Synchronous sensing of three conserved sequences of Zika virus using a DNAs@MOF hybrid: experimental and molecular simulation studies†
Bao-Ping
Xie
b,
Gui-Hua
Qiu
b,
Bin
Sun
 b,
Zi-Feng
Yang
a,
Wen-Hua
Zhang
b,
Zi-Feng
Yang
a,
Wen-Hua
Zhang
 *c,
Jin-Xiang
Chen
*c,
Jin-Xiang
Chen
 *b and
Zhi-Hong
Jiang
*a
*b and
Zhi-Hong
Jiang
*a
aState Key Laboratory of Respiratory Diseases, Institute of Integrated Traditional Chinese Medicine and Western Medicine, Guangzhou Medical University, Guangzhou, China. E-mail: zhihongjiang@gmail.com
bGuangdong Provincial Key Laboratory of New Drug Screening, School of Pharmaceutical Sciences, Southern Medical University, Guangzhou 510515, China. E-mail: jxchen@smu.edu.cn
cCollege of Chemistry, Chemical Engineering and Materials Science, Soochow University, Suzhou 215123, China. E-mail: whzhang@suda.edu.cn
First published on 13th November 2018
Abstract
A metal–organic framework of Cu(II) supported by a zwitterionic pyridinium dicarboxylate and 4,4′-bipyridine mixed ligands was prepared and impregnated with three dye-labeled DNA sequences with fixed wavelength differences. This hybrid material formed is capable of the synchronous detection of three conserved Zika virus RNA sequences with high sensitivity and selectivity, and low detection limits of 0.56 ± 0.02, 0.16 ± 0.04, and 0.19 ± 0.05 nM, without any cross-reactions, as confirmed by both experimental and molecular simulation studies.
The Zika virus (ZIKV), an arthropod-borne flavivirus (family Flaviviridae), was first identified in rhesus monkeys from the Zika forest in Uganda in 1947 and was reported in humans in 1952.1 ZIKV then remained largely neglected until 2007 when the first noteworthy outbreak was found in the Yap Island in Micronesia.2 Patients experienced fever, skin rash, arthralgia, and conjunctivitis. This epidemic subsequently attracted global concern in 2015 when ZIKV was suspected to be responsible for cases of microcephaly in fetuses and babies in northeast Brazil.3 Epidemiological studies of ZIKV are challenging because it is an insidious disease and those who have it show either no or only common symptoms such as fever.4
There is currently no widely deployed test for ZIKV because the cross-reactions among ZIKV, yellow fever virus (YFV), Dengue virus (DENV) and West Nile virus (WNV) make it difficult to distinguish ZIKV by serological methods.5 In addition, it is difficult to definitively confirm the ZIKV infection during the incubation period by immunological analysis techniques.6 However, early diagnosis by the detection of ZIKV nucleic acids looks promising.7 Three conserved target sequences, 5′-CCCCAGGAGAAGCUGGGAAACCAAGCUC-3′ (denoted as T1), 5′-AGCAUAUUGACGUGGGAAAGAC-3′ (denoted as T2), and 5′-GGACUAGUGGUUAGAGGAGACCC-3′ (denoted as T3), in the 3′-noncoding regions of ZIKV RNA have been confirmed,8 and a synchronous detection of these three conserved sequences would lead to an accurate diagnosis of the ZIKV disease and provide a convincing alert.
