Open Access Article
Open Access ArticleVarious antibacterial mechanisms of biosynthesized copper oxide nanoparticles against soilborne Ralstonia solanacearum†
Juanni Chen,
Shuyu Mao,
Zhifeng Xu and
Wei Ding *
*
Laboratory of Natural Product Pesticides, College of Plant Protection, Southwest University, Chongqing 400715, China. E-mail: dingw@swu.edu.cn; Fax: +86-23-6825-0953; Tel: +86-23-6825-0953
First published on 29th January 2019
Abstract
The substantial antimicrobial efficacy of nanoparticles against phytopathogens has been extensively investigated for advanced agricultural applications. However, few reports have focused on soilborne pathogenic bacteria. The aim of this study was to obtain sustainably synthesized copper oxide nanoparticles (CuONPs) using papaya leaf extracts and investigate the bactericidal activity of these CuONPs against Ralstonia solanacearum, the cause of bacterial wilt, under laboratory and greenhouse conditions. The results showed that CuONPs possessed strong antibacterial activity and that all R. solanacearum were killed after exposure to 250 μg mL−1 CuONPs. CuONPs could interact with bacterial cells to prevent biofilm formation, reduce swarming motility and disturb ATP production. Ultrastructural observations by transmission electron microscopy (TEM) revealed that after interactions with CuONPs, bacterial cells suffered significantly from nanomechanical damage to the cytomembrane, accompanied by the absorption of multiple nanoparticles. In addition, molecular studies identified the downregulation mechanism of a series of genes involving pathogenesis and motility. The control efficiency of CuONPs in tobacco bacterial wilt disease management under greenhouse conditions was verified by root irrigation application, demonstrating that as-prepared CuONPs significantly reduced the disease occurrence and disease index. Our studies focused on developing biosynthesized nanoparticles as a biocompatible alternative for soilborne disease management.
1 Introduction
Soilborne bacterial wilt disease, caused by Ralstonia solanacearum, a Gram-negative bacterium, can infect as many as 200 plant families, including cucumbers, bananas, peanuts, and especially nightshade family crops.1 The disease severely reduces crop harvests worldwide due to the genetic diversity and wide geographic distribution of the causal pathogen. Another important reason is the strong survival of R. solanacearum and versatile methods of transmission, such as water, soil and remaining crops from the previous season. This bacterium can produce a diverse array of pathogenic factors, including plant cell wall-degrading enzymes, adhesin-like proteins, phytohormones and reactive oxygen species (ROS), which are mainly regulated by the action of type II and type III protein secretion systems.2 These proteins are also called type II (T2E) and type III effector (T3E) proteins.3 Molecular studies have proven that these factors play crucial roles in the pathogenesis of bacteria and wilt symptom development, thus causing a long-lasting, persistent infection of the hosts.3 In recent years, it has been difficult to completely control the disease by existing agricultural measures, such as the massive use of chemical pesticides, which not only induces bacterial resistance but also causes environmental pollution. This challenge has prompted scientists to search for effective alternatives to bactericides to combat obstinate pathogens.Nanotechnology is currently undergoing rapid progress with substantial success, and its potential application in various fields has been widely developed, especially in agricultural science.4 Many studies have shown the significant use of nanotechnologies as pesticides, nanoscale fertilizers, and pathogen and disease detectors in plant protection and crop production.5 Extraordinarily, metal and metal oxide nanoparticles, such as ZnO, Ag and TiO2, have been well documented as effective bactericides and fungicides, inhibiting the bacterial growth and colony formation of Bipolaris sorokiniana and Magnaporthe grisea in in vitro assays.6–8 Similarly, copper-based nanoparticles have been reported to be noxious to Escherichia coli (E. coli), predominantly due to the formation of a copper(I)–peptide complex from cuprous oxide (Cu2O) and free radical generation from cupric oxide (CuO).9 In addition, Kanhed et al. investigated Cu nanoparticles that could inhibit the hyphal growth of four major plant pathogenic fungi (Fusarium oxysporum, Curvularia lunata, Alternaria alternata, and Phoma destructive) and found that the antifungal activity was mainly due to their large surface area to volume ratio.10
However, whether nanoparticles can effectively manage plant diseases in agroecosystems has rarely been reported. In one study, CuONPs significantly suppressed the growth of Botrytis cinerea, showing the greatest effectiveness among six tested nanomaterials in preventing gray mold disease on rose petals, a notorious worldwide fungal disease.11 Elmer et al. demonstrated that the foliar application of nano-CuO, MnO, and ZnO decreased tomato Fusarium wilt and Verticillium wilt disease in a greenhouse without any physiological toxicity.12 Most importantly, CuZn bimetallic nanoparticles (BNPs) displayed no phytotoxic effects on tomato plants, as shown by assessing the photosystem II (PSII) efficiency of plants after 3 h of exposure to BNPs that exhibited antifungal activity against Saccharomyces cerevisiae.13 As a result, we can infer that CuONPs have the potential to be used as an alternative tool to combat pathogenic microbes and exhibit the notable properties of low effective concentrations and low toxicity.
The sustainable synthesis of metal and metal oxide nanoparticles, such as nano-Ag, CuONPs and nano-ZnO, using plant extracts has received considerable attention as an environmentally benign process.14 Papaya (Carica papaya L.), a type of tropical fruit tree, has been applied for medical treatment and food science applications, but its leaves have attracted particular attention due to their biological antioxidant activities.15 The potential role of components in the leaves as reductants for the preparation of nanoparticles has been reported.16 Therefore, in this study, we used these leaves to sustainably obtain CuONPs. The toxic effects of nanoparticles, including copper-based nanomaterials, are generally attributed to the production of lipid-based peroxides and DNA damage resulting from oxidative stress caused by ROS.9,17 These effects are reported to be associated with intricate toxicity mechanisms. Cell structure deformation and cell plasma membrane damage induced by physical stress, which has been connected to nanoparticle–cell interactions, have also been identified as key sources of toxicity.18–20 However, few studies have evaluated the role of CuONPs in bacterial metabolism, physical motility and molecular modes of action on gene expression.
The main aim of this study was to assess the antibacterial effects of CuONPs biosynthesized using C. papaya L. leaves, characterized by dynamic light scattering (DLS), transmission electron microscopy (TEM), X-ray diffraction (XRD) and X-ray photoelectron spectroscopy (XPS), against soilborne R. solanacearum by investigating bacterial cell viability, ATP production, bacterial swarming mobility and biofilm inhibition. TEM imaging was applied to observe changes in the cytomembrane and ultrastructure of R. solanacearum upon exposure to CuONPs, and the transcription levels of a series of genes involving pathogenesis and motility were quantified using reverse transcription-polymerase chain reaction (RT-PCR). A further in vivo investigation of the control efficacy of CuONPs against bacterial wilt caused by R. solanacearum was conducted on tobacco plants using a root irrigation method under greenhouse conditions.
