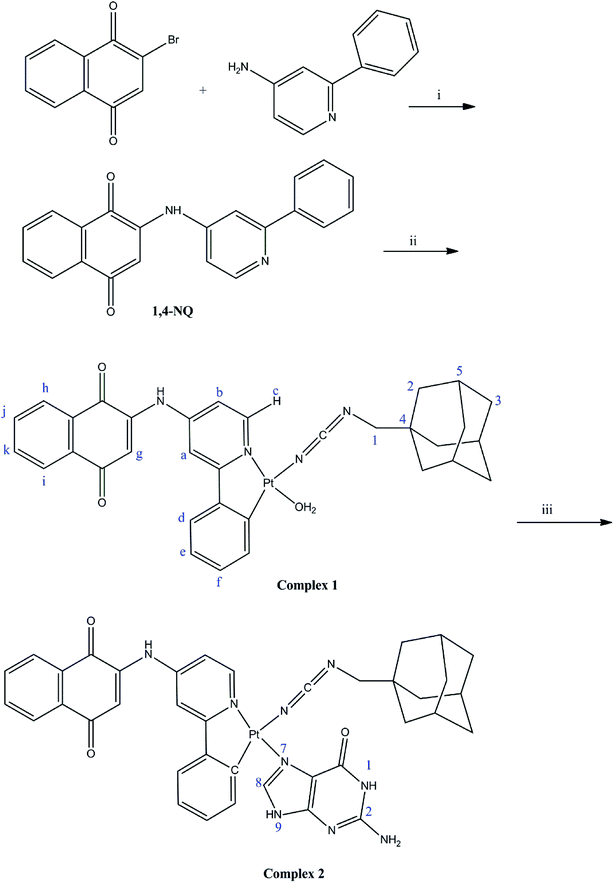
Open Access Article
Open Access ArticleExperimental and theoretical investigation of cyclometallated platinum(II) complex containing adamantanemethylcyanamide and 1,4-naphthoquinone derivative as ligands: synthesis, characterization, interacting with guanine and cytotoxic activity†
Leila Tabrizi‡
 *a,
Bachir Zouchoune‡*bc and
Abdallah Zaiterc
*a,
Bachir Zouchoune‡*bc and
Abdallah Zaiterc
aSchool of Chemistry, National University of Ireland Galway, University Road, Galway, Ireland H91 TK33. E-mail: LEILA.TABRIZI@nuigalway.ie
bLaboratoire de Chimie appliquée et Technologie des Matériaux, Université Larbi Ben M'Hidi – Oum El Bouaghi, 04000 Oum El Bouaghi, Algeria. E-mail: bzouchoune@gmail.com
cUnité de Recherche de Chimie de l'Environnement et Moléculaire Structurale, Université Constantine (Mentouri), 25000 Constantine, Algeria
First published on 2nd January 2019
Abstract
A new cyclometallated platinum(II) complex with 1-adamantanemethylcyanamide (1-ADpcydH) and 2-[amino(2-phenylpyridine)]-1,4-naphthoquinone (1,4-NQ) ligands with the formula cis-Pt(1,4-NQ)(1-ADpcyd)(H2O) was synthesized and fully characterized. Cellular uptake, DNA platination, and cytotoxicity against human MCF-7 breast, HepG-2 liver hepatocellular carcinoma, and HT-29 colon cancer cell lines were evaluated. The interaction of guanine (G) with cis-Pt(1,4-NQ)(1-ADpcyd)(H2O) was studied by 195Pt NMR and mass spectroscopy. Furthermore, DFT calculations were performed on the complexes cis-Pt(1,4-NQ)(1-ADpcyd)(H2O) 1 and cis-Pt(1,4-NQ)(1-ADpcyd)(G) 2 using the BP86-D and B3LYP functionals, in order to gain deeper insights into the molecular and electronic structures. Decomposition energy analysis gave a clear understanding of the bonding within both complexes, showing that the interactions were governed by two-third ionic and one-third covalent characters, which were stronger between the guanine and the Pt(II) center than those between water and the Pt(II).
1. Introduction
Recently, cyclometallated platinum complexes containing N-donor ligands have been the object of much research attention due to interest in their biological activities.1–9 The antitumor activity of different platinum(II) complexes is associated with the platination of DNA, generally through binding to guanine. Hence, anticancer platinum(II) complexes have been investigated with guanine derivatives in theoretical and experimental studies and they have shown a high selectivity for binding to N7 of guanine.10–221,4-Naphthoquinone and its derivatives are attracting great attention as they show important properties that would be useful in many biological processes.23–27 Recently, naphthoquinone–platinum(II) complexes were studied as anticancer drugs and showed significant biological activity and DNA-binding as a result of coordination of the quinone to Pt(II).28
Adamantane derivatives are usually regarded as a ‘lipophilic bullet’ for improving the lipophilicity in biological activities with potential anticancer activity.29–35 Recently, we focused attention on the development of Ni(II), Co(II), Ru(II), Ir(III), Pt(II), Pd(II), and triorganotin(IV) complexes containing phenylcyanamide derivatives as monodentate ligands with biological properties.36–43 Specifically, we synthesized and used 1-adamantanemethylcyanamide (1-ADpcydH) as a monodentate ligand for a platinum(II) complex.
We here report the synthesis and characterization of a new cyclometallated platinum(II) complex of 1-adamantanemethylcyanamide (1-ADpcydH) and 2-[amino(2-phenylpyridine)]-1,4-naphthoquinone (1,4-NQ) ligands with the formula cis-Pt(1,4-NQ)(1-ADpcyd)(H2O). The cellular uptake, DNA platination, and cytotoxicity against human MCF-7 breast, HepG-2 liver hepatocellular carcinoma, and HT-29 colon cancer cell lines were evaluated. Moreover, this complex was studied by theoretical calculations and in an experimental evaluation for its interaction with guanine. The theoretical study was performed on both complexes cis-Pt(1,4-NQ)(1-ADpcyd)(H2O), 1, and cis-Pt(1,4-NQ)(1-ADpcyd)(G), 2, in order to obtain information about their molecular structures by analyzing their molecular orbitals, electronic structures, natural charges, Wiberg bond indices (WBI), and interaction energies using the BP86-D and B3LYP methods,44–52 which have proved their reliability for metallic complexes in many previous studies. We endeavor herein to compare our results with those available experimentally in the literature of related systems.
2. Experimental
2.1. Materials and methods
All reagents and solvents were commercially available and used without further purification. The 1-adamantanemethylcyanamide (1-ADpcydH) ligand was synthesized as previously reported.53 Cis-Pt(DMSO)2Cl2 was purchased from Sigma-Aldrich and used without further purification. 1H, 13C {1H} nuclear magnetic resonance (NMR) spectra were recorded on a Bruker-400 MHz spectrometer at ambient temperature in DMSO-d6. 195Pt NMR spectra were recorded on a Varian 500 AR spectrometer in DMF-d7. ESI mass spectra were recorded on a Waters LCT Premier XE spectrometer in positive- or negative-ion mode using MeOH/H2O as the solvent. Elemental analyses were performed with an EA 3000 CHNS. Fourier transform infrared spectra were recorded on a FT-IR JASCO FT-IR Jasco 680 Plus spectrometer in the region of 4000–400 cm−1 using KBr pellets. pH measurements were made using a Corning 240 pH meter equipped with an Aldrich micro-combination electrode calibrated with Aldrich standard buffer solutions of pH 4, 7, and 10.2.2. Synthesis of the ligand 1-adamantanemethylcyanamide (1-ADpcydH)
The 1-adamantanemethylcyanamide (1-ADpcydH) ligand was synthesized from 1-adamantanemethylamine as previously reported.53 The pure compound was obtained via re-crystallization from an acetonitrile: pentane (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5, volume ratio) solution as yellow needles (0.138 g, 72.63% yield, and 1 mmol). Anal. calc. (%) for C12H18N2 (190.2847): C, 75.74; H, 9.53; N, 14.72; found (%): C, 75.69; H, 9.51; N, 14.73. TOF-MS: 191.2927 [M + H]+. FT-IR: ν 2238 (m, NCNH) cm−1. 1H NMR (DMSO-d6): δ 5.59 (s, 1H, H–NCNH), 2.93 (d, 2H, H-1, 3J 7.2), 1.99 (broad, 3H, H-5), 1.78 (q, 6H, H-3, 3J 7.2), 1.41 (d, 6H, H-2, 3J 7.2). 13C NMR (DMSO-d6): δ 28.5 (C-5), 33.0 (C-4), 35.8 (C-3), 41.1 (C-2), 50.9 (C-1), 94.5 (HNCN).
5, volume ratio) solution as yellow needles (0.138 g, 72.63% yield, and 1 mmol). Anal. calc. (%) for C12H18N2 (190.2847): C, 75.74; H, 9.53; N, 14.72; found (%): C, 75.69; H, 9.51; N, 14.73. TOF-MS: 191.2927 [M + H]+. FT-IR: ν 2238 (m, NCNH) cm−1. 1H NMR (DMSO-d6): δ 5.59 (s, 1H, H–NCNH), 2.93 (d, 2H, H-1, 3J 7.2), 1.99 (broad, 3H, H-5), 1.78 (q, 6H, H-3, 3J 7.2), 1.41 (d, 6H, H-2, 3J 7.2). 13C NMR (DMSO-d6): δ 28.5 (C-5), 33.0 (C-4), 35.8 (C-3), 41.1 (C-2), 50.9 (C-1), 94.5 (HNCN).