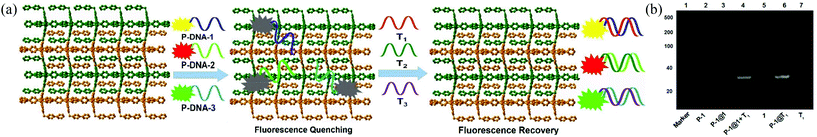
Gold nanoparticles (AuNPs),9 carbon nanotubes (CNTs),10 graphene oxide (GO),11 and metal–organic frameworks (MOFs)12 are promising platforms for nucleic acid detection by means of fluorescence. Of these, MOFs are particularly attractive because of their structural diversity, resulting from a principally unlimited combination of metal ions and organic ligands, facile preparation and characterization, and low cost. Chen and co-workers reported the first case of nucleic acid detection with a Cu(II)-based MOF in 2013.13 Subsequently, several other MOFs, including MIL-101,14 MIL-88,15 and UiO-66-NH2,16 have been employed for similar purposes. In a typical scenario, a MOF weakly interacts with an emissive dye-tagged probe DNA sequence (denoted as P-DNA) and results in emission quenching by photoinduced electron/energy transfer.16 The subsequent introduction of the targeted DNA/RNA sequence triggers a stronger interaction with the P-DNA via base pair matching, serving to detach the P-DNA from the MOF with a concomitant recovery of dye emission.17
Herein, we extended the above described strategy to synchronously detect three conserved sequences in the 3′-noncoding regions of ZIKV RNA using a water-stable two-dimensional (2D) MOF of [Cu(Dcbb)(bipy)(OH)]n (1, H2DcbbBr = 1-(3,5-dicarboxybenzyl)-4′-bipyridiniumbromide; bipy = 4,4-bipyridine, Fig. 1a). MOF 1 comprises paramagnetic Cu2+, polar zwitterionic pyridinium carboxylates, and conjugated phenyl and pyridyl moieties, and is stable in biological media (Tris-HCl, pH 7.4). To eliminate the signal overlap, we selected three dyes with fixed wavelength differences (Δλ) between excitation and emission, viz carboxyfluorescein (FAM, Ex = 494 nm, Em = 518 nm, Δλ = 24 nm), 5(6)-carboxyrhodamine triethylammonium salt (ROX, Ex = 578 nm, Em = 604 nm, Δλ = 26 nm), and cyanine 5 (Cy5, Ex = 649 nm, Em = 670 nm, Δλ = 21 nm) to label three P-DNAs (complementary sequences of ZIKV RNAs). MOF 1 simultaneously absorbed the three P-DNAs to form a hybrid P-DNAs@1 sensing platform which rapidly detects the three conserved sequences of ZIKV RNA with high sensitivity and selectivity at low detection limits of (0.56 ± 0.02) nM, (0.16 ± 0.04) nM, and (0.19 ± 0.05) nM.
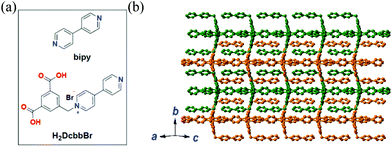 | ||
| Fig. 1 (a) The structures of bipy and H2DcbbBr. (b) The 2D structure of MOF 1 extended within the ac plane showing the complementary stacking of adjacent layers (differentiated by green and orange). | ||
We selected MOF 1 as a sensing platform because of the validated capability of the Cu(II)-based MOF to induce fluorescence quenching upon nucleic acid association. The MOF–nucleic acid hybrid formed thus in turn functions as a sensitive platform for complementary nucleic acids as it contrasts a near-dark background fluorescence.17b,d,e In addition, MOFs also exhibit the key advantages of facile preparation and low cost (e.g. as compared to the Au nanoparticles and carbon nanotubes). Furthermore, 2D MOFs, such as 1, exhibit flat planar structures with a high functionality exposure, and may exhibit a quick fluorescence response as the analytes can be readily absorbed/rejected by the layer.