2 Experimental section
2.1 Biosynthesis and characterization of CuONPs
CuONPs were synthesized using completely clean C. papaya L. leaves. The plant leaves were crushed by a grinder after being dried in a shaded area for 15 days. Ten milliliters of leaf extract was mixed with 90 mL of 5 mM cupric sulfate (CuSO4·5H2O) solution (Sigma, USA) to reduce the precursor. The mixture was continuously reacted at room temperature until a black color was observed. The morphological structures of the synthesized CuONPs were characterized by TEM (Tecnai F12, USA). A Malvern Zetasizer (Nano ZS90, UK) instrument was used to analyze the particle size and zeta potential. The crystal structure of the nanoparticles was analyzed by X-ray diffraction (XRD) using a D/MAX-2004 X-ray powder diffractometer (Rigaku Corporation, Japan) with Cu Kα radiation (λ = 1.5406 Å) at 56 kV and 182 mA. X-ray photoelectron spectroscopy (XPS) measurements were conducted on an ESCA LAB250 spectrometer equipped with an Al Kα X-ray radiation source (1486.6 eV).162.2 Antibacterial activity test
A strain of R. solanacearum (Biovar 3, phylotype I, CQPS-1) with strong pathogenicity, isolated from diseased tobacco plants by our research group, was used in this study.21 The antibacterial properties of the CuONPs were evaluated by measuring bacterial growth and viability. In brief, R. solanacearum was inoculated into fresh, sterile NB culture medium (1.0 g yeast extract, 3.0 g beef extract, 5.0 g peptone, 10.0 g glucose, and 1000 mL deionized water, pH 7.0) and grown overnight at 30 °C until the exponential growth phase (OD = 1.0). The cultures were harvested by centrifugation, and the bacterial cakes were then washed at least three times with sterilized deionized water to eliminate residual medium and diluted to an approximate concentration of 107 CFU mL−1. A typical colony count method, also referred to as the spread plate count method, was applied to evaluate the antibacterial activity of CuONPs. Fifty microliters of R. solanacearum bacteria was incubated on the surface of untreated and CuONP-containing agar plates. Each treatment was tested in triplicate. Bacterial viability was determined based on the ratio of the number of colony-forming units in control to that in CuONPs-treated group.2.3 Biofilm inhibition assay under static conditions
Bacteria were incubated with different concentrations of CuONPs in the same volume, and the mixtures were transferred into sterile 96-well polystyrene plates. The wells at the edge of the plates were excluded to avoid external disturbances. Then, the plates were placed in an incubator at 30 °C in the dark. After 12, 24 and 72 h of incubation, the content in the wells was gently aspirated by a pipette and discarded. Then, 200 μL of distilled water was used to carefully wash the wells. At this time, the bacterial biofilm formed on the walls of the wells was stained with 220 μL of 0.1% crystal violet and subjected to 30 min of static incubation at room temperature. Then, the purple-stained biofilm was washed three times until the residual color in the washing solution was eliminated through natural drying. The final step was the addition of 200 μL of 95% ethanol to dissolve the dye, and R. solanacearum biofilm development was determined by measuring the absorbance at a wavelength of 490 nm, which was recorded using a microplate reader (Multiskan MK3, USA).22 The wells without CuONPs were used as a negative control. The data for each condition were averaged from at least three replicate samples.2.4 Bacterial motility tests
To observe the motility of bacterial cells after treatment with the CuONPs, R. solanacearum cells were grown in a semisolid medium (0.35% agar) (SMM) containing different concentrations of CuONPs (50, 125, and 250 μg mL−1).23 First, the medium was sterilized in an autoclave at 121 °C for 20 min, and 15 mL of CuONP-containing agar medium was poured into sterile Petri dishes (polystyrene, 90 mm diameter). Then, bacterial suspensions were adjusted to 108 CFU mL−1, and 5 μL portions were dropped in the middle of the prepared culture plates. Then, the plates were placed horizontally in an incubator at 30 °C for 72 h. This procedure was carefully conducted to avoid disturbing the semisolid medium. All treatments were tested in triplicate. Swarming diameters were used as a measure of motility and were calculated as follows:| swarming motility diameter = measured diameter (mm) − initial inoculation diameter. |
2.5 ATP measurement
ATP levels were determined to assess the effects of CuONPs on bacterial metabolism. R. solanacearum cells were prepared as described above and dispersed in 50 mM sodium HEPES buffer (pH 7.0) containing 5 mM glucose. Untreated bacterial cells and cells treated with CuONPs for 5 h were subjected to 1% trichloroacetic acid (TCA) in the presence of 2 mM EDTA. Following incubation in an ice bath for 30 min, the cells were neutralized with 0.1 M Tris acetate (pH 7.8) to remove residual TCA. After centrifugation, all supernatants were used to determine ATP levels using an ATP Assay Kit (Beyotime Biological Co., Ltd., China).202.6 TEM observations
The ultrastructural changes of R. solanacearum cells interacting with CuONPs were visualized using TEM as previously described.24 Briefly, bacterial cells were collected in the log phase using the abovementioned procedures and pelleted. Then, 0.5 mL of cells was treated with the same volume of CuONPs at the tested concentration for 2 h and centrifuged. Following fixation overnight with 2.5% glutaraldehyde, all samples were postfixed with 1% osmium tetroxide for 1 h and subsequently dehydrated in graded concentrations of ethanol (15, 25, 55, 80, and 100%) and embedded in epoxy resin using a standard protocol. Thin sections were deposited on bare grids (200 mesh), which were stained with uranyl acetate for 5 min and observed by TEM (JEOL JEM-1230, Japan) at 80 kV after drying.2.7 Gene expression analysis
Typically, RNA was extracted using TRIzol reagent (Sigma, USA). Cultures of R. solanacearum cells were grown overnight in nutrient broth (NB) medium at 30 °C with shaking at 180 rpm to obtain bacteria in the exponential phase and at their most active. Then, the cells were harvested by centrifugation at 5000 rpm and washed three times with PBS at pH 6.8. The pellets were transferred into PBS solution and adjusted to an optical density of 1.0 at a wavelength of 600 nm. Then, 500 μL of 125 μg mL−1 CuONPs and sterilized water (control) were pipetted into RNase-free centrifuge tubes, and the same volume of bacterial cell suspension was poured into the tubes. After incubation for 2 h at 30 °C and 180 rpm, untreated and CuONP-treated R. solanacearum cells were immediately centrifuged at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 10 min at 4 °C. To ensure the full release of RNA from the bacterial cells, lysozyme was included in the mixture. As described in the manufacturer's instructions, 1 mL of TRIzol (Invitrogen, USA) was added to the tested samples and allowed to incubate for five minutes, after which chloroform was introduced to the tubes for separation. Following centrifugation at 4 °C at 10
000 rpm for 10 min at 4 °C. To ensure the full release of RNA from the bacterial cells, lysozyme was included in the mixture. As described in the manufacturer's instructions, 1 mL of TRIzol (Invitrogen, USA) was added to the tested samples and allowed to incubate for five minutes, after which chloroform was introduced to the tubes for separation. Following centrifugation at 4 °C at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm, the upper aqueous phase was removed and transferred into an RNase-free tube. Then, 0.5 mL of isopropanol was gently added for RNA precipitation at room temperature. The supernatant was discarded, and the remaining RNA was washed twice with cold ethanol. The final RNA product was resuspended in ddH2O and purified as previously described to remove DNA fragments.20 Two microliters of RNA was used as the template for cDNA synthesis according to the manufacturer's instructions of the iScript™ cDNA Synthesis Kit (Bio-Rad, USA) and a Bio-Rad real-time PCR system.
000 rpm, the upper aqueous phase was removed and transferred into an RNase-free tube. Then, 0.5 mL of isopropanol was gently added for RNA precipitation at room temperature. The supernatant was discarded, and the remaining RNA was washed twice with cold ethanol. The final RNA product was resuspended in ddH2O and purified as previously described to remove DNA fragments.20 Two microliters of RNA was used as the template for cDNA synthesis according to the manufacturer's instructions of the iScript™ cDNA Synthesis Kit (Bio-Rad, USA) and a Bio-Rad real-time PCR system.