2.3. Synthesis of the ligand 2-[amino(2-phenylpyridine)]-1,4-naphthoquinone (1,4-NQ)
A solution of 2-phenyl-pyridin-4-ylamine (0.171 g, 1 mmol) and triethylamine (697 μL, 5 mmol) in 20 mL tetrahydrofuran was added dropwise to a solution of 2-bromo-1,4-naphthoquinone (0.237 g, 1 mmol) in 20 mL tetrahydrofuran. The orange solution was stirred for 24 h at room temperature. The resulting precipitate was removed by filtration and washed with diethylether. The resulting material was purified by silica gel column chromatography (1% methanol/dichloromethane) to provide the product as orange solids (0.221 g, 67.78% yield, and 1 mmol). Anal. calc. (%) for C21H14N2O2 (326.3481): C, 77.29; H, 4.32; N, 8.58; found (%): C, 77.22; H, 4.30; N, 8.54. TOF-MS: 327.1134 [M + H]+. FT-IR: ν 1712, 1656 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretch) cm−1. 1H NMR (DMSO-d6): δ 9.99 (s, 1H, NH), 8.30 (d, 1H, H-b, 3J = 7.2 Hz), 7.94–8.00 (m, 4H, H-h,i,e), 7.81 (t, 1H, H-k, 3J = 7.2 Hz), 7.74 (t, 1H, H-j, 3J = 7.2 Hz), 7.44–7.50 (m, 3H, H-d,f), 6.97 (s, 1H, H-a), 6.45 (d, 1H, H-c, 3J = 7.2 Hz), 5.83 (s, 1H, H-g). 13C NMR (DMSO-d6): δ 182.0 (C
O stretch) cm−1. 1H NMR (DMSO-d6): δ 9.99 (s, 1H, NH), 8.30 (d, 1H, H-b, 3J = 7.2 Hz), 7.94–8.00 (m, 4H, H-h,i,e), 7.81 (t, 1H, H-k, 3J = 7.2 Hz), 7.74 (t, 1H, H-j, 3J = 7.2 Hz), 7.44–7.50 (m, 3H, H-d,f), 6.97 (s, 1H, H-a), 6.45 (d, 1H, H-c, 3J = 7.2 Hz), 5.83 (s, 1H, H-g). 13C NMR (DMSO-d6): δ 182.0 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 180.0 (C
O), 180.0 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 158.0 (Ar), 153.5 (Ar), 149.9 (Ar), 138.0 (Ar), 134.5 (Ar), 132.1 (Ar), 128.9 (Ar), 126.8 (Ar), 124.9 (Ar), 123.0 (Ar), 108.8 (C-b), 106.0 (C-e), 102.5 (C-g).
O), 158.0 (Ar), 153.5 (Ar), 149.9 (Ar), 138.0 (Ar), 134.5 (Ar), 132.1 (Ar), 128.9 (Ar), 126.8 (Ar), 124.9 (Ar), 123.0 (Ar), 108.8 (C-b), 106.0 (C-e), 102.5 (C-g).
2.4. Synthesis of the complex 1, cis-Pt(1,4-NQ)(1-ADpcyd)(H2O)
The ligands of 1-ADpcydH (190 mg, 1 mmol) and 1,4-NQ (326 mg, 1 mmol) in 20 mL ethanol/water (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, volume ratio) were added to a stirred solution of cis-[PtCl2(DMSO)2] (423 mg, 1 mmol) in 10 mL ethanol. A saturated solution of KOH in water was added dropwise to the solution until the pH value was 8.5. The resulting solution was stirred for 72 h at room temperature. Then, the reaction mixture was filtered and the resulting solution was evaporated under reduced pressure and dissolved in acetonitrile (10 mL). The addition of n-pentane (40 mL) caused the precipitation of a brown solid. The solid was washed with diethyl ether (3 × 10 mL) and dried under vacuum (0.409 g, 56.31% yield, and 1 mmol). Anal. calc. (%) for C33H32N4O3Pt (727.7162): C, 54.47; H, 4.43; N, 7.70; found (%): C, 54.43; H, 4.41; N, 7.66. TOF-MS: 750.7059 [M + Na]+. FT-IR: ν 1689, 1632 (C
1, volume ratio) were added to a stirred solution of cis-[PtCl2(DMSO)2] (423 mg, 1 mmol) in 10 mL ethanol. A saturated solution of KOH in water was added dropwise to the solution until the pH value was 8.5. The resulting solution was stirred for 72 h at room temperature. Then, the reaction mixture was filtered and the resulting solution was evaporated under reduced pressure and dissolved in acetonitrile (10 mL). The addition of n-pentane (40 mL) caused the precipitation of a brown solid. The solid was washed with diethyl ether (3 × 10 mL) and dried under vacuum (0.409 g, 56.31% yield, and 1 mmol). Anal. calc. (%) for C33H32N4O3Pt (727.7162): C, 54.47; H, 4.43; N, 7.70; found (%): C, 54.43; H, 4.41; N, 7.66. TOF-MS: 750.7059 [M + Na]+. FT-IR: ν 1689, 1632 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretch), 2146 (m, NCN) cm−1. 1H NMR (DMSO-d6): δ 9.89 (s, 1H, NH), 8.49 (d, 1H, H-c, 3J = 7.2 Hz), 8.20 (d, 1H, H-b, 3J = 7.2 Hz), 8.00–8.09 (m, 4H, H-h,i,e), 7.75–7.80 (m, 2H, H-k,j), 7.24–7.29 (m, 2H, H-d,f), 6.81 (s, 1H, H-a), 5.79 (s, 1H, H-g), 3.64 (coordinated water), 2.79 (d, 2H, H-1, 3J 7.2), 1.91 (broad, 3H, H-5), 1.67 (q, 6H, H-3, 3J 7.2), 1.13 (d, 6H, H-2, 3J 7.2). 13C NMR (DMSO-d6): δ 181.1 (C
O stretch), 2146 (m, NCN) cm−1. 1H NMR (DMSO-d6): δ 9.89 (s, 1H, NH), 8.49 (d, 1H, H-c, 3J = 7.2 Hz), 8.20 (d, 1H, H-b, 3J = 7.2 Hz), 8.00–8.09 (m, 4H, H-h,i,e), 7.75–7.80 (m, 2H, H-k,j), 7.24–7.29 (m, 2H, H-d,f), 6.81 (s, 1H, H-a), 5.79 (s, 1H, H-g), 3.64 (coordinated water), 2.79 (d, 2H, H-1, 3J 7.2), 1.91 (broad, 3H, H-5), 1.67 (q, 6H, H-3, 3J 7.2), 1.13 (d, 6H, H-2, 3J 7.2). 13C NMR (DMSO-d6): δ 181.1 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 179.9 (C
O), 179.9 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 158.1 (Ar), 153.4 (Ar), 150.0 (Ar), 138.5 (Ar), 135.0 (Ar), 132.9 (Ar), 130.0 (Ar), 127.0 (Ar), 124.0 (Ar), 122.0 (Ar), 112.0 (NCN), 109.9 (C-b), 105.8 (C-e), 103.2 (C-g), 47.91 (C-1), 40.9 (C-2), 34.1 (C-3), 31.0 (C-4), 27.0 (C-5). 195Pt{1H} NMR (107.6 MHz, DMF-d7): δ −2794 ppm.
O), 158.1 (Ar), 153.4 (Ar), 150.0 (Ar), 138.5 (Ar), 135.0 (Ar), 132.9 (Ar), 130.0 (Ar), 127.0 (Ar), 124.0 (Ar), 122.0 (Ar), 112.0 (NCN), 109.9 (C-b), 105.8 (C-e), 103.2 (C-g), 47.91 (C-1), 40.9 (C-2), 34.1 (C-3), 31.0 (C-4), 27.0 (C-5). 195Pt{1H} NMR (107.6 MHz, DMF-d7): δ −2794 ppm.