MOF 1 is moisture- and water-stable. Its powder X-ray diffraction (PXRD) patterns, as well as those of its fresh powder immersed in H2O for 48 h, are in general agreement with those of the simulated one from the single crystal X-ray diffraction, indicating its bulk phase purity and water stability (Fig. S1, ESI†). The slight peak difference in the low-angle region may ascribe to the fact that the simulated patterns are based on the SQUEEZED data. Each Cu(II) center in MOF 1 is coordinated by a pair of N atoms from two bipy ligands and a pair of O atoms from two monodentate carboxylates of two Dcbb ligands (Fig. S2a, ESI†). The structure propagates along the a and c directions via monodentate carboxylate and bipy ligands, respectively, yielding a 2D layer structure (Fig. 1b). Notably, the ancillary bipy ligand and the un-coordinated dipyridyl site of the Dcbb ligand from the adjacent layers are parallel to each other, forming strong and complementary π–π interactions (b direction, Fig. S2b, ESI†).
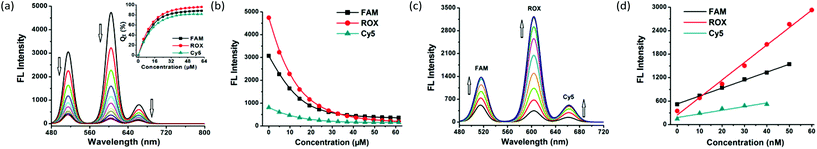
Upon gradual introduction of MOF 1 (2 mM) into a mixed solution of FAM-, ROX-, and Cy5-tagged P-DNAs of constant concentrations (all with 50 nM), the intensity of the diagnostic fluorescence of FAM (518 nm), ROX (604 nm) and Cy5 (670 nm) proportionately decreased (Fig. 2a and b). The FAM and Cy5 fluorescence is nearly stabilized when the MOF 1 concentration reaches 41.3 μM, resulting in the maximum quenching efficiencies (QE) of (88 ± 2)% and (80 ± 4)% for FAM and Cy5. Meanwhile, the ROX fluorescence decreased until the concentration reached 57.5 μM, corresponding to a QE of (96 ± 3)% for ROX. Considering the sensitivity of the assay, 41.3 μM of MOF 1 was selected for further experiments. The three QE values (average: 88% for FAM, 80% for Cy5 and 96% for ROX) are slightly higher than those reported for MOF N,N-bis(2-hydroxy-ethyl)dithiooxamidato copper(II) (H2dtoaCu) for HIV and HBV RNA sequences (85% for FAM and 91% for ROX),18 MOF {[Cu(Cmdcp)(phen)(H2O)]2·9H2O}n (H3CmdcpBr = N-carboxymethyl-(3,5-dicarboxyl)pyridinium bromide, phen = phenanthroline) for ebolavirus conserved RNA sequences and the ebolavirus-encoded miRNA-like fragment (80% for FAM and 95% for ROX),17h and MOF [Cu(Dcbcp)(bpe)]n (Dcbcp = N-(3,5-dicarboxylbenzyl)-(3-carboxyl) pyridinium, bpe = 1,2-bis(4-pyridyl)ethylene) for Dengue and Zika virus RNA sequences (82% for FAM and 92% for ROX).17e
MOF 1 powders can be fully dissolved in water to give a homogeneous solution during the loading of P-DNAs. In order to investigate the stability of the solution of MOF 1, we repeated the above experiment after its preparation for 10, 15, 20 and 48 h, then treated with three P-DNAs and obtained similar QE values. Dynamic light scattering analysis (DLS, Fig. S3, ESI†) determination shows good hydration particle size distributions of both MOF 1 (PDI = 0.021) and P-DNAs@1 (PDI = 0.261) with the average sizes of 482.5 nm and 630.0 nm, which further confirms the successful loading of the P-DNAs by MOF 1.
The fluorescence of FAM, ROX, and Cy5 recovers upon introducing the target ZIKV RNA conservative sequences T1, T2 and T3. The hybridization between nucleic acid complements resulted in fluorescence recovery within (12.0 ± 2.4) min for FAM, (2.3 ± 0.5) min for ROX, and (3.0 ± 0.7) min for Cy5, at a concentration of 25 nM for T1, T2, and T3 (Fig. S4, ESI†).