The selected pathogenesis-related genes were tested. The primers were designed by the Beijing Genomics Institute, referring to the R. solanacearum GMI1000 genome, as shown in Table 1. The 16S rRNA was chosen as the reference gene to ensure the amplification efficiency of each primer. RT-qPCR was conducted using a 20 μL final reaction system, including 1 μL of cDNA template, 7 μL of RNase-free water, 7 μL of Power SsoFast™ EvaGreen® Supermix (Bio-Rad Laboratories, USA), and 1 μL of each forward and reverse primer in a CFX96TM Real-Time System (Bio-Rad, USA). The PCR amplification procedure began with denaturation at 95 °C for 3 min, followed by one cycle of 95 °C for 10 s and 54 °C for 20 s and then 40 cycles of 60 °C for 30 s and 95 °C for 1 min.20 The relative expression of each RNA was quantitatively calculated using the 2ΔΔ−CT method, where ΔΔCT = ΔCT (treated sample) − ΔCT (untreated sample), ΔCT = CT (target gene) − CT (16S rRNA), and CT is the threshold cycle value for the amplified gene.
| Tested genes | Function or protein encoded | Primer | Sequence (5′–3′) |
|---|---|---|---|
| Mobility genes | |||
| pilT | Fimbrial type-4 assembly membrane transmembrane protein | Forward | GGTACCTCTAGACATCGTGGCACTCCGGAG |
| Reverse | GGTACCTCTAGACATCGTGGCACTCCGGAGC | ||
| egl | Endoglucanase precursor (endo-1,4-beta-glucanase) (cellulase) | Forward | AAATCCAGATATCGAATTGCCAA |
| Reverse | GCGTGCCGTACCAGTTCTG | ||
| filA | Pilus subunit | Forward | TTCGTAGTACAGCAGTTGGCT |
| Reverse | GCAGTTCGAGTTCTATGCCG | ||
| flgM | Negative regulator of flagellin synthesis (anti-sigma-28 factor) protein | Forward | GGCCATCACGCCGACCAA |
| Reverse | GCGAGACCTGCTGCACCGA | ||
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||
| Pathogenesis genes | |||
| phcA | Virulence transcription regulator protein | Forward | TTGTAGGTCTCGCACACCAG |
| Reverse | GCTCGCTCGATCAGTACCTC | ||
| xpsR | Transcription regulator | Forward | AGATCGACATAGCGCTGCTT |
| Reverse | TTACTTTGCGGACCTGCTCT | ||
| epsE | EPS I polysaccharide export inner membrane protein | Forward | CTGGATAAAGCCACGCAAAG |
| Reverse | CAGTGGTACATCGCCATCAC | ||
| pehC | Polygalacturonase | Forward | CTCAGCAACGTCAACTTCCA |
| Reverse | CCGTAGTTCAGGCAATCGTT | ||
| hrpB | Transcriptional activator of hrp genes cluster; type III secretion system protein | Forward | AATACGCAAATGCGGTTTTC |
| Reverse | CTTCTTCCGCTTCTTCATCG | ||
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||
| Reference genes | |||
| 16S rRNA | 16S rRNA | Forward | AGAGTTTGATCCTGGCTCAG |
| Reverse | ACGGCTACCTTGTTA | ||
2.8 Greenhouse experiments
Tobacco seeds were sown in plastic pots (6.5 cm × 7 cm) filled with a peat substrate and placed in a manual climatic incubator at 25 °C with a light density of approximately 110 μmol m−2 s−1 and a light/dark photoperiod of 14 h/10 h. After 40 days of growth until the development of the fourth leaf, dozens of uniform tobacco seedlings were prepared for greenhouse experiments. Each group consisted of twenty plants. The experiments included four groups (0, 50, 125, 250 μg mL−1 of CuONPs) and were conducted by the posttreatment method. In other words, 10 mL of R. solanacearum at OD600 = 0.1 (≈107 CFU mL−1) was applied via noninjured root inoculation to plant. Twenty-four hours later, 10 mL suspensions containing different concentrations of CuONPs were applied by root irrigation. Deionized water was used as a control group. All inoculated tobacco seedlings were incubated in the growth chamber at 30 °C under the same culture conditions and watered once every two days. After approximately 7 days of growth, bacterial wilt disease symptoms appeared. Then, the disease occurrence in the tobacco seedlings and the disease index were investigated at 7, 9, 13 and 19 days. The bacterial wilt disease classification of the tobacco plants in the laboratory was evaluated and recorded according to the “Protocols of Disease Investigation and Classification”. Disease indexes were calculated using the following equation:| disease index = Σ(ni × vi)/N × 4 × 100 |
2.9 Statistical analysis
Statistical analysis was conducted using the statistical software SPSS 11.0 (SPSS Inc., Chicago, IL, USA). The data in all groups were analyzed by one-way analysis of variance (ANOVA). A variance test analysis was applied to evaluate the differences between the treatments. The results were considered statistically significant and extremely significant when the p-values were <0.05 and <0.01, respectively.3 Results and discussion
3.1 Characterization of prepared CuONPs
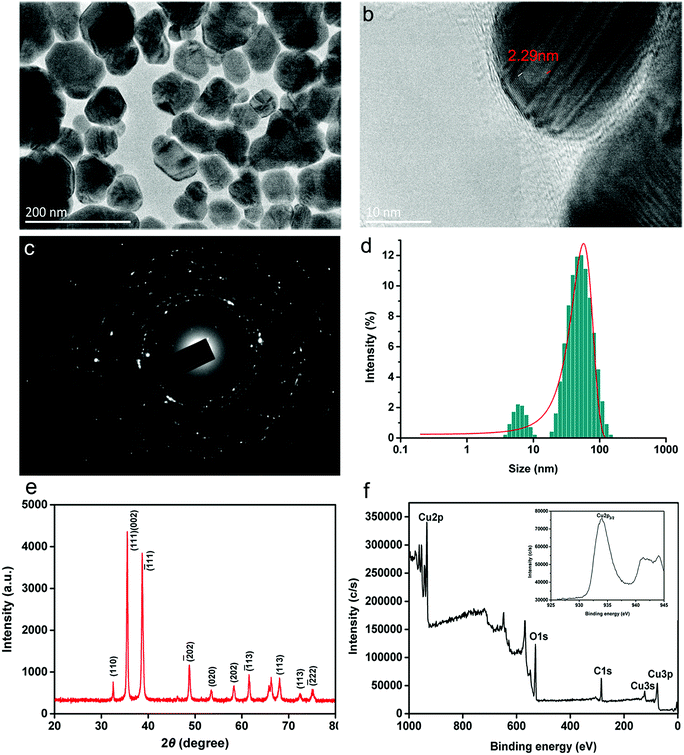
CuONPs were sustainably synthesized through the reduction of the precursor by the bioactive compounds extracted from C. papaya L. leaves. The typical morphology of the obtained CuONPs measured by TEM is illustrated in Fig. 1a. The nanomaterials consisted of irregular spherical particles and agglomerations of various sizes with a clear crystal lattice structure of 2.29 nm (Fig. 1b). In addition, the representative selected area electron (SAED) pattern for CuONPs displayed various diffraction rings for monoclinic CuONPs (Fig. 1c). The particle size distribution of CuONP particles presented in Fig. 1c indicates that the majority of nanoparticles, approximately 78%, were in the range of 40–80 nm. The sizes of the particles observed in previous reports prepared by other methods were in the range of 4.8–7.8 nm.25 We inferred that the size differences are attributed to the types of organic extracts, which play key roles in the synthesis of metal nanoparticles. In previous study, improved dispersibility of extracts possibly contributes to more favorable contact with metal salts, forming smaller nanoparticle sizes.25Fig. 1e shows the X-ray diffraction pattern of the as-prepared CuONPs, which illustrates that CuO is single-phase with a crystalline structure (JCPDS-05-0661).26 Several sharp diffraction peaks at 2θ = 32.53, 35.49, 38.72, 48.7, 53.47, 58.32, 61.56 and 68.13 were identified, which was in accordance with previous results.27 The two representative peaks positioned at 2θ values of 35.49 and 38.72, which were assigned to the (111, 002) and (1−11) crystallographic planes, confirmed the dominant presence of CuO in the prepared powder. The XPS spectrum is typically applied for measuring surfaces and metal oxidation. As shown in Fig. 1f, the prominent peaks in broad XPS survey scans were mainly focused on Cu2p, C1s and O1s, proving the existence of Cu, O and C in the sample. The elemental composition was determined by high-resolution scans, as shown in the inset plot in Fig. 1f. A strong Cu2p core level spectrum was observed, representing a characteristic peak with a binding energy of 933.9 eV, which corresponds to Cu2p3/2.28 Two other peaks at 121.1 and 77.2 eV associated with Cu3s and Cu3p, respectively, were also observed.29 In addition, O1s high-resolution scans were located at low binding energy peaks at 529.8 eV, indicating the presence of oxygenated groups in CuO (Fig. S1a†). Typically, the C1s peak with a binding energy of 284.8 eV, shown in Fig. S1b,† which is assigned to C–C, C–O, and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O functional groups, is ascribed to the low amount of amorphous carbon adsorbed onto the catalyst surface.25 These results showed that the prepared powder consists mainly of CuO as well as nanocrystalline nanoparticles with functional groups, which is in agreement with the results of a previous study.28