2.5. Synthesis of the complex 2, cis-Pt(1,4-NQ)(1-ADpcyd)(G): interaction of cis-Pt(II) complex 1 with guanine
Complex 1 (1 mmol) and guanine (5 mmol) were dissolved in 5 mL DMF and stirred at 40 °C for 24 h. The reaction was followed by l95Pt NMR spectroscopy. The 195Pt NMR spectra were recorded in DMF with an insert D2O tube. The pH of the reaction mixtures was monitored during the reaction and varied from 6.4 to 7.2. After 24 h, the unreacted guanine was removed by filtration. The solution was rotary-evaporated to dryness to give the crude product. The residue was dissolved in acetonitrile (10 mL) and n-hexane (50 mL) was added to produce the precipitation of light brown solids (0.754 g, 87.65% yield, and 1 mmol). Anal. calc. (%) for C38H35N9O3Pt (860.8270): C, 53.02; H, 4.10; N, 14.64; found (%): C, 52.98; H, 4.08; N, 14.61. TOF-MS: 883.8168 [M + Na]+. 1H NMR (DMSO-d6): δ 11.29 (s, 1H, NH1), 10.90 (s, 1H, NH9), 9.89 (s, 1H, NH), 8.49 (d, 1H, H-c, 3J = 7.2 Hz), 8.34 (s, 1H, H-8), 8.20 (d, 1H, H-b, 3J = 7.2 Hz), 8.00–8.09 (m, 4H, H-h,i,e), 7.75–7.80 (m, 2H, H-k,j), 7.24–7.29 (m, 2H, H-d,f), 6.92 (s, 2H, H–NH2), 6.81 (s, 1H, H-a), 5.79 (s, 1H, H-g), 2.79 (d, 2H, H-1, 3J 7.2), 1.91 (broad, 3H, H-5), 1.67 (q, 6H, H-3, 3J 7.2), 1.13 (d, 6H, H-2, 3J 7.2). 13C NMR (DMSO-d6): δ 181.0 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 178.6 (C
O), 178.6 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 158.0 (Ar), 156.1, 154.0 (Ar), 151.9 (Ar), 149.9 (Ar), 138.6 (Ar), 136.6 (Ar), 134.9 (Ar), 132.1 (Ar), 129.9 (Ar), 126.6 (Ar), 123.5 (Ar), 122.0 (Ar), 112.1 (NCN), 109.9 (C-b), 107.2, 105.5 (C-e), 102.9 (C-g), 48.0 (C-1), 41.0 (C-2), 34.2 (C-3), 31.0 (C-4), 27.1 (C-5). 195Pt{1H} NMR (107.6 MHz, DMF-d7): δ −3229 ppm.
O), 158.0 (Ar), 156.1, 154.0 (Ar), 151.9 (Ar), 149.9 (Ar), 138.6 (Ar), 136.6 (Ar), 134.9 (Ar), 132.1 (Ar), 129.9 (Ar), 126.6 (Ar), 123.5 (Ar), 122.0 (Ar), 112.1 (NCN), 109.9 (C-b), 107.2, 105.5 (C-e), 102.9 (C-g), 48.0 (C-1), 41.0 (C-2), 34.2 (C-3), 31.0 (C-4), 27.1 (C-5). 195Pt{1H} NMR (107.6 MHz, DMF-d7): δ −3229 ppm.
2.6. Stability tests
The stability of complex 1 was tested by dissolving it in PBS buffer/0.5% DMSO and keeping for 72 h at 37 °C. Briefly, 10 μL of the solution was injected into an HPLC system (Thermo, USA) connected to a UV/Vis spectrophotometer. A Hypersil Gold Dim (100 × 2.1 mm, Thermo, USA) reversed-phase column was used at a flow rate of 0.7 mL min −1.2.7. Cell culture
Human MCF-7 breast, HepG-2 liver hepatocellular carcinoma, and HT-29 colon cancer cell lines were obtained from the American Type Culture Collection (ATCC, USA). All reagents and cell culture media were purchased from Gibco Company (Germany). The cells were maintained in Dulbecco's Modified Eagle Medium (DMEM) supplemented with 10% FBS, 100 IU mL−1 of penicillin, 100 μg mL−1 of streptomycin, and 2 mM of Glutamax at 37 °C in a humidified incubator at 5% CO2. The adherent cultures were grown as monolayers and were passaged once in 4–5 days by trypsinizing them with 0.25% trypsin–EDTA.2.8. Cytotoxicity
The growth inhibitory effect toward tumor cells was evaluated by means of the MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) tetrazolium reduction assay.54 Stock solutions of free ligands and complex 1 were prepared by dissolving the compounds in DMSO (0.008 mM) that for cisplatin was prepared by dissolving in RPMI-1640 medium. The stocks were further diluted with the respective medium containing 10% FBS (0.02–10 μM) before addition to the cells. Cells were trypsinized with 0.25% trypsin–EDTA and counted with 0.4% trypan blue. The cells were seeded at a concentration of 7500 cells/100 μL per well and grown for 24 h at 37 °C in a humidified incubator. After 24 h, the medium was removed and replaced with fresh medium containing the compound to be studied at the appropriate concentration. Triplicate cultures were established for each treatment. After 72 h, each well was treated with 10 μL of a 5 mg mL−1 MTT saline solution, and, following 5 h of incubation, 100 μL of a sodium dodecylsulfate solution in HCl (0.01 M) was added. The optical absorbance of each well (96-well plates) was quantified using EnVision multilabel plate readers (PerkinElmer, Waltham, MA, USA) at 450 nm wavelength. The mean absorbance values were then calculated and exported to Graphpad Prism version 6.0 for Windows and plotted in the logarithmic form to determine the IC50 values. The IC50 value was calculated as the concentration reducing the proliferation of the cells by 50% and is presented herein as the mean (±SE) of three independent experiments each with triplicates. P < 0.05 was considered to be statistically significant.2.9. Cellular platinum accumulation
Cellular accumulation of complex 1 and cisplatin was measured in HT-29 cells. 1 × 106 cells were seeded in 60 mm tissue culture dishes. After incubation overnight at 37 °C, the cells were treated with complex 1 or cisplatin for 1 h at a final concentration of 10 μM at 0.5% DMSO. The cell monolayers at the end of the incubation with complex 1 or cisplatin were washed twice with PBS (phosphate buffered saline), trypsinized, and harvested into appropriate media. Cell suspensions were centrifuged, the pellets were digested with 12 M HCl, and the platinum content was analyzed by ICP-MS. All the experiments were performed in triplicate.2.10. DNA platination
DNA platination was performed by using complex 1 and cisplatin (10 μM) in HT-29 cells as previously reported.392.11. Computational details
Density functional theory (DFT) calculations were carried out using the 2014.01 version of the Amsterdam Density Functional (ADF) program55 developed by Baerends and co-workers.56–60 Electron correlation was treated within the local density approximation (LDA) in the Vosko–Wilk–Nusair parametrization.61 The DFT-D method consisting of BP86-D62 was used for all calculations to compensate for the incapacity of the generalized gradient approximation BP86 (ref. 63–66) functional to describe the dispersion effects correctly besides the hybrid-type B3LYP.67,68 The atom electronic configurations were described by a triple-ζ Slater-type orbital (STO) basis set for H 1s, C 2s and 2p, N 2s and 2p, O 2s and 2p augmented with a 3d single-ζ polarization for C, N, and O atoms and with a 2p single-ζ polarization for H atoms. A triple-ζ STO basis set was used for the Pt 5d and 6s augmented with a single-ζ 6p polarization function for Pt. For the heavy Pt atom, the scalar relativistic zero-order regular approximation (ZORA) was used, with the associated optimized valence basis set. The BP86-D calculations were performed assuming the frozen-core approximation up to 1s for C, 3p for the first-row metals, and 4f for Pt.56–60 Full geometry optimizations were carried out using the analytical gradient method implemented by Versluis and Ziegler.69 Vibrational frequency calculations were performed on all the optimized geometries to verify that these structures were true minima on the potential energy surface.70,71 All the energy values reported in the ESI† include zero-point energy correction. Representations of the orbitals and of the molecular structures were done by using the ADF-GUI55 and the MOLEKEL4 programs,72 respectively. The natural population-based and Wiberg bond indices (NPAI and WBI, respectively)73 were obtained from calculations implemented in the NBO 6.0 program.74,753. Results and discussion
3.1. Synthesis and characterization
Complex 1 was prepared in high yield by the 1-ADpcydH and 1,4-NQ ligands and complex cis-[Pt(DMSO)2Cl2] in ethanol at pH ∼8.5 (Scheme 1). The cis-Pt(II) complex 1 was a brown solid and was soluble in acetonitrile, ethanol, chloroform, acetone, DMF, and DMSO.The 1-ADpcydH and 1,4-NQ ligands and cis-Pt(II) complex 1 were characterized by elemental analyses, and IR, multinuclear NMR (1H, 13C, and 195Pt), and mass spectrometry. The ESI-TOF MS spectra of 1-ADpcydH and 1,4-NQ showed peaks centered at m/z = 191.2927 and 327.1134, corresponding to [M + H]+. Furthermore, the ESI-TOF MS spectra of cis-Pt(II) complex 1 showed peaks centered at m/z = 727.7162 with the typical platinum isotope pattern, corresponding to [M + Na]+ (Fig. S1–S3†).