A gradual increase in the fluorescence intensity of the three probes was detected with increasing target concentrations in the range of 0–100 nM (Fig. 2c and Fig. S5a, ESI†). The high fluorescence recovery efficiencies RE were (2.0 ± 0.1) for T1, (8.4 ± 0.2) for T2, and (2.5 ± 0.1) for T3 (Fig. S5b, ESI†). Linear relationships between the fluorescence intensity and the concentration of the three targets were observed (Fig. 2d) in the ranges of 0–50 nM for FAM, 0–60 nM for ROX and 0–40 nM for Cy5, giving detection limits of (0.56 ± 0.02) nM for T1, (0.16 ± 0.04) nM for T2 and (0.19 ± 0.05) nM for T3 with a signal-to-noise ratio of 3. These observations collectively suggest the successful formation of hybrid duplexes between the P-DNAs and target RNAs, mediated by the unique electronic structure of MOF 1. The detection limits of the three targets are comparable with MOF H2dtoaCu for HIV RNA (0.87 nM) and HBV RNA (0.22 nM) sequences,18 MOF {[Cu(Cmdcp)(phen)(H2O)]2·9H2O}n for ebolavirus conserved RNA sequences (0.16 nM) and the ebolavirus-encoded miRNA-like fragment (0.11 nM),17h and MOF [Cu(Dcbcp)(bpe)]n for Dengue (0.18 nM) and Zika virus (0.12 nM) RNA sequences.17e
Cross-reactions are an important concern when evaluating assay specificity and reliability of a multiplex detection. The sensing specificity of the P-DNAs@1 system toward each target RNA was therefore investigated. Mixing T1 alone with P-DNAs@1 only caused the fluorescence enhancement of FAM (Fig. S6, ESI†). Similar observations were also made when mixing T2 or T3 with P-DNAs@1 that exclusively triggered the fluorescence storage of ROX or Cy5. These are definitive pieces of evidence that there is no cross-reaction among the three probes in the multiplex detection process.
The sensitive detection as a consequence of selective fluorescence quenching and restoration is related to the structural characteristics of MOF 1. The zeta potential of +11.7 mV for MOF 1 indicates that it is capable of interacting electrostatically with DNA sequences bearing negatively charged phosphate backbones. MOF 1 also consists of conjugated ligands capable of π–π stacking with the nucleobases. The carboxylates and uncoordinated N atoms in MOF 1 are also suitable hydrogen-bonding acceptors to further promote interactions with the DNA sequences. These structural characteristics suggest that MOF 1 is ideal to form stable hybrids with the probe DNAs to facilitate the photoinduced electron transfer (PET)16 and quench the FAM, ROX and Cy5 emissions. Nevertheless, if MOF 1 bound too tightly to the P-DNA or to the more rigid, duplex P-DNA@RNA, the fluorescence would either not recover or recover slowly. The anticipated switchable degrees of interactions between the relatively rigid MOF 1 and flexible, single-stranded P-DNA, as well as the rigid double-stranded P-DNA@RNA were further validated by fluorescence titration, fluorescence anisotropy (FA), and polyacrylamide gel electrophoresis (PAGE) experiments.
The weaker interactions of DNA@RNA duplexes with MOF 1 compared to those of the P-DNAs corroborate their binding constants. MOF 1 binds P-DNAs through electrostatic, dipolar and dispersion forces and the fluorescence quenching can thus be considered as static.19 The binding constants (Kb) of P-DNA and hybrid duplex DNA@RNA to MOF 1 were calculated from log[(F0 − F)/F] = n![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log[Q] + log
log[Q] + log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Kb,20 and featured 1.7 × 103 M−1 for P-DNA-1@T1, 6.8 × 104 M−1 for P-DNA-2@T2, and 5.2 × 103 M−1 for P-DNA-3@T3. These values are marginally lower than the Kb values of P-DNAs to MOF 1 (4.4 × 104 M−1 for FAM-tagged P-DNA-1, 3.2 × 105 M−1 for ROX-tagged P-DNA-2, and 2.1 × 104 M−1 for Cy5-tagged P-DNA-3), suggesting that the formed DNA@RNA hybrid duplexes exhibit much lower affinity towards MOF 1 than the P-DNAs and so are detached from the MOF 1 surface, leading to fluorescence recovery.