O functional groups, is ascribed to the low amount of amorphous carbon adsorbed onto the catalyst surface.25 These results showed that the prepared powder consists mainly of CuO as well as nanocrystalline nanoparticles with functional groups, which is in agreement with the results of a previous study.28
A plot of the zeta potential of green synthesized CuONPs versus pH (4.0–9.0) is presented in Fig. S2.† Because the interaction between CuONPs and bacterial cells proceeded in culture medium in the following experiments, the zeta potential was measured in B medium using a Malvern Zetasizer (Nano ZS90, UK) instrument. The zeta potential of the CuONPs decreased with increasing pH. The zeta potential had a positive value below pH 8 due to the high level of hydrogen ions, indicating that the nanoparticles were stable in the bacterial culture medium (pH 6.5–7.0). The dispersion possessed a negative charge when the pH was greater than 8.0, which was possibly associated with the adsorption of carboxy groups on the surface.
3.2 Reduction in bacterial cell viability
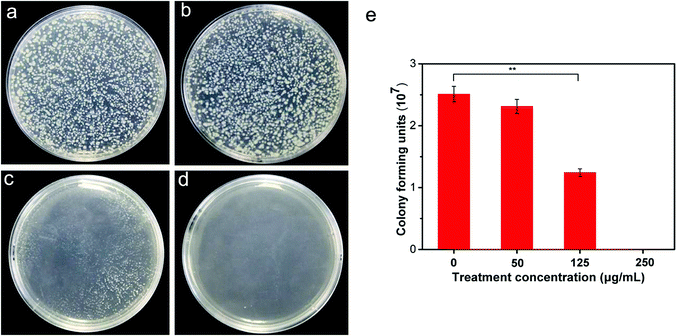
To investigate the antibacterial activity of CuONPs, bacteria were treated for 2 h with CuONP suspensions at various concentrations ranging from 50 to 250 μg mL−1. Due to the irregular agglomeration caused by van der Waals and capillary forces, the nanoparticle–bacteria interactions were facilitated under moderate vibration (180 rpm in this experiment) to ensure adequate dispersion of the nanoparticles. Cell viability was evaluated by measuring the number of colony-forming units (CFU) on NB agar plates (Fig. 2e). Each treatment was conducted at least in triplicate. As presented in Table S1,† in comparison to the control, untreated bacterial cells yielded 2.51 × 107 CFU, whereas the viability of CuONP-treated bacterial cultures decreased to at least 92.03% and 49.48% following exposure to nanoparticle concentrations of 50 and 125 μg mL−1, respectively. After treatment with CuONPs at a concentration of 250 μg mL−1, the cells showed complete inactivation, with no viability. The data clearly suggested that the CuONPs displayed a concentration-dependent antibacterial effect, similar to the effects of other metal oxide nanoparticles, including nano-MgO and ZnO, which have been reported for other pathogenic bacteria and fungi.30,31 Fig. 2a–d shows photographs of bacterial colonies on agar medium. Notably, metal and metal oxide nanoparticles, such as nano-Fe and ZnO, exhibit metal cytotoxicity on human cells and microorganisms only at relatively high concentrations.32 At the appropriate dose, these metal ions could participate in important metabolic processes in bacteria, serving as coenzymes, cofactors and catalysts, and structural stabilizers of several enzymes.33 Roselli et al. have shown that E. coli can metabolize Zn2+ as an oligoelement.34 This utility explains the results of the antibacterial test, which showed that low concentrations of CuONP-treated bacteria still had high cell viability. Even carbon-based nanomaterials (SWCNTs, MWCNTs and graphene) are known to kill most bacteria at a dose of 250 μg mL−1 under the same experimental conditions.19,24 These common phenomena are likely associated with the increased opportunity for direct contact with bacterial cells.3.3 Inhibition of bacterial biofilm formation
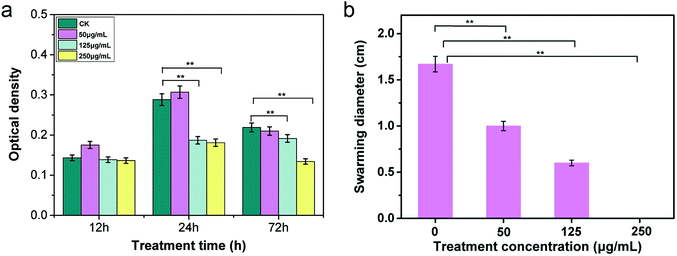
In nature, bacteria live in aggregates and communicate with each other by intimate contact, forming architecturally well-organized cooperating communities, referred to as biofilms.35,36 Biofilm formation and invasion into host cells are vital processes during pathogenic colonization and infection. As previously reported, Gram-negative phytopathogenic bacteria produce biofilm-like aggregates that are conducive to the virulence of bacteria by blocking the xylem vessels of host plants and increasing microbial resistance to antibacterial compounds.37 We further investigated the antibiofilm activity of CuONPs with the aim of determining the antimicrobial effectiveness of nanoparticles against R. solanacearum. For this purpose, 100 μL of R. solanacearum cultures were inoculated into sterile 96-well plates containing 200 μL NB medium to develop biofilms. Various concentrations of CuONPs were added to the wells and incubated at 30 °C for 12, 24, and 72 h under anaerobic conditions. The wells without CuONPs were used as controls. After staining and washing, the biofilm bacterial population on the well wall was directly measured by determining the optical density at a wavelength of 600 nm. As shown in Fig. 3a, we found that treatments with all tested nanoparticle suspensions had no effect on the bacterial cells biofilms after an initial 12 h incubation. However, after incubation periods of 24 h and 72 h, compared to the untreated samples, significant inhibition of bacterial cells was observed in the presence of 125 and 250 μg mL−1 CuONPs, which caused more than a 35% and 37% reduction in biofilms at 24 h and a 12% and 38% reduction in biofilms at 72 h, respectively (Fig. 3a).Several nanoparticles, such as ZnO, gold and silver nanoparticles, have been reported to possess strong antibiofilm properties against Streptococcus pneumonia, E. coli and Pseudomonas aeruginosa.38,39 They have also been widely exploited as novel bacteria-resistant products for the long-term disinfection of root canals, preventing the formation of E. faecalis biofilms.40 Most importantly, biofilm-associated R. solanacearum cells can produce a variety of extracellular polymeric substances (EPS) along with various proteins, which are known to be key pathogenic factors when a pathogen infects host roots in the rhizosphere.41 Thus, although there was no change in biofilm formation during the 12 h treatment period, our results clearly demonstrated that the application of CuONPs could effectively exert antibacterial activity against R. solanacearum by disturbing the invasion course after a longer incubation time.