The ν (NCN) band of 1-ADpcydH ligand was observed at 2238 cm−1, which was shifted to lower frequency, 2146 cm−1, upon the coordination of cyanamide through the cyano nitrogen to Pt(II) in the cis-Pt(II) complex 1. Deprotonation of the cyanamide group shifted ν (NCN) to lower frequencies, such as carbodiimides, meaning form B was the main involvement with the cyanamide anion group. The resonance form A had a nitrile stretching frequency similar to the neutral cyanamide (Scheme 2).39 Complex 1 showed ν(OH) and γ(H2O) bands at 3446 and 1141 cm−1, respectively, due to the coordinated water.76
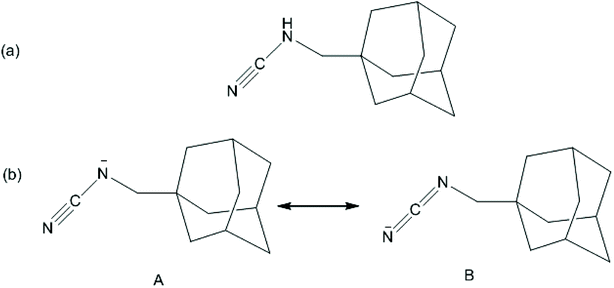 | ||
| Scheme 2 (a) The structure of 1-adamantanemethylcyanamide (1-ADpcydH). (b) Resonance structures of the 1-adamantanemethylcyanamide anionic form. | ||
The 1HNMR spectra of the free ligands and cis-Pt(II) complex 1 are shown in Fig. S4–S6.† The NH proton of 1-ADpcydH ligand at 5.59 ppm disappeared in the cis-Pt(II) complex 1 due to the coordination of the cyanamide group to Pt(II) in complex 1. The proton H–C of 1,4-NQ resonances underwent the largest shift from 6.45 ppm in the free ligand 1,4-NQ to 8.49 ppm in Pt(II) complex 1, indicating a strong relationship with the geometry of the molecule. Such a quite large downfield shift originated from the shielding exerted on H–C by the N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C
C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N bond of 1-ADpcydH and this was possible only in the cis isomer (Scheme 1). Therefore, the cis configuration of the Pt(II) complex 1 could be carefully inferred. In the 1HNMR spectrum of the cis-Pt(II) complex 1, the other signals were shifted, with respect to the free ligands, which confirmed the formation of Pt(II) complex.
N bond of 1-ADpcydH and this was possible only in the cis isomer (Scheme 1). Therefore, the cis configuration of the Pt(II) complex 1 could be carefully inferred. In the 1HNMR spectrum of the cis-Pt(II) complex 1, the other signals were shifted, with respect to the free ligands, which confirmed the formation of Pt(II) complex.
In the 13C spectra of the cis-Pt(II) complex 1, the signals were shifted compare to the free ligands (Fig. S7–S9†).
The 195Pt NMR of cis-Pt(II) complex 1 revealed a singlet at −2794 ppm, indicating the presence of a single solution species, and this deduction was individually confirmed by 1H and 13C NMR spectroscopy. Moreover, this chemical shift range was assignable to the platinum(II) complexes having N^N^C^O as ligands (Fig. S10†).77
3.2. Interaction cis-Pt(II) complex 1 with guanine (G)
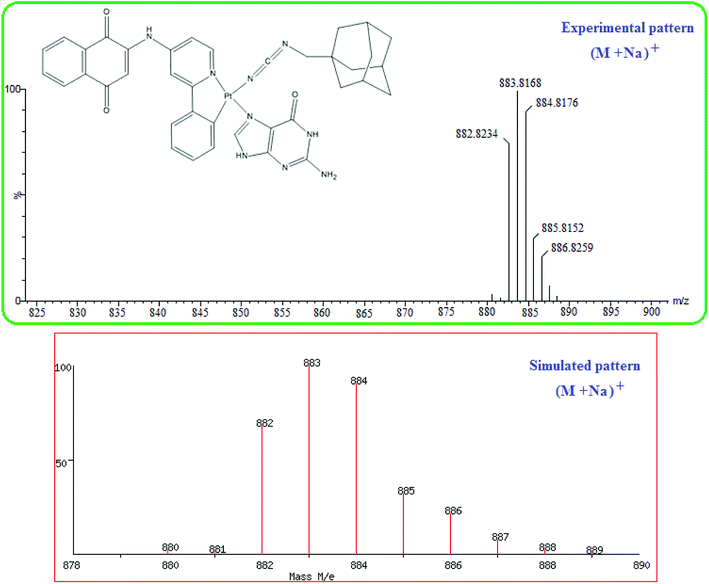
The activity of cisplatin is mostly, in terms of DNA damage, affected through its ability to bind to the N7 position of guanine bases. Therefore, the reaction of complex 1 at micromolar concentrations with 5 equiv. of guanine was studied. During the reaction, the pH of the unbuffered reaction mixtures differed between 6.4 and 7.2, approximately close to physiological conditions. The reaction was followed by 195Pt NMR spectroscopy (Fig. 1). Comparison of the 195Pt NMR spectra obtained during the reaction with that at t = 0 confirmed the binding of the Pt(II) complex 1 to the N7 guanine. At t = 0, the 195Pt NMR spectra of 1 revealed one peak at −2796 ppm, indicating the presence of a single solution species. As the reaction proceeded, 195Pt NMR of reaction showed two solution species at −2796 and −3230 ppm, where the signal at −2796 ppm decreased and the new signal at −3230 ppm increased in intensity. Furthermore, the signal at −3230 ppm could be assigned to the chemical shift range of platinum(II) complexes having N^N^N^C as ligands.78 The chemical shift was consistent with substitution of O of H2O with N7 of guanine. After 24 h, the final product showed only one signal at −3230 ppm. The final product, cis-Pt(1,4-NQ)(1-ADpcyd)(G), 2, was also confirmed by mass (Fig. 2) and 1H and 13C NMR spectra (Fig. S11 and S12†). The mass spectrum exhibited peaks centered at m/z = 883.8168 with the typical platinum isotope pattern corresponding to [M + Na]+. The 1H spectrum of complex 2 revealed one major aromatic H8 guanine as a singlet at 8.34 ppm and two singlets at 10.90 and 11.29 ppm for NH9 and NH1 guanine, respectively. The 195Pt NMR of cis-Pt(II) complex 2 revealed a singlet at −3229 ppm (Fig. S13†).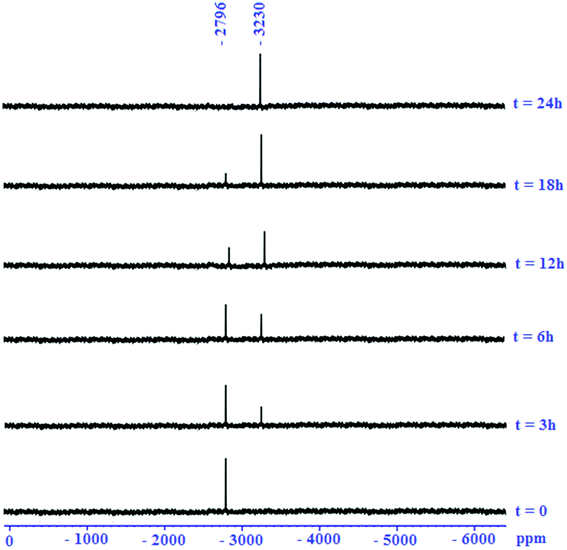 | ||
| Fig. 1 Reaction of cis-Pt(II) complex 1 with guanine as followed by l95Pt NMR spectroscopy (conditions as mentioned in the Experimental section). | ||
3.3. Stability of complex 1
The stability of the cis-Pt(II) complex 1 in PBS/DMSO (0.5%) solution was studied by HPLC-UV at 37 °C. The results confirmed that complex 1 was stable in physiological conditions over 72 h (Fig. S14†).3.4. In vitro cytotoxicity studies
The effect of the free ligands 1-ADpcydH and 1,4-NQ and also complex 1 on a panel of human cancer cell lines, including MCF-7 breast, HepG-2 liver hepatocellular carcinoma, and HT-29 colon adenocarcinoma, was examined using the MTT assay. The cytotoxicity parameters, in terms of IC50 obtained after 72 h exposure, are presented in Table 1 and Fig. S15.† Complex 1 exhibited lower IC50 values (IC50 0.56–7.25 μM) against MCF-7, HepG-2, and HT-29 than the corresponding 1-ADpcydH and 1,4-NQ ligands and also than cisplatin. Remarkably, the lowest IC50 value for complex 1 against HT-29 was 0.56 μM, which displayed approximately a 4.5-fold increase compared with cisplatin, thus indicating its higher potency. The cytotoxicity of complex 1 against the HepG-2 was higher than that of the platinum(II) complexes with 8-hydroxyquinoline (IC50 7.79–18.54 μM).79| Compounds | IC50 (μM) | ||
|---|---|---|---|
| HepG-2 | HT-29 | MCF-7 | |
| a The compounds were dissolved in 0.1% of DMSO and diluted with water, while cisplatin was dissolved in water. Cells were treated for 72 h with increasing concentrations of the tested compounds. IC50 values were calculated as mean values obtained from three independent experiments and presented with their standard deviations. | |||
| >100 | >100 | >100 | 1-ADpcydH |
| >100 | >100 | >100 | 1,4-NQ |
| 7.25 ± 0.05 | 0.56 ± 0.03 | 1.52 ± 0.05 | 1 |
| 8.32 ± 0.08 | 2.52 ± 0.05 | 7.35 ± 0.07 | Cisplatin |
3.5. Intracellular platinum accumulation in HT-29 cells
The cellular accumulation of the cis-Pt(II) complex 1 was studied and compared to that of cisplatin. The amount of Pt associated was determined using ICP-MS with HT-29 cells incubated after 1 h with complex 1 or cisplatin. It was found that complex 1 was highly accumulated when compared to cisplatin in HT-29 cell lines. The accumulation of complex 1 in HT-29 cells was 12.25 ± 0.10 fmol of Pt(II), which was greater than that of cisplatin, containing 0.26 ± 0.05 fmol of Pt(II).3.6. DNA platination
As the main target for the Pt(II) complexes within the cell is the nuclear DNA, so the DNA binding of complex 1 in HT-29 cell line was studied and compared with cisplatin (Fig. 3). In effect, complex 1 showed a high degree of DNA platination in the HT-29 cell line compared with cisplatin, which was in agreement with the accumulation data.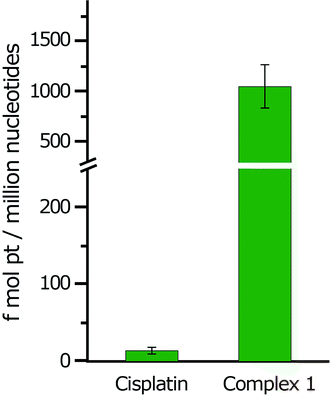 | ||
| Fig. 3 Intracellular DNA platination of cisplatin and complex 1 in the HT-29 cell line after 1 h incubation at 10 μM final concentration. | ||
Overall, the high levels of complex 1 bound to the DNA in HT-29 cell and interaction with guanine suggest that the direct attack of nucleotides on cis-Pt(II) complex 1 may be of importance in the mechanism of action for this Pt(II) complex.