Kb,20 and featured 1.7 × 103 M−1 for P-DNA-1@T1, 6.8 × 104 M−1 for P-DNA-2@T2, and 5.2 × 103 M−1 for P-DNA-3@T3. These values are marginally lower than the Kb values of P-DNAs to MOF 1 (4.4 × 104 M−1 for FAM-tagged P-DNA-1, 3.2 × 105 M−1 for ROX-tagged P-DNA-2, and 2.1 × 104 M−1 for Cy5-tagged P-DNA-3), suggesting that the formed DNA@RNA hybrid duplexes exhibit much lower affinity towards MOF 1 than the P-DNAs and so are detached from the MOF 1 surface, leading to fluorescence recovery.
FA also features as a measurement of the rotational motion of fluorophore-labelled DNA, providing evidence on the strength of the P-DNA attached to the surface of MOF 1.21 The addition of MOF 1 into P-DNA solutions led to an increase in the fluorescence anisotropy of the dyes by factors of 0.26 for FAM-tagged P-DNA-1, 0.29 for ROX-tagged P-DNA-2, and 0.32 for Cy5-tagged P-DNA-3. In sharp contrast, introducing MOF 1 into solutions of P-DNA@T duplexes caused a negligible FA change of the latter (Fig. S7, ESI†). These results confirm the stronger interactions of MOF 1 with the P-DNA single strands relative to the duplex DNA@RNA.
The gel image captured the banding patterns of FAM-tagged P-DNA-1 (denoted as P-1), P-1@1, P-1@1 + T1, and P-1@T1, with T1 and MOF 1 as controls (Fig. 3b). MOF 1 fails to give any light band upon UV illumination (lane 5) as it is too large to move from the origin under the PAGE conditions. P-1 features one light band (lane 2), but no band was observed after P-1 was incubated with MOF 1 (lane 3), diagnostic of their strong interactions.22 The hybrid duplex P-1@T1 exhibits a light band (lane 6) of higher molecular weight than T1 (lane 7). The same light band is visible when introducing T1 into the P-1@1 sensing system (lane 4), indicating that the interaction of MOF 1 with the duplex P-1@T1 is weaker than its corresponding interaction with P-1. Similar observations were also made for the other two P-DNA strands and DNA@RNA duplexes (Fig. S8, ESI†).
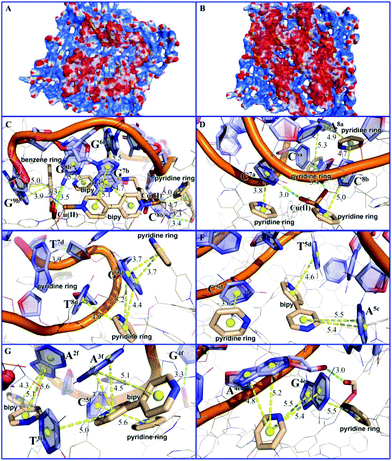
Molecular simulation studies using the Molecular Operating Environment (MOE) package23 indicated that the surface area of MOF 1 is largely positively charged, while the P-DNAs exhibit negative and neutral surfaces, resulting in good electrical complementation (Fig. 4A). Because of their structural flexibility, P-DNAs are fully stretched and attached to the surface of MOF 1. In contrast, the P-DNA@RNA duplexes have a small contact surface with MOF 1 due to their own helical structure, showing a “middle uplift and contact at both ends” model (Fig. 4B). As shown in Fig. 4C–H, the P-DNAs and P-DNA@RNA duplexes bind to MOF 1 mainly through the π–π stacking from the nucleobase with pyridinium, benzene, and bipy rings. There are 53 π–π stacking interactions between the ring centroids of MOF 1 and P-DNAs with a distance in the range of 3.2 to 6.0 Å, compared to 42 π–π stacking interactions between MOF 1 and P-DNA@RNAs (ring centroids in the range of 3.5 to 5.8 Å). In addition, there also exist 7 hydrogen-bonding interactions between the carboxylate/coordination water of MOF 1 and nitrogen atoms of P-DNAs, and 4 hydrogen-bonding interactions between that with P-DNA@RNAs.