3.4 Reduction in bacterial swarming motility
Biofilm formation is commonly mediated by flagella-induced motility in the infection process. Researchers have found that swarming and swimming motility are required for efficient invasion and colonization in host plants for most species of soilborne bacteria.42 The motility of R. solanacearum is regulated by one to four polar flagella, which assist the bacteria in moving toward the host rhizosphere, attaching to host roots, and invading root tissue, especially in the early stage of infection.43 Therefore, it is essential to understand whether CuONPs affect the swarming characteristics of R. solanacearum bacteria. To investigate the effects of CuONPs on swarming ability during the course of treatment, the bacterial swarming diameter was measured on CuONP-containing NB agar plates. Fig. 3b shows that the displacement distances in the presence of various concentrations of CuONPs were significantly shortened, reflecting a substantial decrease in motility. In addition, no movement of R. solanacearum cells was observed in the 250 μg mL−1 CuONP-treated samples, which can be attributed to the bactericidal activity of the nanoparticles, as shown in Fig. 3b. It is observed that interesting forms of toxicity toward pathogens play roles in the antibacterial mechanism of CuONPs, physically preventing the movement of R. solanacearum. MgONPs were also reported to repress the swimming mobility and twitching motility of R. solanacearum after attachment on the cell, but the effect remains insufficient to kill the cells.31 The substantially decreased transcriptional expression of motility-related genes in R. solanacearum induced by nanomaterials might explain these behaviors.20 To date, there is still a lack of detailed research regarding the effects of metal oxide nanoparticles on the mobility of the bacteria.3.5 ATP level measurement
Many substances, such as proteins and biomolecules, are translocated into or out of bacterial cells across the cell membrane and organelles through transporters belonging to the ATP-binding cassette (ABC) superfamily, whose function is coupled to the energy released from ATP hydrolysis.44 All behaviors are used to maintain the physiological metabolism of bacteria. According to the abovementioned results, CuONPs can directly interact with cells and damage the cytomembrane, which contains several crucial energy-transducing enzymes, undoubtedly interfering in ATP production. As hypothesized, the results showed that the ATP level of bacteria exposed to CuONPs gradually decreased with increasing treatment time (Fig. 4). There was little ATP production in R. solanacearum in the presence of 250 μg mL−1 CuONPs after incubation for 5 h, which implied that the CuONPs disturbed the metabolism of bacterial cells. These findings were similar to previous reports demonstrating that silver nanoparticles depleted ATP content in bacterial cells within 5 min, with no evidence of ATP leakage.45 Chen et al. also confirmed that the underlying mechanism of GO involved disturbances to energy metabolism processes and the occurrence of lipid oxidation.20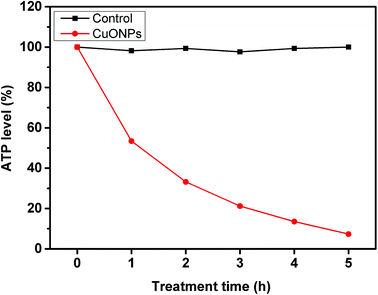 | ||
| Fig. 4 ATP levels of R. solanacearum cells after 5 h of incubation with water (control) and 250 μg mL−1 CuONPs. ATP levels were determined at timed intervals. | ||
3.6 Morphological observations by TEM
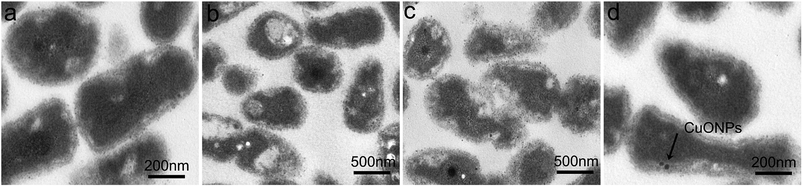
We further investigated the morphological structure of R. solanacearum using TEM after CuONP treatment with the purpose of assessing the direct toxicity to bacteria. Visually, after incubation for 3 h under moderate vibration, in the case of CuONPs, moderately specific damage was observed on the bacterial cell membrane compared to that in the unexposed control group, which possessed an ordered and intact cell envelope (Fig. 5a). Moreover, from the perspective of intracellular structure, as shown in Fig. 5b, CuONPs could cause cytoplasmic condensation and vacuolization in bacteria, especially at high doses. In addition, the absorption of CuONPs by bacteria was observed in TEM images (Fig. 5d). These direct antimicrobial effects have also been observed in several previous studies regarding the impact of copper-based nanoparticle toxicity on cell lines, bacteria and even fungi.46,47 Other types of nanoparticles, such as graphene,19 carbon nanotubes.48 and metal oxide nanoparticles (ZnO, MgO, and NiO),31,38 have also shown excellent stimulatory effects on R. solanacearum at corresponding sublethal exposures in experiments. It was clear in these studies that the NPs could pass through the bacterial cell wall and then reach or pass through the cellular membrane.For nearly a decade, many researchers have examined the complex mechanisms by which metal nanoparticles cause toxicity to bacteria and other microorganisms. The results demonstrated that metal oxide nanoparticles may exert substantial antimicrobial effects through cellular oxidative stress and facile penetration of the cell wall or cell membrane, subsequently undergoing endocytosis, accumulating in the cytoplasm and eventually causing cell lysis and death.49,50 It has been speculated that these phenomena could be explained by increased contact opportunity and particle-specific interactions due to the effects imposed by their nanometer scale, such as a higher surface area.18 A large number of glycoprotein receptors on the surface of bacteria are known as adhesins and contribute to nanoparticle–bacteria interaction.51 When nanoparticles interact with bacterial cells, the electrostatic interaction between positively charged CuONPs and the negatively charged bacterial membrane is likely to play a key role in the adsorptive action (Fig. S2†).31 However, carbon nanotubes (CNTs) directly induced aggregation interactions with pathogens on account of their supportive function or functionalization with sugar-based ligands.52 The toxic behaviors observed in this experiment were largely in agreement with previous reports. In addition, copper (Cu2+) ions released from or adsorbed onto CuONP coatings could essentially account for the nanotoxicity toward biological cells.46,53
3.7 Gene expression profile
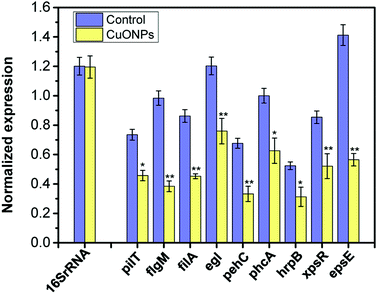
Moreover, to further understand the molecular behavior of CuONPs against the phytopathogen R. solanacearum in vitro, a set of genes involved in bacterial motility (pilT, flgM, and filA), biofilm formation, and pathogenesis (pehC, phcA and hrpB) were selected for genotoxicity evaluation, as listed in Table S1.† After exponentially grown bacterial cells were collected and treated with 125 μg mL−1 CuONPs for 2 h, the transcription levels of the selected genes were determined using RT-qPCR. The gene for 16S rRNA was used as a reference gene for normalization in the transcription analysis of gene expression. As shown in Fig. 6, as hypothesized, genes with different functions were all downregulated in R. solanacearum after exposure to CuONPs at 125 μg mL−1, while the reference gene remained unchanged regardless of the treatment. In particular, the transcription levels of flgM, filA and epsE were extremely significantly reduced compared to those of genes in untreated bacterial cells.For R. solanacearum, flagella-driven swimming motility and biofilm formation significantly contribute to invasive virulence in plants, which is modulated by the pilT, filA and flgM genes.1,54 Furthermore, all of these genes play important roles in the flagella in initial bacteria-to-surface interactions with the host for Pseudomonas.55 Liu et al. demonstrated that the inactivation of pilT eliminated twitching motility and reduced the virulence of R. solanacearum.56 Additionally, in R. solanacearum, PhcA, a member of the LysR family of transcriptional regulators, plays a vital role in the regulatory system of pathopoiesis, which is considered the central element of the intricate system.57 It can positively regulate the expression of several types of pathogenicity factors, such as exopolysaccharides (EPS).57 The type III secretion system (T3SS) is another of the most remarkably regulated networks in phytopathogens, allowing the vectorial secretion of effector proteins across the bacterial envelope into the host, where they modulate host defense responses and physiology, which is encoded by a cluster of hrp genes.8 TTSS is controlled by hrpB in R. solanacearum, an AraC family transcriptional regulator. The analysis of gene expression showed that the transcription levels of pathogenicity-related genes (phcA, hrpB, xpsR and pehC) were significantly decreased when exposed to CuONPs.