3.7. Theoretical investigation
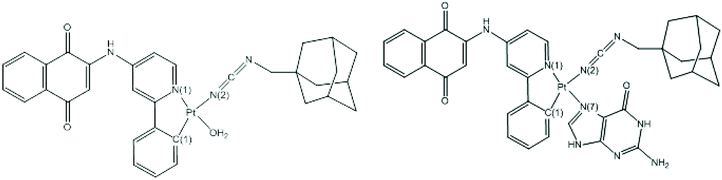
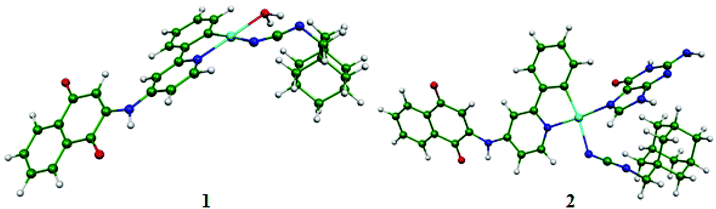
The full geometry optimizations without symmetry constraints were carried out on the neutral 1 and 2 complexes having 16-MVE (metal valence electrons) in their singlet and triplet spin state to complement the experimental studies. The triplet structures were higher in energy than their corresponding singlet structures by about 40 kcal mol−1; thus they are not discussed within this study. The Pt–N(1), Pt–N(2) and Pt–C(1) bond distances in both investigated complexes were comparable (Table 2). Indeed, for complex 1, the Pt–O(H2O) bond length of 2.253 (BP86-D) or 2.124 Å (B3LYP), respectively, was nearly similar to the experimental ones of 2.149 Å, while the Pt–N(1), Pt–N(2) and Pt–C(1) were 2.108, 2.286, and 2.059 Å (BP86-D) or 2.003, 2.223, and 1.995 Å (B3LYP), which were longer (BP86-D) and comparable (B3LYP) to those of the experimental values of related Pt(II) systems.80–83 For complex 2, the Pt–N(7) bond length was 2.146 (BP86-D) or 2.052 Å, where the B3LYP value was comparable to the experimental ones reported in the literature, inversely to the case of the BP86-D one, which was calculated to be longer.81 For both structures, the C(1)-Pt-N(2) (174°) and N(1)–Pt–N(7) (178°) or N(1)–Pt–O (177°) bond angles were quite linear around the Pt(II) center. The computed geometric parameters place emphasis on a slightly deformed square planar ML4 geometry having the 16-MVE configuration, as witnessed by the N(1)–N(2)–O–C(1) and N(1)–N(2)–N(7)–C(1) dihedral angles of about 0° for complexes 1 and 2, respectively. Relatively, the deformation around the metal cation of complex 1 was more marked than that of complex 2.
| 1 | 2 | |||
|---|---|---|---|---|
| BP86-D | B3LYP | BP86-D | B3LYP | |
| HOMO–LUMO gap | 0.59 | 2.31 | 0.23 | 1.94 |
| Pt–N(1) | 2.108 | 2.003 | 2.123 | 2.028 |
| Pt–C(1) | 2.059 | 1.995 | 2.077 | 2.012 |
| Pt–N(2) | 2.286 | 2.223 | 2.209 | 2.134 |
| Pt–O | 2.253 | 2.124 | — | — |
| Pt–N(7) | — | — | 2.146 | 2.052 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||
| Dihedral angle (°) | ||||
| N(1)–N(2)–N(7)–C(1) | — | — | 2.0 | 4.0 |
| N(1)–N(2)–O–C(1) | 3.0 | 1.0 | — | — |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||
| WBI (Wiberg bond index) | ||||
| Pt–N(1) | 0.481 | 0.488 | 0.432 | 0.432 |
| Pt–C(1) | 0.720 | 0.727 | 0.674 | 0.676 |
| Pt–N(2) | 0.238 | 0.235 | 0.288 | 0.290 |
| Pt–N(7) | — | — | 0.367 | 0.378 |
| Pt–O | 0.278 | 0.279 | — | — |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||
| Natural charge (NBO) | ||||
| Pt | 0.662 | 0.728 | 0.679 | 0.744 |
| N(1) | −0.371 | −0.401 | −0.385 | −0.415 |
| C(1) | −0.127 | −0.149 | −0.128 | −0.156 |
| N(2) | −0.668 | −0.714 | −0.739 | −0.770 |
| N(7) | — | — | −0.413 | −0.421 |
| O | −0.898 | −0.917 | — | — |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||
| Natural electronic configuration | ||||
| Pt | 6s0.555d8.786p0 | 6s0.535d8.736p0 | 6s0.555d8.776p0 | 6s0.525d8.726p0 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||
| Frontier orbital occupations of H2O and guanine | ||||
| σ | 1.87 | 1.85 | 1.91 | 1.92 |
| π* | 0.0 | 0.0 | 0.07 | 0.06 |
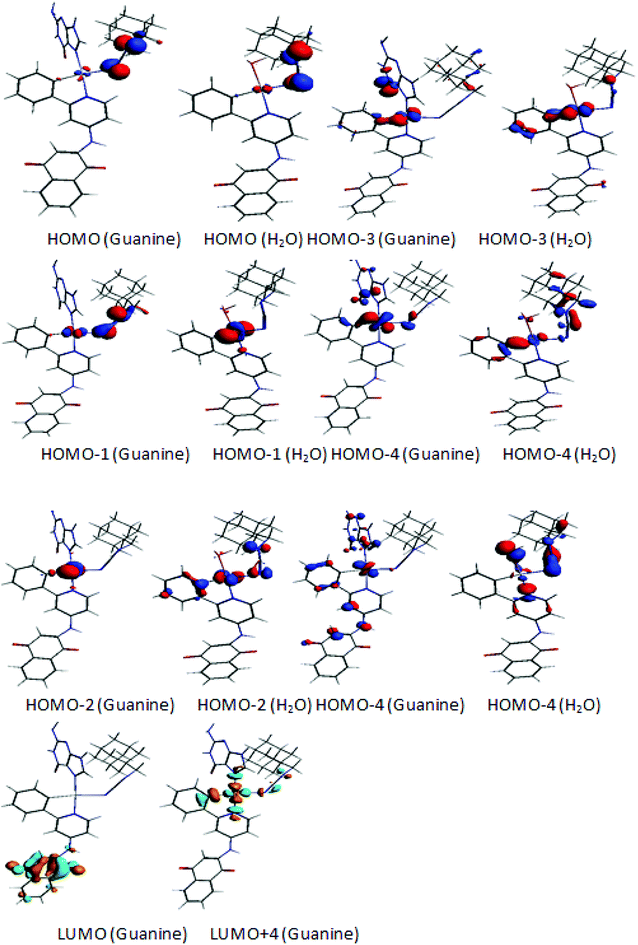
A Mulliken population analysis of the MO plots for complex 2 showed that the two highest HOMO and HOMO-1 were metal–ligand based with a small metallic contribution as presented in Fig. 5. However, the HOMO was purely metallic. The LUMO was chiefly a ligand orbital, thus the reduction should not affect its molecular structure. Furthermore, the HOMO-3 and the HOMO-4 showed σ and π Pt–C(1) bonding, respectively.
For complex 1, the HOMO was principally a ligand orbital, while the HOMO-1 was purely metallic and the LUMO was principally orbital, as sketched out in Fig. 5. It is worth noting that there was no participation of the water molecules in these crucial orbitals. Accordingly, oxidation and reduction of the neutral should not affect the molecular structure.