The electronic energies by UFF force field simulation studies revealed that the electronic energy of MOF 1 with single chain P-DNAs (ΔGP-DNAs@MOF) is much lower than that with the P-DNA@RNA duplexes (ΔGMOF+P-DNA@RNAs). The electronic energy difference (ΔΔG) between MOF 1 with three kinds of single chain fluorescence-tagged P-DNAs and that with DNA@RNA duplexes is evaluated to be −117.12 kcal mol−1, further validating that MOF 1 binds more tightly to P-DNAs than to P-DNA@RNAs, and corroborate the experimental evidence discussed above.
In summary, we have demonstrated for the first time a synchronous detection of three conserved ZIKV RNA sequences using a Cu(II)-based MOF hybridized with dye-tagged DNA sensors. The detection is specific and fast with the detection limit in the nano-molar range and with no cross-reactions during the detection process. The strategy presented herein is thus universal to the early diagnosis of challenging viruses with high accuracy. We are currently exploring such potential using designer MOFs for the early diagnosis of various epidemic diseases, including Zika, Ebola, Dengue, Chikungunya, and others.
Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
This work was financially supported by the Natural Science Foundation of Guangdong (2018A030313456) and the Natural Science Foundation of China (21671143, 21874064, and 21871203).References
- D. Musso, T. Nhan, E. Robin, C. Roche, D. Bierlaire, K. Zisou, A. S. Yan, V. M. Cao-Lormeau and J. Broult, Euro. Surveill., 2014, 19, 20761 Search PubMed.
- R. S. Lanciotti, O. L. Kosoy, J. J. Laven, J. O. Velez, A. J. Lambert, A. J. Johnson, S. M. Stanfield and M. R. Duffy, Emerging Infect. Dis., 2008, 14, 1232 CrossRef CAS PubMed.
- G. S. Campos, A. C. Bandeira and S. I. Sardi, Emerging Infect. Dis., 2015, 21, 1885 CrossRef PubMed.
- B. D. Foy, K. C. Kobylinski, J. L. C. Foy, B. J. Blitvich, A. T. da Rosa, A. D. Haddow, R. S. Lanciotti and R. B. Tesh, Emerging Infect. Dis., 2011, 17, 880 CrossRef PubMed.
- (a) W. Dejnirattisai, P. Supasa, W. Wongwiwat, A. Rouvinski, G. Barba-Spaeth, T. Duangchinda, A. Sakuntabhai, V. M. Cao-Lormeau, P. Malasit, F. A. Rey, J. Mongkolsapaya and G. R. Screaton, Nat. Immunol., 2016, 17, 1102 CrossRef CAS PubMed; (b) K. Stettler, M. Beltramello, D. A. Espinosa, V. Graham, A. Cassotta, S. Bianchi, F. Vanzetta, A. Minola, S. Jaconi, F. Mele, M. Foglierini, M. Pedotti, L. Simonelli, S. Dowall, B. Atkinson, E. Percivalle, C. P. Simmons, L. Varani, J. Blum, F. Baldanti, E. Cameroni, R. Hewson, E. Harris, A. Lanzavecchia, F. Sallusto and D. Corti, Science, 2016, 14, 8505 Search PubMed.
- C. J. Haug, M. P. Kieny and B. Murgue, N. Engl. J. Med., 2016, 374, 1801 CrossRef CAS PubMed.
