018, 46, 256–267 CrossRef CAS.
- J. Zhang, K. Kobert, T. Flouri and A. Stamatakis, Bioinformatics, 2014, 30, 614–620 CrossRef CAS PubMed.
- R. C. Edgar, Bioinformatics, 2010, 26, 2460–2461 CrossRef CAS PubMed.
- J. G. Caporaso, J. Kuczynski, J. Stombaugh, K. Bittinger, F. D. Bushman, E. K. Costello, N. Fierer, A. G. Pena, J. K. Goodrich, J. I. Gordon, G. A. Huttley, S. T. Kelley, D. Knights, J. E. Koenig, R. E. Ley, C. A. Lozupone, D. McDonald, B. D. Muegge, M. Pirrung, J. Reeder, J. R. Sevinsky, P. J. Turnbaugh, W. A. Walters, J. Widmann, T. Yatsunenko, J. Zaneveld and R. Knight, Nat. Methods, 2010, 7, 335–336 CrossRef CAS PubMed.
- C. Lozupone and R. Knight, Appl. Environ. Microbiol., 2005, 71, 8228–8235 CrossRef CAS PubMed.
- L. Zhang, N. Li, Z. Z. Ma and P. F. Tu, J. Agric. Food Chem., 2011, 59, 8754–8760 CrossRef CAS PubMed.
- Y. F. Zhu, J. J. Chen, X. M. Ji, X. Hu, T. J. Ling, Z. Z. Zhang, G. H. Bao and X. C. Wan, Food Chem., 2015, 170, 110–117 CrossRef CAS PubMed.
- W. Wang, L. Zhang, S. Wang, S. Shi, Y. Jiang, N. Li and P. Tu, Food Chem., 2014, 152, 539–545 CrossRef CAS PubMed.
- X. P. Gu, Z. Wu, F. Y. Jin, B. Pan, Y. F. Zhao, J. Li, J. Zheng and P. F. Tu, China J. Chin. Mater. Med., 2018, 43, 2339–2344 Search PubMed.
- E. Mukwevho, T. A. Kohn, D. Lang, E. Nyatia, J. Smith and E. O. Ojuka, Am. J. Physiol.: Endocrinol. Metab., 2008, 294, E582–E588 CrossRef CAS PubMed.
- A. Arya, M. M. Al-Obaidi, N. Shahid, M. I. Bin Noordin, C. Y. Looi, W. F. Wong, S. L. Khaing and M. R. Mustafa, Food Chem. Toxicol., 2014, 71, 183–196 CrossRef CAS PubMed.
- S. M. Henning, J. Yang, M. Hsu, R. P. Lee, E. M. Grojean, A. Ly, C. H. Tseng, D. Heber and Z. Li, Eur. J. Nutr., 2018, 57, 2759–2769 CrossRef CAS PubMed.
- Q. Nie, J. Hu, H. Gao, L. Fan, H. Chen and S. Nie, Food Hydrocolloids, 2019, 86, 34–42 CrossRef CAS.
- H. Wu, E. Esteve, V. Tremaroli, M. T. Khan, R. Caesar, L. Manneras-Holm, M. Stahlman, L. M. Olsson, M. Serino, M. Planas-Felix, G. Xifra, J. M. Mercader, D. Torrents, R. Burcelin, W. Ricart, R. Perkins, J. M. Fernandez-Real and F. Backhed, Nat. Med., 2017, 23, 850–858 CrossRef CAS PubMed.
- D. E. Roopchand, R. N. Carmody, P. Kuhn, K. Moskal, P. Rojas-Silva, P. J. Turnbaugh and I. Raskin, Diabetes, 2015, 64, 2847–2858 CrossRef CAS PubMed.
- P. J. Turnbaugh, R. E. Ley, M. A. Mahowald, V. Magrini, E. R. Mardis and J. I. Gordon, Nature, 2006, 444, 1027–1031 CrossRef PubMed.
- M. K. W. Crusell, T. H. Hansen, T. Nielsen, K. H. Allin, M. C. Ruhlemann, P. Damm, H. Vestergaard, C. Rorbye, N. R. Jorgensen, O. B. Christiansen, F. A. Heinsen, A. Franke, T. Hansen, J. Lauenborg and O. Pedersen, Microbiome, 2018, 6, 89 CrossRef PubMed.
- M. C. Simon, K. Strassburger, B. Nowotny, H. Kolb, P. Nowotny, V. Burkart, F. Zivehe, J. H. Hwang, P. Stehle and G. Pacini, Diabetes Care, 2015, 38, 1827–1834 CrossRef CAS PubMed.
- X. Wei, J. Tao, S. Xiao, S. Jiang, E. Shang, Z. Zhu, D. Qian and J. Duan, Sci. Rep., 2018, 8, 3685 CrossRef PubMed.
- P. Kovatcheva-Datchary, A. Nilsson, R. Akrami, Y. S. Lee, F. De Vadder, T. Arora, A. Hallen, E. Martens, I. Bjorck and F. Backhed, Cell Metab., 2015, 22, 971–982 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8ra09259a |
| This journal is © The Royal Society of Chemistry 2019 |