The Pt(II) charges in both complexes 1 and 2 were comparable to that computed in previous works, particularly those obtained by B3LYP.84,85 There was an increase in negative charge at the nitrogen N(7) atom of guanine from −0.39 (free guanine) to −0.42 (complex 2), corresponding to an increase in electron density at this nitrogen center contained in complex 2. This electron density enhancement was in accord with the electron transfer from the Pt metal occupied orbitals responsible for the backdonation into the vacant π* antibonding orbital of guanine as illustrated by the orbital occupations in Table 2. Whereas, there was an important decrease in negative charge at the oxygen atom of H2O encountered in complex 1 from −0.96 (BP86-D) to −0.89, corresponding to a slight decrease in electron density at this center in agreement with the σ-donor character of the H2O ligand as illustrated by the σ-donation (Table 2), where the lone pair localized on the oxygen has lost 0.13e (its occupation is 1.87 after interaction as shown in Table 2). The natural electronic configuration of the Pt center (Table 2) obtained by both BP86-D and B3LYP methods showed that the major electronic transfers arose from back-donation from the occupied 5d orbitals of the Pt, which were significantly depopulated compared to the donation from the σ lone pair of the oxygen in 1 and the lone pair of nitrogen in 2 to the Pt 6s orbital. As a result, the Pt(center) NAO configuration exhibited significant 5d depopulation (Table 2) stemming essentially from the 5dxz orbital, which was more responsible for the major back-donation to the vacant π* orbitals of the neighboring ligands, displaying a loss of 0.82e and 0.84e in 1 and 2, respectively, leading to a significant positive atomic charge of +0.662 and 0.728 (BP86-D). Furthermore, the Wiberg bond indices (WBI) gathered in Table 2 provide an in-depth look into the bonding between the Pt(II) center and its surrounding atoms. Indeed, the large Pt–C(1) WBI values of 0.720 and 0.674 (BP86-D) or 0.727 and 0.676 (B3LYP) for 1 and 2, respectively, correspond to double bonding, while the different WBI of Pt–N values of 0.481 and 0.488 (BP86-D) or 0.432 and 0.432 (B3LYP) for 1 and 2, respectively, are comparable describing a dative σ-bonding. The Pt–O WBI of 0.278 (BP86-D) or 0.279 (B3LYP) also describe a dative σ-bond between the oxygen lone pair and they are somewhat weaker than those between the Pt center and the different nitrogen atoms.
| ΔEint = ΔEpauli + ΔEelstat + ΔEorb | (1) |
| ΔEint = ΔEpauli + ΔEelstat + ΔEorb + ΔEdisp | (2) |
The bonding was stronger in the guanine case than in the water one. Hence, the energy decomposition analysis showed an important interaction energy amount of 52.1 (BP86-D) or 45.9 kcal mol−1 (B3LYP) for Pt–N(7) (guanine) compared to that for Pt–O (H2O) of 38.5 (BP86-D) or 37.8 kcal mol−1 (B3LYP) (Table 3).
| Energy decomposition | |||||
|---|---|---|---|---|---|
| ΔEint | ΔEpauli | ΔEelec | ΔEorb | ΔEdisp | |
| Complex 1 | |||||
| BP86-D | −1.67(−38.5) | 3.73 | −2.93 | −2.09 | −0.38 |
| B3LYP | −1.64(−37.8) | 3.81 | −3.41 | −2.10 | — |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||||
| Complex 2 | |||||
| BP86-D | −2.26(−52.1) | 4.89 | −4.13 | −1.94 | −1.08 |
| B3LYP | −1.99(−45.9) | 5.20 | −4.86 | −2.39 | — |
Table 4 shows that the values of ΔEelstat were more important than those of ΔEorb in both the water and guanine cases, where the ΔEelstat corresponds to two-thirds of the stabilization contributions to the attractive interaction energy (ΔEelstat + ΔEorb).
| 1 | 2 | |||||||
|---|---|---|---|---|---|---|---|---|
| BP86-D | B3LYP | BP86-D | B3LYP | |||||
| ρ(r) | ∇2ρ(r) | ρ(r) | ∇2ρ(r) | ρ(r) | ∇2ρ(r) | ρ(r) | ∇2ρ(r) | |
| Pt–N(1) | 0.103 | 0.015 | 0.131 | 0.037 | 0.099 | 0.014 | 0.098 | 0.015 |
| Pt–C(1) | 0.129 | 0.004 | 0.146 | 0.007 | 0.125 | 0.004 | 0.125 | 0.005 |
| Pt–N(2) | 0.066 | 0.004 | 0.076 | 0.007 | 0.078 | 0.007 | 0.077 | 0.007 |
| Pt–N(7) | — | — | — | — | 0.091 | 0.013 | 0.089 | 0.013 |
| Pt–O | 0.065 | 0.006 | 0.084 | 0.016 | — | — | — | — |
The interactions within the complex 2 were governed by one-third covalent (32% BP86-D or 33% B3LYP) and two-thirds electrostatic characters (68% BP86-D or 67% B3LYP), giving rise to a more pronounced ionic character than covalent one; while for the water case, the covalent character increase depends on the ionic one. Indeed for complexes 1 and 2, the ionic character makes a greater contribution than the covalent character in the stabilization contributions to the attractive interaction energy (ΔEelstat + ΔEorb).
The variation of the ionic character is correlated to the NAO charges calculated for the anionic fragment gathered in Table 2, which shows the progressive enhancement of the positive charges of the Pt(II) center of +0.662 (BP86) or +0.728 (B3LYP) (water) and of +0.679 (BP86) or +0.744 (B3LYP) for (guanine), in agreement with the strengthening of the ΔEelstat electrostatic contribution according to the following order: H2O < guanine (Table 3).
Furthermore, the donor–acceptor strength of the different ligands were classified according with their relative amounts of σ-donation and π-backdonation occurring between the ligand L and the residual fragment, where the decreasing orders of σ-donation and π-backdonation were established as follows, respectively: H2O > guanine. With respect to the electronic transfers, the H2O ligand can be considered as the strongest σ-donor.
The H2O is only an σ-donor with an σ-donation amount of 0.13 (BP86-D), and is considered as a relatively strong σ-donor compared to the guanine amount of 0.09 (BP96-D). The guanine ligand was revealed to be an σ-donor rather than a π-acceptor (0.07) with BP86-D, in agreement with its interaction with metal fragments (Table 3). For the guanine case, the π-acceptor character was more important than the σ one, in agreement with the σ-donation and π-accepting amounts.
The values of the charge density ρ(r) distribution and the corresponding Laplacian ∇2ρ(r) at the bond critical point (BCP) allowed this characterization as defined in the literature,89,91,92 and provided information about the bonding schemes, as illustrated in Scheme 1. It is clear from the results on model systems that QTAIM is a very useful tool to characterize the nature of bonding. Both BP86 and B3LYP gave comparable values of ρ(r) and ∇2ρ(r) as illustrated in Table 4. These values were indicative of an intermediate chemical bonding between the ionic and covalent characters, confirming the same trend obtained within the energy decomposition.
4. Conclusions
A new platinum(II) complex cis-Pt(1,4-NQ)(1-ADpcyd)(H2O), (1-ADpcydH: 1-Adamantanemethylcyanamide, 1,4-NQ: 2-[amino(2-phenylpyridine)]-1,4-naphthoquinone), was synthesized and characterized. Structural characterization by FT-IR, elemental analyses, multinuclear NMR (1H, 13C, and 195Pt), and mass spectrometry showed that the ligands 1-ADpcydH, 1,4-NQ, and one H2O molecules were coordinated to the Pt(II) center in a cis-arranged form around the Pt(II) center. Interaction of the cis-Pt(II) complex 1 with guanine was followed by 195Pt NMR spectroscopy and the final product, cis-Pt(1,4-NQ)(1-ADpcyd)(G), was confirmed by its mass spectrum. Both 195Pt NMR and the mass spectrum confirmed the binding of the Pt(II) complex 1 to the N7 guanine. The free ligands 1-ADpcydH and 1,4-NQ and complex 1 on MCF-7, HepG-2, and HT-29 cancer cell lines were tested. Here, complex 1 was more active than free ligands and cisplatin in the three cancer cell lines. The lowest IC50 value for complex 1 against HT-29 was a 4.5-fold increase compared with cisplatin. Complex 1 showed a higher level of bonding to the DNA in HT-29 cell compared to cisplatin. Both DNA platination and interaction with guanine suggested the importance of the direct attack of nucleotides on cis-Pt(II) complex 1 in the mechanism of action for the Pt(II) complex. DFT calculations showed that both complexes 1 and 2, cis-Pt(1,4-NQ)(1-ADpcyd)(L) (L = H2O (1), guanine (2)), adopted a square planar geometry, in agreement with the 16-MVE configuration of the Pt(II) cation. The decomposition energy analysis revealed an important interaction energy amount for Pt–N(7) (guanine) compared to that for Pt–O(H2O). For both complexes 1 and 2, the electrostatic interactions were more important than those stemming from the orbital ones due to the polarity between the Pt(II) positive charge and its surrounding atoms negative charge. The present experimental and computational studies may hopefully contribute an important effort toward the development of DNA-targeted Pt(II) metallodrugs.Author contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.Conflicts of interest
The authors declare no competing financial interest.Abbreviations
| 1,4-NQ | 2-[Amino(2-phenylpyridine)]-1,4-naphthoquinone |
| 1-ADpcydH | 1-Adamantanemethylcyanamide |
| G | Guanine |
| DMSO | Dimethyl sulfoxide |
| MeOH | Methanol |
| THF | Tetrahydrofuran |
| NMR | Nuclear magnetic resonance |
| FT-IR | Fourier-transform infrared |
| MVE | Metal valence electrons |
| ADF | Amsterdam Density Functional |
| DFT | Density functional theory |
| STO | Slater-type orbital |
| ZORA | Zero-order regular approximation |
| LDA | Local density approximation |
| NAO | Natural atomic orbital |
| HOMO | Highest occupied molecular orbital |
| LUMO | Lowest unoccupied molecular orbital |
| NBO | Natural bond orbital |
| EDA | Energy decomposition analysis |
| QTAIM | Quantum theory of atoms in molecules |
| BCP | Bond critical point |
Acknowledgements
The authors acknowledge general support from the School of Chemistry, National University of Ireland (NUI), Galway. BZ is grateful to the Algerian MESRS (Ministère de l'Enseignement Supérieur et de la Recherche Scientifique) and DGRSDT (Direction Générale de la Recherche Scientifique et du Développement Technologique) for Financial support.References
- R. Cortés, M. Crespo, L. Davin, R. Martín, J. Quirante, D. Ruiz, R. Messeguer, C. Calvis, L. Baldomà and J. Badia, et al., Eur. J. Med. Chem., 2012, 54, 557–566 CrossRef PubMed.