- (a) A. C. Gourinat, O. O'Connor, E. Calvez, C. Goarant and M. Dupont-Rouzeyrol, Emerging Infect. Dis., 2015, 21, 84 CrossRef CAS PubMed; (b) D. Musso, C. Roche, T. X. Nhan, E. Robin, A. Teissier and V. M. Cao-Lormeau, J. Clin. Virol., 2015, 68, 53 CrossRef PubMed.
- G. Kuno and G. J. J. Chang, Arch. Virol., 2007, 152, 687 CrossRef CAS PubMed.
- F. Degliangeli, P. Kshirsagar, V. Brunetti, P. P. Pompa and R. Fiammengo, J. Am. Chem. Soc., 2014, 136, 2264 CrossRef CAS PubMed.
- L. Y. Wang, Y. Q. Cheng, H. Wang and Z. P. Li, Analyst, 2012, 137, 3667 RSC.
- H. F. Dong, J. Zhang, H. X. Ju, H. T. Lu, S. Y. Wang, S. Jin, K. H. Hao, H. W. Du and X. J. Zhang, Anal. Chem., 2012, 84, 4587 CrossRef CAS PubMed.
- (a) G. Y. Wang, C. Song, D. M. Kong, W. J. Ruan, Z. Chang and Y. Li, J. Mater. Chem. A, 2014, 2, 2213 RSC; (b) F. L. Hu, Y. Mi, C. Zhu, B. F. Abrahams, P. Braunstein and J. P. Lang, Angew. Chem., Int. Ed., 2018, 57, 12696 CrossRef CAS PubMed; (c) F. L. Hu, Y. X. Shi, H. H. Chen and J. P. Lang, Dalton Trans., 2015, 44, 18795 RSC; (d) H. X. Li, W. Zhao, H. Y. Li, Z. L. Xu, W. X. Wang and J. P. Lang, Chem. Commun., 2013, 14, 4259 RSC.
- X. Zhu, H. Y. Zheng, X. Wei, Z. Y. Lin, L. H. Guo, B. Qiu and G. N. Chen, Chem. Commun., 2013, 49, 1276 RSC.
- J. M. Fang, F. Leng, X. J. Zhao, X. L. Hu and Y. F. Li, Analyst, 2014, 139, 801 RSC.
- Y. L. Liu, W. L. Fu, C. M. Li, C. Z. Huang and Y. F. Li, Anal. Chim. Acta, 2015, 861, 55 CrossRef CAS PubMed.
- H. T. Zhang, J. W. Zhang, G. Huang, Z. Y. Du and H. L. Jiang, Chem. Commun., 2014, 50, 12069 RSC.
- (a) L. Qin, L. X. Lin, Z. P. Fang, S. P. Yang, G. H. Qiu, J. X. Chen and W. H. Chen, Chem. Commun., 2016, 52, 132 RSC; (b) S. P. Yang, S. R. Chen, S. W. Liu, X. Y. Tang, L. Qin, G. H. Qiu, J. X. Chen and W. H. Chen, Anal. Chem., 2015, 87, 12206 CrossRef CAS PubMed; (c) H. Q. Zhao, G. H. Qiu, Z. Liang, M. M. Li, B. Sun, L. Qin, S. P. Yang, W. H. Chen and J. X. Chen, Anal. Chim. Acta, 2016, 922, 55 CrossRef CAS PubMed; (d) H. Q. Zhao, S. P. Yang, N. N. Ding, L. Qin, J. X. Chen, W. H. Zhang, W. H. Chen and T. S. A. Hor, Dalton Trans., 2016, 45, 5092 RSC; (e) B. P. Xie, Q. H. Qiu, P. P. Hu, Z. Liang, Y. M. Liang, B. Sun, L. P. Bai, Z. H. Jiang and J. X. Chen, Sens. Actuators, B, 2018, 254C, 1133 CrossRef; (f) P. P. Hu, N. Liu, K. Y. Wu, L. Y. Zhai, B. P. Xie, B. Sun, W. J. Duan, W. H. Zhang and J. X. Chen, Inorg. Chem., 2018, 57, 8382 CrossRef CAS PubMed; (g) B. Sun, Z. Liang, B. P. Xie, L. Z. Li, R. T. Li, Z. H. Jiang, L. P. Bai and J. X. Chen, Talanta, 2018, 179, 658 CrossRef CAS PubMed; (h) G. H. Qiu, Z. H. Weng, P. P. Hu, W. J. Duan, B. P. Xie, B. Sun, X. Y. Tang and J. X. Chen, Talanta, 2018, 180, 396 CrossRef CAS PubMed; (i) L. Qin, Z. Y. Sun, K. Cheng, S. W. Liu, J. X. Pang, W. H. Chen, Z. Cheng and J. X. Chen, ACS Appl. Mater. Interfaces, 2017, 9, 41378 CrossRef CAS PubMed; (j) F. L. Li, S. P. Yang, W. H. Zhang, Q. Liu, H. Yu, J. X. Chen and J. P. Lang, ChemistrySelect, 2016, 1, 2979 CrossRef CAS.