These results clearly indicated that CuONPs negatively affected bacterial motility and virulence, potentially acting as T3SS inhibitors. These results provide substantial verification regarding the response of R. solanacearum cells to CuONP stress obtained in the experiments mentioned above from the molecular level, which would be useful for evaluating and predicting the antibacterial potency of nanomaterials. We can conclude that the antibacterial mechanism of CuONPs is very likely through decreased expression levels of virulence factors related to cell motility and pathogenicity. Similar alterations in the gene expression of R. solanacearum have been observed under exposure to graphene oxide.20 Transcriptomic analysis has been widely used to reveal the molecular mechanisms of nanoparticle toxicity as antibacterial agents. Several previous studies have also revealed the biocidal effects of nano-TiO2, ZnO and CuNPs, likely through the decreased gene transcription of a variety of genes involving cellular functions, such as metabolic activities, stress response, pathogenesis, and toxin production.17,58
3.8 High efficacy of CuONPs in controlling bacterial wilt in tobacco
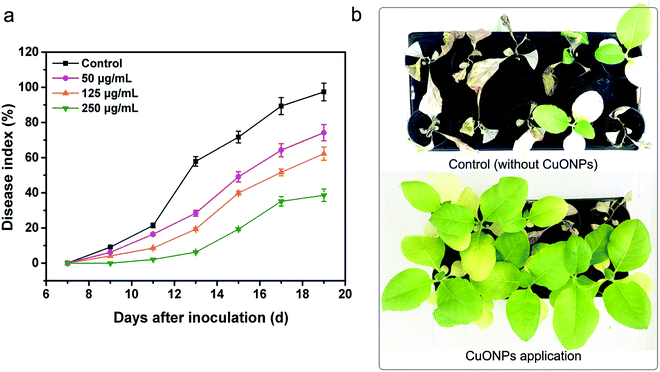
In the following study, experiments on potted plants were performed under greenhouse conditions to evaluate whether CuONPs, regarded as an agricultural antibacterial agent, could manage bacterial wilt disease. As shown in Fig. 7a and S3,† overall, treating tobacco seedlings with CuONPs reduced the severity and occurrence of bacterial wilt disease caused by R. solanacearum. Moreover, there was a clearly positive correlation between the treatment concentration and disease index. The disease on untreated plants was particularly severe after incubation for 20 days, and the disease index reached up to 97.3%, while the disease indexes of the tobacco seedlings exposed to CuONPs at concentrations of 50, 125, and 250 μg mL−1 were reduced to 74.2%, 62.2% and 38.1%, respectively, which clearly showed different disease symptoms (Fig. 7b). The corresponding disease occurrence in treated plants was 76.5%, 59.4% and 32.5% in comparison to 98.5% in the untreated samples (Fig. S3†). In particular, the foliar spraying of CuONPs on tomato plants and eggplants at higher concentrations appeared to substantially control Fusarium wilt disease development during the vegetative period without phytotoxicity, whether in soilless medium or in soil.12 Fig. 7b shows the disease symptoms after the root irrigation application of CuONPs for 20 days. Tobacco seedlings without nanoparticle treatment were nearly wilted and dead, while a few withering seedlings were observed when the plants were treated with CuONPs. Hence, our results demonstrate the potential application of this nanoparticle in the future management of bacterial diseases of vegetable crops.Over the past decade, there have been many studies regarding the antibacterial activity of metal or metal oxide nanoparticles against phytopathogenic bacteria and fungi at lower concentrations than commercial bactericides. For instance, ZnO and CuO nanoparticles have been reported to exhibit toxicity to Vibrio fischeri,6 Xanthomonas axonopodis pv. punicae,59 and Phytophthora infestans.60 The dose-dependent antifungal activity of copper-based nanoparticles was also demonstrated in fungal strains, such as Venturia inaequalis,61 F. oxysporum,62 Alternaria alternata and B. cinerea63 CuONPs also have antifungal activities and prevent B. cinerea infections in rose plants.11 The potential toxicity mechanisms were related to mechanical damage, oxidative injury and gene toxicity. Our results also suggested that CuONPs exert excellent antibacterial activity against R. solanacearum due to different mechanisms in combination with physical and chemical damage and gene expression inhibition (Fig. 8).
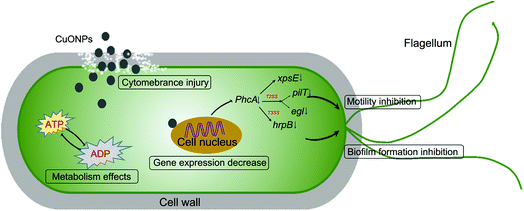 | ||
| Fig. 8 The different underlying antibacterial mechanisms of CuONPs against soilborne phytopathogenic R. solanacearum. | ||
Most importantly, using plant extracts for the synthesis of nanoparticles makes them more biocompatible by eliminating some toxic chemical substances during the reaction process.14 To date, a large number of organisms, including bacteria, fungi and plants, have been investigated for the synthesis of metallic nanoparticles, which could act as nontoxic and superior antibacterial agents for diagnostic applications and cancer therapies in medical science fields.64 Valodkar et al. investigated sustainable synthesis methods for the production of Cu and CuO nanoparticles using natural materials, such as magnolia leaf extract and stem latex of Euphorbia nivulia.65 In vitro assays indicated that these nanoparticles exhibited substantial antibiotic properties against various pathogens, such as E. coli, Bacillus subtilis, and Staphylococcus aureus.16 Though CuONPs exhibited phytotoxic on some plants, most of the studies reported above demonstrated the effects of nanoparticle only in hydroponic conditions, where metal availability and root structure are greatly different from growth in a solid matrix.66 Moreover, plant responses to metal/metal oxide nanoparticle are dependent on the dose, the plant species, and the application site (e.g., root or shoot). Exposure of mung bean to Ag NPs (2000 mg kg−1) in soil results in growth effects compared to significant reductions observed in agar at 10–40 mg L−1.67
Notably, these current antimicrobial assays are limited to laboratory conditions (i.e., in vitro) and lack practical applications in the field or in greenhouse conditions. In particular, silver nanoparticles have been investigated by Jo et al. for effectively reducing the severity of two fungal diseases (Bipolaris sorokiniana and Magnaporthe grisea) on perennial ryegrass (Lolium perenne) in growth chamber inoculation assays.7 Ocsoy et al. demonstrated that 100 ppm Ag@dsDNA@GO composites effectively reduced the severity of bacterial spot disease in tomato transplants.68 Similar results were obtained by Chen et al., demonstrating that GO–Ag composites had a significant effect in controlling leaf spot disease infected by F. graminearum.69 One study showed that CuONPs did not inhibit Fusarium wilt fungus in in vitro assays; however, tomato disease estimates were decreased by 31%, indicating that the host defense could be manipulated.12 Therefore, the underlying persistent control mechanisms and the complicated interaction between nanoparticles and plants in agroecosystems still need to be explored in further studies. Considering the advantages of nanoparticles over bactericides, our results indicated that CuONPs have great promise for their application in the management of bacterial diseases in tobacco plants.