- P. Chellan, K. M. Land, A. Shokar, A. Au, S. H. An, C. M. Clavel, P. J. Dyson, C. de Kock, P. J. Smith and K. Chibale, et al., Organometallics, 2012, 31, 5791–5799 CrossRef CAS.
- J. Quirante, D. Ruiz, A. González, C. López, M. Cascante, R. Cortés, R. Messeguer, C. Calvis, L. Baldomà and A. Pascual, et al., J. Inorg. Biochem., 2011, 105, 1720–1728 CrossRef CAS PubMed.
- J. Ruiz, V. Rodríguez, N. Cutillas, A. Espinosa and M. J. Hannon, J. Inorg. Biochem., 2011, 105, 525–531 CrossRef CAS PubMed.
- J. Ruiz, C. Vicente, C. de Haro and A. Espinosa, Inorg. Chem., 2011, 50, 2151–2158 CrossRef CAS PubMed.
- P. Wang, C.-H. Leung, D.-L. Ma, R. W.-Y. Sun, S.-C. Yan, Q.-S. Chen and C.-M. Che, Angew. Chem., Int. Ed., 2011, 50, 2554–2558 CrossRef CAS PubMed.
- H. Samouei, M. Rashidi and F. W. A. Heinemann, J. Organomet. Chem., 2011, 696, 3764–3771 CrossRef CAS.
- J. Albert, R. Bosque, M. Crespo, J. Granell, C. López, R. Cortés, A. González, J. Quirante, C. Calvis and R. Messeguer, et al., Bioorg. Med. Chem., 2013, 21, 4210–4217 CrossRef CAS PubMed.
- M. Crespo, Inorganics, 2014, 2, 115–131 CrossRef CAS.
- N. Farrell, in Platinum-based Drugs in Cancer Therapy, ed. L. R. Kelland and N. P. Farrell, Humana Press, Totowa, NJ, 2000, pp. 321–338 Search PubMed.
- E. R. Jamieson and S. J. Lippard, Chem. Rev., 1999, 99, 2467–2498 CrossRef CAS PubMed.
- N. J. Wheate, B. J. Evison, A. J. Herlt, D. R. Phillips and J. G. Collins, Dalton Trans., 2003, 3486–3492 RSC.
- N. Farrell, Y. Qu, L. Feng and B. Van Houten, Biochemistry, 1990, 29, 9522–9531 CrossRef CAS PubMed.
- N. Farrell, Y. Qu and P. Hacker, J. Med. Chem., 1990, 33, 2179–2184 CrossRef CAS.
- J. D. Roberts, B. van Houten, Y. Qu and N. P. Farrell, Nucleic Acids Res., 1989, 17, 9719–9733 CrossRef CAS PubMed.
- Z. D. Bugarcic, T. Soldatovic, R. Jelic, B. Alguero and A. Grandas, Dalton Trans., 2004, 3869–3877 RSC.
- U. Frey, J. D. Ranford and P. J. Sadler, Inorg. Chem., 1993, 32, 1333–1340 CrossRef CAS.
- A. Gelasco and S. J. Lippard, Anticancer activity of cisplatin and related complexes, in Metallopharmaceuticals I. Topics in Biological and Inorganic Chemistry, ed. M. J. Clarke and P. J. Sadler, Springer-Verlag, Berlin, 1999, vol. 1, pp. 1–43 Search PubMed.
- J. Reedijk, Chem. Rev., 1999, 99, 2499–2510 CrossRef CAS PubMed.
- (a) K. J. Barnham, Z. J. Guo and P. J. Sadler, J. Chem. Soc., Dalton Trans., 1996, 2867–2876 RSC; (b) K. J. Barnham, M. I. Djuran, P. D. Murdoch and P. J. Sadler, J. Chem. Soc., Chem. Commun., 1994, 721–722 RSC; (c) K. J. Barnham, M. I. Djuran, P. D. Murdoch, J. D. Ranford and P. J. Sadler, J. Chem. Soc., Dalton Trans., 1995, 3721–3726 RSC; (d) J. M. Teuben and J. Reedijk, J. Biol. Inorg Chem., 2000, 5, 463–468 CrossRef CAS PubMed.
- H.-C. Tai, R. Brodbeck, J. Kasparkova, N. J. Farrer, V. Brabec, P. J. Sadler and R. J. Deeth, Inorg. Chem., 2012, 51, 6830–6841 CrossRef CAS PubMed.
- F. Sebesta and J. V. Burda, J. Inorg. Biochem., 2017, 172, 100–109 CrossRef CAS PubMed.
- E. N. da Silva Jr, R. F. S. Menna-Barreto, M. C. F. R. Pinto, R. S. F. Silva, D. V. Teixeira, M. C. B. V. de Souza, C. A. de Simone, S. L. de Castro, V. F. Ferreira and A. V. Pinto, Eur. J. Med. Chem., 2008, 43, 1774–1780 CrossRef PubMed.
- S. L. Castro, F. S. Emery and E. N. da Silva Jr, Eur. J. Med. Chem., 2013, 69, 678–700 CrossRef PubMed.
- G. A. M. Jardim, W. J. Reis, M. F. Ribeiro, F. M. Ottoni, R. J. Alves, T. L. Silva, M. O. F. Goulart, A. L. Braga, R. F. S. Menna-Barreto, K. Salomão, S. L. Castro and E. N. da Silva Jr, RSC Adv., 2015, 5, 78047–78060 RSC.
- (a) L. Lin, Q. Shen, G.-R. Chen and J. Xie, Bioorg. Med. Chem., 2008, 16, 9757 CrossRef CAS PubMed; (b) B. D. Bala, S. Muthusaravanan, T. S. Choon, M. A. Ali and S. Perumal, Eur. J. Med. Chem., 2014, 85, 737–746 CrossRef PubMed.
- W. S. Nascimento, C. A. Camara and R. N. de Oliveira, Synthesis, 2011, 20, 3220–3224 Search PubMed.
- A. P. Neves, M. X. G. Pereira, E. J. Peterson, R. Kipping, M. D. Vargas, F. P. Silva Jr, J. W. M. Carneiro and N. P. Farrell, J. Inorg. Biochem., 2013, 119, 54–64 CrossRef CAS PubMed.
- M. Chinnapattu, K. I. Sathiyanarayanan and P. S. Iyer, Bioorg. Med. Chem. Lett., 2015, 25, 952–955 CrossRef CAS PubMed.
- L. Wanka, K. Iqbal and P. R. Schreiner, Chem. Rev., 2013, 113, 3516–3604 CrossRef CAS PubMed.
- S. Y. Sun, P. Yue, W. K. Hong and R. Lotan, Cancer Res., 2000, 60, 7149–7155 CAS.
- M. Horvat, L. Uzelac, M. Marjanovic, N. Cindro, O. Frankovic, K. Mlinaric-Majerski, M. Kralj and N. Basaric, Evaluation of Antiproliferative Effect of N-(alkyladamantyl)phthalimides In vitro, Chem. Biol. Drug Des., 2012, 79, 497–506 CrossRef CAS PubMed.
- J. D. Parkes, R. C. Baxter, C. D. Marsden and J. E. Rees, J. Neurol., Neurosurg. Psychiatry, 1974, 37, 422–426 CrossRef CAS.
- J. Beigel and M. Bray, Antiviral Res., 2008, 78, 91–102 CrossRef CAS PubMed.
- A. Garcia, R. C. Machado, R. M. Grazul, M. T. P. Lopes, C. C. Corrêa, H. F. Dos Santos, M. V. de Almeida and H. Silva, J. Biol. Inorg Chem., 2016, 21, 275–292 CrossRef CAS PubMed.