- T. Ye, Y. F. Liu, M. Luo, X. H. Ji, G. H. Zhou and Z. K. He, Analyst, 2014, 139, 1721 RSC.
- M. Ganeshpandian, R. Loganathan, E. Suresh, A. Riyasdeen, M. A. Akbarshade and M. Palaniandavar, Dalton Trans., 2014, 43, 1203 RSC.
- (a) I. J. Joye, G. Davidov-Pardo, R. D. Ludescher and D. J. McClements, Food Chem., 2015, 185, 261 CrossRef CAS PubMed; (b) D. Thomsson, R. Camacho, Y. X. Tian, D. Yadav, G. Sforazzini, H. L. Anderson and I. G. Scheblykin, Small, 2013, 9, 2619 CrossRef CAS PubMed.
- (a) M. E. McCarroll, F. H. Billiot and I. M. Warner, J. Am. Chem. Soc., 2001, 123, 3173 CrossRef CAS PubMed; (b) M. J. Zou, Y. Chen, X. Xu, H. D. Huang, F. Liu and N. Li, Biosens. Bioelectron., 2012, 32, 148 CrossRef CAS PubMed; (c) X. Y. Wang, M. J. Zou, H. D. Huang, Y. Q. Ren, L. M. Li, X. D. Yang and N. Li, Biosens. Bioelectron., 2013, 41, 569 CrossRef CAS PubMed.
- J. M. Goldberg, S. Batjargal, B. S. Chen and E. J. Petersson, J. Am. Chem. Soc., 2013, 135, 18651 CrossRef CAS PubMed.
- Molecular Operating Environment (MOE), 2014.09, Chemical Computing Group Inc., 1010 Sherbooke St. West, Suite #910, Montreal, QC, Canada, H3A 2R7, 2014 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Experimental section, synthesis, X-ray crystallography, crystallographic data, selected bond distances (Å) and angles (°), and PXRD patterns for 1. Synchronous fluorescence detection of T1, T2, and T3 using MOF 1 and molecular simulation studies. The coordination geometry for Cu in 1 and the two-dimensional structure of 1. DLS measurement of MOF 1 and P-DNAs@1. Influence of the incubation time between P-DNAs@1 and the target RNAs on the fluorescence intensity. Fluorescence changes at 518 nm, 604 nm and 670 nm of P-DNAs versus concentrations of T1, T2, and T3. The recovery efficiencies RE of P-DNAs in the presence of T1, T2 and T3. Fluorescence anisotropy of P-DNA and P-DNA@T before and after the addition of MOF 1. Polyacrylamide gel electrophoresis. CCDC 1568167. For ESI and crystallographic data in CIF or other electronic format see DOI: 10.1039/c8qi01031e |
| This journal is © the Partner Organisations 2019 |