4 Conclusions
In conclusion, this research evaluated the antibacterial activity of CuONPs against soilborne phytopathogenic R. solanacearum. The experimental results proved that CuONPs displayed dose-dependent bacteriostatic activity, remarkably reducing bacterial viability and killing bacteria at high concentrations. Antibacterial tests demonstrated that the interaction of CuONPs with bacterial cells resulted in decreased bacterial motility and biofilm formation and disturbed ATP production in cells. TEM images revealed clear damage to the cell morphological structure and absorption of nanoparticles due to direct contact with nanoparticles. Furthermore, the downregulated expression of different genes in CuONP-treated cells indicated that reduced motility and bacterial virulence, which are necessary for the infection of host plants, could be responsible for the strong antibiotic properties of CuONPs. Furthermore, CuONPs were found to be an efficient antibacterial alternative for preventing bacterial wilt disease under greenhouse conditions. Our results suggest that this nanoparticle could serve as a new management strategy for plant protection in the future.Conflicts of interest
The authors declare no competing financial interest.Acknowledgements
This work was supported by the Research Fund for the Doctoral Program of Southwest University (SWU117012), National Natural Science Foundation of China (31572041) and the Fundamental Research Funds for the Central Universities (XDJK2017C066).References
- S. Cunnac, A. Occhialini, P. Barberis, C. Boucher and S. Genin, Mol. Microbiol., 2004, 53, 115–128 CrossRef CAS PubMed.
- M. Poueymiro and S. Genin, Curr. Opin. Microbiol., 2009, 12, 44–52 CrossRef CAS PubMed.
- H. Liu and T. P. Denny, Phytopathology, 2007, 97, S66–S67 CrossRef PubMed.
- L. R. Khot, S. Sankaran, J. M. Maja, R. Ehsani and E. W. Schuster, Crop Prot., 2012, 35, 64–70 CrossRef CAS.
- A. Servin, W. Elmer, A. Mukherjee, R. De la Torre-Roche, H. Hamdi, J. C. White, P. Bindraban and C. Dimkpa, J. Nanopart. Res., 2015, 17, 92 CrossRef.
- M. Heinlaan, A. Ivask, I. Blinova, H. C. Dubourguier and A. Kahru, Chemosphere, 2008, 71, 1308–1316 CrossRef CAS PubMed.
- Y. K. Jo, B. H. Kim and G. Jung, Plant Dis., 2009, 93, 1037–1043 CrossRef CAS.
- J. Vasse, S. Genin, P. Frey, C. Boucher and B. Brito, Mol. Plant-Microbe Interact., 2000, 13, 259–267 CrossRef CAS PubMed.
- S. Meghana, P. Kabra, S. Chakraborty and N. Padmavathy, RSC Adv., 2015, 12293–12299 RSC.
- P. Kanhed, S. Birla, S. Gaikwad, A. Gade, A. B. Seabra, O. Rubilar, N. Duran and M. Rai, Mater. Lett., 2014, 115, 13–17 CrossRef CAS.
- Y. Hao, X. Q. Cao, C. X. Ma, Z. T. Zhang, N. Zhao, A. Ali, T. Q. Hou, Z. Q. Xiang, J. Zhuang, S. J. Wu, B. S. Xing, Z. Zhang and Y. K. Rui, Front. Plant Sci., 2017, 8, 1322 CrossRef PubMed.
- W. H. Elmer and J. C. White, Environ. Sci.: Nano, 2016, 3, 1072–1079 RSC.
- O. Antonoglou, J. Moustaka, I. D. S. Adamakis, I. Sperdouli, A. A. Pantazaki, M. Moustakas and C. Dendrinou-Samara, ACS Appl. Mater. Interfaces, 2018, 10, 4450–4461 CrossRef CAS PubMed.
- S. Iravani, Green Chem., 2011, 13, 2638–2650 RSC.
- N. Otsuki, N. H. Dang, E. Kumagai, A. Kondo, S. Iwata and C. Morimoto, J. Ethnopharmacol., 2010, 127, 760–767 CrossRef PubMed.
- R. Sankar, P. Manikandan, V. Malarvizhi, T. Fathima, K. S. Shivashangari and V. Ravikumar, Spectrochim. Acta, Part A, 2014, 121, 746–750 CrossRef CAS PubMed.
- Y. P. Xie, Y. P. He, P. L. Irwin, T. Jin and X. M. Shi, Appl. Environ. Microbiol., 2011, 77, 2325–2331 CrossRef CAS PubMed.
- F. K. Alsammarraie, W. Wang, P. Zhou, A. Mustapha and M. Lin, Colloids Surf., B, 2018, 171, 398–405 CrossRef CAS PubMed.
- J. N. Chen, H. Peng, X. P. Wang, F. Shao, Z. D. Yuan and H. Y. Han, Nanoscale, 2014, 6, 1879–1889 RSC.
- J. N. Chen, S. L. Li, J. X. Luo, Y. Q. Zhang and W. Ding, J. Nanosci. Nanotechnol., 2017, 17, 186–195 CrossRef CAS PubMed.
- Y. Liu, D. S. Wu, Q. P. Liu, S. T. Zhang, Y. M. Tang, G. F. Jiang, S. L. Li and W. Ding, Eur. J. Plant Pathol., 2017, 147, 541–551 CrossRef CAS.
- X. G. Li, Z. Yan and J. P. Xu, Microbiology-Sgm, 2013, 149, 353–362 CrossRef PubMed.
- C. O'may and N. Tufenkji, Appl. Environ. Microbiol., 2011, 77, 3061–3067 CrossRef PubMed.
- S. B. Liu, T. H. Zeng, M. Hofmann, E. Burcombe, J. Wei, R. R. Jiang, J. Kong and Y. Chen, ACS Nano, 2011, 5, 6971–6980 CrossRef CAS PubMed.
- V. V. T. Padil and M. Cernik, Int. J. Nanomed., 2013, 8, 889–898 Search PubMed.
- M. Vaseem, A. Umar, S. H. Kim and Y. B. Hahn, J. Phys. Chem. C, 2008, 112, 5729–5735 CrossRef CAS.
- S. Patil, A. Sandberg, E. Heckert, W. Self and S. Seal, Biomaterials, 2007, 28, 4600–4607 CrossRef CAS PubMed.
- Z. S. Hong, Y. Cao and J. F. Deng, Mater. Lett., 2002, 52, 34–38 CrossRef CAS.
- M. Durando, R. Morrish and A. J. Muscat, J. Am. Chem. Soc., 2008, 130, 16659–16668 CrossRef CAS PubMed.
- J. Z. Xue, Z. H. Luo, P. Li, Y. P. Ding, Y. Cui and Q. S. Wu, Sci. Rep., 2014, 4, 5408 CrossRef CAS PubMed.