- L. Tabrizi and H. Chiniforoshan, RSC Adv., 2017, 7, 34160–34169 RSC.
- L. Tabrizi and H. Chiniforoshan, Dalton Trans., 2017, 46, 2339–2349 RSC.
- L. Tabrizi and H. Chiniforoshan, Dalton Trans., 2016, 45, 18333–18345 RSC.
- L. Tabrizi and H. Chiniforoshan, BioMetals, 2017, 30, 59–70 CrossRef CAS PubMed.
- L. Tabrizi and H. Chiniforoshan, Invest. New Drugs, 2016, 34, 723–732 CrossRef CAS PubMed.
- L. Tabrizi, P. McArdle, A. Erxleben and H. Chiniforoshan, Inorg. Chim. Acta, 2015, 438, 94–104 CrossRef CAS.
- M. Jazestani, H. Chiniforoshan, L. Tabrizi and P. McArdle, J. Biomol. Struct. Dyn., 2017, 35, 2055–2065 CrossRef CAS PubMed.
- M. Jazestani, H. Chiniforoshan, L. Tabrizi, P. McArdle and B. Notash, Inorg. Chim. Acta, 2016, 450, 402–410 CrossRef CAS.
- A. Zaiter and B. Zouchoune, Struct. Chem., 2018, 29, 1307–1320 CrossRef CAS.
- N. Naili and B. Zouchoune, Struct. Chem., 2018, 29, 725–739 CrossRef CAS.
- B. Zouchoune, S. M. Zendaoui and J. Y. Saillard, J. Organomet. Chem., 2018, 858, 47–52 CrossRef CAS.
- A. Saiad and B. Zouchoune, Can. J. Chem., 2015, 93, 1096–1108 CrossRef CAS.
- N. Benhamada, R. Bouchene, S. Bouacida and B. Zouchoune, Polyhedron, 2015, 91, 59–67 CrossRef CAS.
- S. Drideh, B. Zouchoune, S.-M. Zendaoui and J.-Y. Saillard, Theor. Chem. Acc., 2018, 137, 99 Search PubMed.
- F. Chekkal, S. M. Zendaoui, B. Zouchoune and J. Y. Saillard, New J. Chem., 2013, 37, 2293–2302 RSC.
- A. Benmachiche, S. M. Zendaoui, S. E. Bouaoud and B. Zouchoune, Int. J. Quantum Chem., 2013, 113, 985–996 CrossRef CAS.
- M. S. Zendaoui, J. Y. Saillard and B. Zouchoune, ChemistrySelect, 2016, 5, 940–948 CrossRef.
- R. J. Letcher, W. Zhang, C. Bensimon and R. J. Crutchley, Inorg. Chim. Acta, 1993, 210, 183–191 CrossRef CAS.
- M. C. Alley, D. A. Scudiero, A. Monks, M. L. Hursey, M. J. Czerwinski, D. L. Fine, B. J. Abbott, J. G. Mayo, R. H. Shoemaker and M. R. Boyd, Cancer Res., 1988, 48, 589 CAS.
- ADF2007.01., SCM Theoretical Chemistry, Vrije Universiteit, Amsterdam, The Netherlands, http://www.scm.com Search PubMed.
- E. J. Baerends, D. E. Ellis and P. Ros, Chem. Phys., 1973, 2, 41–51 CrossRef CAS.
- G. te Velde and E. J. Baerends, J. Comput. Phys., 1992, 99, 84–98 CrossRef CAS.
- C. Fonseca Guerra, J. G. Snijders, G. te Velde and E. Baerends, Theor. Chem. Acc., 1998, 99, 391–403 Search PubMed.
- F. M. Bickelhaupt and E. Baerends, J. Rev. Comput. Chem., 2000, 15, 1–86 Search PubMed.
- G. te Velde, F. M. Bickelhaupt, E. J. Baerends, C. F. Guerra, S. J. A. van Gisbergen, J. G. Snijders and T. Ziegler, J. Comput. Chem., 2001, 22, 931–967 CrossRef CAS.
- S. D. Vosko, L. Wilk and M. Nusair, Can. J. Phys., 1980, 58, 1200–1211 CrossRef CAS.
- S. Grimme, J. Comput. Chem., 2006, 27, 1787–1799 CrossRef CAS.
- A. D. Becke, J. Chem. Phys., 1986, 84, 4524–4529 CrossRef CAS.
- A. D. Becke, Phys. Rev. A, 1988, 38, 3098 CrossRef CAS.
- J. P. Perdew, Phys. Rev. B: Condens. Matter Mater. Phys., 1986, 33, 8822–8824 CrossRef.
- J. P. Perdew, Phys. Rev. B: Condens. Matter Mater. Phys., 1986, 34, 7406 CrossRef.
- A. D. Becke, J. Chem. Phys., 1993, 98, 5642–5648 CrossRef.
- C. Lee, W. Yang and R. G. Parr, Phys. Rev. B: Condens. Matter Mater. Phys., 1988, 37, 785–789 CrossRef CAS.
- L. Versluis and T. Ziegler, J. Chem. Phys., 1988, 88, 322 CrossRef CAS.
- L. Fan and T. Ziegler, J. Chem. Phys., 1992, 96, 9005–9012 CrossRef CAS.
- L. Fan and T. Ziegler, J. Phys. Chem., 1992, 96, 6937–6941 CrossRef CAS.
- P. Flükiger, H. P. Lüthi, S. Portmann and J. Weber, MOLEKEL, Version 4.3.win32, Swiss Center for Scientific Computing (CSCS), Switzerland, 2000–2001, http://www.cscs.ch/molekel/ Search PubMed.
- K. B. Wiberg, Tetrahedron, 1968, 24, 1083–1084 CrossRef CAS.
- F. Weinhold and C. R. Landis, Valency and Bonding: A Natural Bond Orbital Donor–Acceptor Perspective, Cambridge University Press, Cambridge, UK, 2005 Search PubMed.
- E. D. Glendening, J. K. Badenhoop, A. E. Reed, J. E. Carpenter, J. A. Bohmann, C. M. Morales and F. Weinhold, Natural Bond Orbitals “Analysis Programs”, Theoretical Chemistry Institute, University of Wisconsin, Madison, WI, 2001 Search PubMed.
- S. Chandra and Vandana, Spectrochim. Acta, Part A., 2014, 129, 333–338 CrossRef CAS PubMed.
- V. Mishra and N. Thirupathi, ACS Omega, 2018, 3, 6075–6090 CrossRef CAS.
- G. Arena, L. M. Scolaro, R. F. Pasternack and R. Romeo, Inorg. Chem., 1995, 34, 2994–3002 CrossRef CAS.
- Q.-P. Qin, Z.-F. Chen, J.-L. Qin, X.-J. He, Y.-L. Li, Y.-C. Liu, K.-B. Huang and H. Liang, Eur. J. Med. Chem., 2015, 92, 302–313 CrossRef CAS PubMed.
- D. Michalska and R. Wysokiński, Collect. Czech. Chem. Commun., 2004, 69, 63–72 CrossRef CAS.
- J. V. Burda, J. Šponer and J. Leszczynski, JBIC, J. Biol. Inorg. Chem., 2000, 5, 178–188 CrossRef CAS PubMed.
- X.-P. Zhang, V. Y. Chang, J. Liu, X.-L. Yang, W. Huang, Y. Li, C.-H. Li, G. Muller and X.-Z. You, Inorg. Chem., 2014, 54, 143–152 CrossRef PubMed.
- J. Li, F. Liang, Y. Zhao, X. Liu, J. Fan and L. Liao, J. Mater. Chem. C, 2017, 5, 6202–6209 RSC.
- S. Yao, J. P. Plastaras and L. G. Marzilli, Inorg. Chem., 1994, 33, 6061–6077 CrossRef CAS.
- P. W. Asman, J. Coord. Chem., 2017, 1–20 CrossRef.
- K. Kitaura and K. Morokuma, Int. J. Quantum Chem., 1976, 10, 325 CrossRef CAS.
- K. Morokuma, Acc. Chem. Res., 1977, 10, 294–300 CrossRef CAS.
- T. Ziegler and A. Rauk, Theor. Chem. Acc., 1977, 46, 1–10 Search PubMed.
- R. F. W. Bader, Atoms in Molecules a Quantum Theory, Clanderon Press, Oxford, UK, 1990 Search PubMed.
- R. F. W. Bader, J. Phys. Chem. A, 1998, 102, 7314–7323 CrossRef CAS.
- P. L. A. Popelier and G. Logothetis, J. Organomet. Chem., 1998, 555, 101–111 CrossRef CAS.
- T. S. Thakur and G. R. Desiraju, J. Mol. Struct.: THEOCHEM, 2007, 810, 143–154 CrossRef CAS.
Footnotes |
| † Electronic supplementary information (ESI) available: 1H, 13C NMR spectra of free ligands and 1H, 13C, 195Pt NMR spectra of Pt(II) complex and mass spectra of ligand free ligands and Pt(II) complex. See DOI: 10.1039/c8ra08739c |
| ‡ These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2019 |