- L. Cai, J. N. Chen, Z. W. Liu, H. C. Wang, H. K. Yang and W. Ding, Front. Microbiol., 2018, 9, 790 CrossRef PubMed.
- D. Das, B. C. Nath, P. Phukon and S. K. Dolui, Colloids Surf., B, 2013, 111, 556–560 CrossRef CAS PubMed.
- A. Gaballa and J. D. Helmann, J. Bacteriol., 1998, 180, 5815–5821 CAS.
- M. Roselli, A. Finamore, I. Garaguso, M. S. Britti and E. Mengheri, J. Nutr., 2003, 133, 4077–4082 CrossRef CAS PubMed.
- P. C. Bogino, M. D. Oliva, F. G. Sorroche and W. Giordano, Int. J. Mol. Sci., 2013, 14, 15838–15859 CrossRef PubMed.
- B. E. Ramey, M. Koutsoudis, S. B. von Bodman and C. Fuqua, Curr. Opin. Microbiol., 2004, 7, 602–609 CrossRef CAS PubMed.
- Y. Mori, K. Inoue, K. Ikeda, H. Nakayashiki, C. Higashimoto, K. Ohnishi, A. Kiba and Y. Hikichi, Mol. Plant Pathol., 2016, 17, 890–902 CrossRef CAS PubMed.
- P. Bhattacharyya, B. Agarwal, M. Goswami, D. Maiti, S. Baruah and P. Tribedi, Antonie Van Leeuwenhoek International Journal Of General And Molecular Microbiology, 2018, 111, 89–99 CrossRef CAS PubMed.
- P. Singh, S. Pandit, J. Garnaes, S. Tunjic, V. R. S. S. Mokkapati, A. Sultan, A. Thygesen, A. Mackevica, R. V. Mateiu, A. E. Daugaard, A. Baun and I. Mijakovic, Int. J. Nanomed., 2018, 13, 3571–3591 CrossRef PubMed.
- E. Ertem, B. Gutt, F. Zuber, S. Allegri, B. Le Ouay, S. Mefti, K. Formentin, F. Stellacci and Q. Ren, ACS Appl. Mater. Interfaces, 2017, 9, 34762–34772 CrossRef CAS PubMed.
- E. Saile, J. A. McGarvey, M. A. Schell and T. P. Denny, Phytopathology, 1997, 87, 1264–1271 CrossRef CAS PubMed.
- J. Yao and C. Allen, J. Bacteriol., 2007, 189, 6415–6424 CrossRef CAS PubMed.
- J. Tans-Kersten, D. Brown and C. Allen, Mol. Plant-Microbe Interact., 2004, 17, 686–695 CrossRef CAS PubMed.
- A. L. Davidson and J. Chen, Annu. Rev. Biochem., 2004, 73, 241–268 CrossRef CAS PubMed.
- C. N. Lok, C. M. Ho, R. Chen, Q. Y. He, W. Y. Yu, H. Z. Sun, P. K. H. Tam, J. F. Chiu and C. M. Che, J. Proteome Res., 2006, 5, 916–924 CrossRef CAS PubMed.
- J. D. Moore, A. Avellan, C. W. Noack, Y. S. Guo, G. V. Lowry and K. B. Gregory, Environ. Sci.: Nano, 2017, 4, 321–2335 RSC.
- G. Singh, J. Beddow, C. Mee, L. Maryniak, E. M. Joyce and T. J. Mason, Int. J. Toxicol., 2017, 36, 478–484 CrossRef CAS PubMed.
- H. Q. Chen, B. Wang, D. Gao, M. Guan, L. N. Zheng, H. Ouyang, Z. F. Chai, Y. L. Zhao and W. Y. Feng, Small, 2013, 9, 2735–2746 CrossRef CAS PubMed.
- M. Raffi, S. Mehrwan, T. M. Bhatti, J. I. Akhter, A. Hameed, W. Yawar and M. M. ul Hasan, Ann. Microbiol., 2010, 60, 75–80 CrossRef CAS.
- D. Wang, L. X. Zhao, H. Y. Ma, H. Zhang and L. H. Guo, Environ. Sci. Technol., 2017, 51, 10137–10145 CrossRef CAS PubMed.
- C. V. Bamford, A. H. Nobbs, M. E. Barbour, R. J. Lamont and H. F. Jenkinson, Microbiology-Sgm, 2015, 161, 18–29 CrossRef CAS PubMed.
- L. R. Gu, T. Elkin, X. P. Jiang, H. P. Li, Y. Lin, L. W. Qu, T. R. J. Tzeng, R. Joseph and Y. P. Sun, Chem. Commun., 2005, 874–876 RSC.
- A. Rotini, A. Tornambe, R. Cossi, F. Iamunno, G. Benvenuto, M. T. Berducci, C. Maggi, M. C. Thaller, A. M. Cicero, L. Manfra and L. Migliore, Front. Microbiol., 2017, 8, 2076 CrossRef PubMed.
- M. A. Schell, Annu. Rev. Phytopathol., 2000, 38, 263–292 CrossRef CAS PubMed.
- G. A. O'Toole and R. Kolter, Mol. Microbiol., 1998, 30, 295–304 CrossRef.
- H. Liu, Y. Kang, S. Genin, M. A. Schell and T. P. Denny, Microbiology, 2001, 147, 3215–3229 CrossRef CAS PubMed.
- S. J. Clough, A. B. Flavier, M. A. Schell and T. P. Denny, Appl. Environ. Microbiol., 1997, 63, 844–850 CAS.
- R. Yu, X. H. Fang, P. Somasundaran and K. Chandran, Chemosphere, 2015, 128, 207–215 CrossRef CAS PubMed.
- K. K. Mondal and C. Mani, Ann. Microbiol., 2012, 62, 889–893 CrossRef CAS.
- K. Giannousi, I. Avramidis and C. Dendrinou-Samara, RSC Adv., 2013, 3, 21743–21752 RSC.
- J. Montag, L. Schreiber and J. Schonherr, J. Agric. Food Chem., 2006, 54, 893–899 CrossRef CAS PubMed.
- P. Patra, S. Mitra, N. Debnath and A. Goswami, Langmuir, 2012, 28, 16966–16978 CrossRef CAS PubMed.
- S. M. Ouda, Res. J. Microbiol., 2014, 9, 34–42 CrossRef CAS.
- M. Shah, D. Fawcett, S. Sharma, S. K. Tripathy and G. E. J. Poinern, Materials, 2015, 8, 7278–7308 CrossRef CAS PubMed.
- M. Valodkar, R. N. Jadeja, M. C. Thounaojam, R. V. Devkar and S. Thakore, Mater. Chem. Phys., 2011, 128, 83–89 CrossRef CAS.
- C. O. Dimkpa, J. E. McLean, D. W. Britt and A. J. Anderson, Ind. Biotechnol., 2012, 8, 344–357 CrossRef CAS.
- W. M. Lee, J. I. Kwak and Y. J. An, Chemosphere, 2012, 86, 491–499 CrossRef CAS PubMed.
- I. Ocsoy, M. L. Paret, M. A. Ocsoy, S. Kunwar, T. Chen, M. X. You and W. H. Tan, ACS Nano, 2013, 7, 8972–8980 CrossRef CAS PubMed.
- J. N. Chen, L. Sun, Y. Cheng, Z. C. Lu, K. Shao, T. T. Li, C. Hu and H. Y. Han, ACS Appl. Mater. Interfaces, 2016, 8, 24057–24070 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8ra09186b |
| This journal is © The Royal Society of Chemistry 2019 |